Introduction
Sperm-associated antigen 9 (SPAG9) has
characteristics of a scaffold protein and is involved in the c-Jun
N-terminal kinase JNK signaling pathway, binding to JNK, which
suggests that it is involved in physiological processes, including
apoptosis, survival, proliferation and tumorigenesis (1,2). SPAG9 is
a member of the cancer/testis antigen family expressed from a
single copy gene located on human chromosome 17q21. Proteins in the
cancer/testis antigen family are overexpressed in a variety of
types of cancer (3,4). SPAG9 is also expressed in a variety of
tumors (5–9). SPAG9 has been proposed as a novel
biomarker for early diagnosis of multiple human tumors, including
ovarian, cervical and breast cancers (10–12).
Certain studies have revealed that small interfering RNA inhibits
expression of SPAG9 and inhibits the growth of various types of
tumor cells (13,14). SPAG9 protein has been demonstrated to
be involved in cancer progression; however, the underlying
mechanisms remain unknown (15,16).
Hepatocellular carcinoma (HCC) is the sixth most
common type of cancer globally and is the third most common cause
of cancer-associated mortality (17).
The incidence of HCC is higher in South East Asia and Africa
compared with other regions of the world (18). The crucial etiological factors
involved in the development of HCC include infection with hepatitis
virus, the structural or functional mutation of oncogenes and tumor
suppressor genes (19–21). Long non-coding RNA URHC regulates cell
proliferation and apoptosis by zinc-activated channels via the
extracellular signal-related kinase/mitogen-activated protein
kinase (MAPK) signaling pathway in HCC (22), and microRNA-24 may modify aflatoxin
B1-related HCC prognosis and tumorigenesis (23). Therefore, it is necessary to further
the research in this area in order to improve the diagnosis and
treatment of HCC. In the present study, a series of methods were
used to evaluate the expression level of SPAG9 in human HCC tumor
tissue and its potential underlying mechanisms.
Materials and methods
Cell culture
The QGY human HCC cell line was purchased from
Shanghai Institute of Pharmaceutical Industry (Shanghai, China) and
cultured in RPMI-1640 (HyClone; GE Healthcare Life Sciences, Logan,
UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin (GE Healthcare) at 37°C in
the presence of 5% CO2.
Patient samples
A total of 16 HCC participants were enrolled between
August 2010 and March 2013 at Xiangya Hospital, Central South
University (Changsha, China). Written informed consent was obtained
from all patients prior to enrollment in the present study. The
exclusion criteria of the present study were as follows: i)
Patients had distant metastasis; ii) Patients had received previous
radiotherapy and chemotherapy prior to hepatectomy; iii) Patients
with serious infection or other malignant diseases. The
experimental protocols were approved by the Institutional Review
Board of Xiangya Hospital. Carcinoma and adjacent noncancerous
tissues were obtained from 16 patients with HCC during surgical
tumor resections in accordance with informed consent. The present
study was approved by the Ethics Committee of Hunan Provincial
Second People's Hospital (Changsha, China). HCC was confirmed by
pathobiology. All clinical and biological data are presented in
Table I.
 | Table I.Characteristics of patients with
HCC. |
Table I.
Characteristics of patients with
HCC.
| Samples | Age (years) | Sex | Tumor size (cm × cm ×
cm) | Pathological
diagnosis and classification (Edmondson grading system) (30) |
|---|
| 1 | 42 | Male | 5.0×4.0×3.5 | 2–3grade well
differentiated HCC |
| 2 | 40 | Female | 5.0×5.0×4.5 | 2 grade moderately
differentiated HCC |
| 3 | 39 | Male | 7.0×5.0×5.0 | 2 grade moderately
differentiated HCC |
| 4 | 44 | Female | 3.5×3.0×0.5 | 2–3grade well
differentiated HCC |
| 5 | 45 | Male | 4.0×3.0×3.0 | 2–3grade well
differentiated HCC |
| 6 | 48 | Male | 4.0×3.5×2.0 | 2–3grade well
differentiated HCC |
| 7 | 58 | Male | 6.0×6.0×4.5 | 2 grade moderately
differentiated HCC |
| 8 | 56 | Male | 3.0×2.5×2.0 | 1–2grade well
differentiated HCC |
| 9 | 50 | Male | 3.0×4.0×4.0 | 2–3grade well
differentiated HCC |
| 10 | 43 | Male | 3.5×4.0×2.0 | 2 grade moderately
differentiated HCC |
| 11 | 57 | Female | 5.5×5.0×4.0 | 1–2grade well
differentiated HCC |
| 12 | 51 | Male | 3.5×3.0×3.0 | 2–3grade well
differentiated HCC |
| 13 | 64 | Male | 6.0×3.5×4.0 | 2 grade moderately
differentiated HCC |
| 14 | 58 | Female | 5.0×3.5×3.0 | 2–3grade well
differentiated HCC |
| 15 | 48 | Male | 4.0×5.0×3.5 | 2 grade moderately
differentiated HCC |
| 16 | 54 | Male | 4.0×3.5×3.0 | 1–2grade well
differentiated |
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from HCC tissues and
corresponding non-tumor normal tissues using TRIzol reagent (CWbio,
Beijing, China) and cDNA synthesis was performed using the
RevertAid First Strand cDNA Synthesis kit (CWbio), according to the
manufacture's protocol. The qPCR was performed at 95°C for 10 min,
followed by 40 cycles at 95°C for 15 sec and at 60°C for 60 sec and
qPCR was performed using GoTaq qPCR master mix (Promega
Corporation, Madison, WI, USA); BRYT Green® dye was used
in the GoTaq qPCR master mix. For detection of SPAG9 mRNA
expression levels, GAPDH was amplified in parallel as an internal
control. The sequences of the primers used for qPCR were as
follows: SPAG9 forward, 5′-AGCCGACTTTTCAGCTCCTC-3′ and reverse,
5′-AAAGCCTGCACTCTACCGTC-3′; GAPDH forward,
5′-CAATGACCCCTTCATTGACC-3′ and reverse, 5′-GACAAGCTTCCCGTTCTCAG-3′
and the temperature protocol of reverse transcription was 37°C. The
mRNA expression level was evaluated using evaluated threshold cycle
(Cq) values. The Cq values were normalized to the expression levels
of GAPDH and the relative amount of mRNA specific to each of the
target genes was determined using the 2−ΔΔCq method
(18–22). qPCR was performed with the BIO-RAD
CFK96™ Real-Time System (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The data were analyzed using BIO-RADCFK Manager 2.0 software
(Bio-Rad Laboratories, Inc.) for 40 cycles. Experiments were
performed in triplicate.
Immunohistochemistry (IHC) and
evaluation of staining
IHC was performed using the peroxidase
anti-peroxidase technique as follows: Slides were incubated in
citrate buffer at pH 6 and heated in a microwave for 21 min at (200
W). The antibody for SPAG9 was purchased from Abcam (cat. no.
ab12331; Abcam, Cambridge, MA, USA). Antibody against SPAG9 (1:150;
ab12331; Abcam) was overlaid on HCC and corresponding non-tumorous
normal tissue sections (4 µm) and incubated overnight at 4°C.
Secondary antibody incubation using alkaline phosphatase-conjugated
mouse anti-rabbit immunoglobulin G (cat. no. sc-2358; 1:1,000;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was performed at
room temperature for 30 min.
Tissue sections were blindly evaluated by two
pathologists from the Department of Pathology, Xiangya Hospital,
Central South University (Changsha, China) in an effort to provide
a consensus on staining patterns by light microscopy
(magnification, ×100; Olympus Corporation, Tokyo, Japan). SPAG9
staining was performed according to the methods described by
Kanojia et al (23). Each case
was rated according to a score that added a scale of intensity of
staining to the area of staining. At least 10 high-power fields
were chosen randomly and >1,000 cells were counted for each
section. The intensity of staining was graded on the following
scale: 0, no staining; 1+, mild staining; 2+, moderate staining;
3+, intense staining. The area of staining was evaluated as
follows: 0, no staining of cells in any microscopic fields; 1+,
<30% of tissue stained positive; 2+, 30–60% stained positive;
3+, >60% stained positive. The minimum score when determined
(extension + intensity) was, therefore, 0, and the maximum was 6. A
combined staining score (extension + intensity) of ≤2 was
considered to be negative staining (low staining); a score of 3–4
was considered to be moderate staining; and a score of 5–6 was
considered to be strong staining.
Western blot analysis
The HCC tissues, corresponding non-tumor normal
tissues and QGY cells were lysed in RIPA buffer at 4°C for 5 min
(CWbio) and total protein concentration was determined using a
Pierce® BCA Protein Assay kit (Thermo Fisher Scientific,
Inc.). Extracts containing 50 µg protein were separated in 10%
SDS-PAGE gels and electroblotted onto nitrocellulose membranes
(HyClone; GE Healthcare Life Sciences). The membranes were blocked
using Tris-buffered saline/Tween-20 (25 mM Tris-HCl, 150 mM NaCl,
pH 7.5 and 0.05% Tween-20) supplemented with 5% non-fat milk at
room temperature for 3 h, followed by overnight incubation at 4°C
with primary antibodies (rabbit anti-SPAG9 antibody; Abnova, Tapai,
Taiwan; cat. no. PAB8794; dilution, 1:500). Following three washes
with Tris-buffered saline/Tween-20 (25 mM Tris-HCl, 150 mM NaCl, pH
7.5 and 0.05% Tween-20), the membranes were incubated with mouse
anti-rabbit IgG horseradish peroxidase-conjugated secondary
antibodies (cat. no. sc-2491; Santa Cruz Biotechnology, Inc.;
dilution, 1:5,000) at room temperature for 1 h and the specific
signals were visualized using an ECL detection system. Anti-GAPDH
antibody (Santa Cruz Biotechnology, Inc.; cat. no. Sc-25778;
1:3,000) was used as a loading control at 4°C overnight. The bands
were analyzed using Gel Automated Digitizing System software
(version 4.0; Silk Scientific, Orem, UT, USA). The relative
expression levels (fold) were evaluated by normalizing the
integrated optical density (IOD) for each band to that of the
corresponding GAPDH band.
Design and synthesis of SPAG9
hairpin-like small interfering (si)RNA, and construction of
recombinant eukaryotic expression plasmid
According to the SPAG9 gene sequence
(NM_001130527.2), the oligo DNA single strand 1, 2 was designed and
synthesized by Ambion; Thermo Fisher Scientific, Inc., and was used
as the target of RNAi. The control siRNA oligonucleotides 3 and 4
were also designed and led to the formation of double-stranded DNA
with annealed oligonucleotides. EcoRI and BamH RV restriction sites
were introduced at the end of the terminal. The lentivirus
pWPT-green fluorescent protein (GFP) was used as the carrier; the
viral vector was purchased from Clontech Laboratories, Inc.
(Mountainview, CA, USA). A lentivirus vector containing GFP, SPAG9
RNAi sequences or control siRNA was constructed: SPAG9 siRNA, TCT
GGA AAC GAC ATT TAT GG; control siRNA, TGA AGG TCG GAG TCA ACG GAT
T.
siRNA virus infection of QGY
cells
QGY HCC cells were cultured in RPMI-1640 medium
supplemented with 10% FBS at 37°C in 5% CO2. There were
three groups assessed in the present study: An empty vector group,
a control siRNA group and a SPAG9 siRNA group. At 60% confluence,
the lentivirus vector was added at a multiplicity of infection of
10 at 37°C. Following 48 h, infection efficiency (based on GFP
expression level) was determined using an inverted fluorescence
microscope (Leica DMl3000B; Leica Microsystems GmbH, Wetzlar,
Germany).
Detection of cell growth by MTT
Cells in the logarithmic growth phase were seeded
into 96-well plates at lx103 cells per well in a 200 µl
volume. A total of 20 µl MTT (5 mg/ml) was added to each well and
plates were incubated at 37°C for 4 h. The liquid was removed from
each well and 150 µl dimethyl sulfoxide was added at 37°C for 5
min. Plates were rapidly oscillated at room temperature at 300 × g
on a microplate reader (MK3; Thermo Fisher Scientific, Inc.) for 60
sec to fully dissolve the precipitate, and absorbance of each well
at 490 nm was evaluated.
Cell cycle analysis by flow
cytometry
The QGY cells were cultured in serum-free RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in 5%
CO2 for 24 h and digested with 0.25% trypsin at room
temperature for 2 min, washed once with PBS, centrifuged at 476 × g
for 5 min and resuspended in 70% ethanol. Cell density was adjusted
to lx106 cells/ml and cells were fixed with 4%
formaldehyde at 4°C overnight. Cells were washed with cold PBS and
100 µl RNase A (0.5 mg/ml) was added. After 30 min at 37°C, 400 µl
propidium iodide (10 µl/ml) was added and the samples were
incubated at 4°C in the dark for 30 min. A FACS Canto II flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA) was then used
to detect the cell cycle distribution.
Transwell chamber assay to investigate
the migration ability of cells
Cells in the logarithmic growth phase were digested
with trypsin at room temperature for 2 min. A total of 600 µl 10%
FBS supplemented with RPMI-1640 was added to the lower Transwell
chamber and 3×105 cells in 300 µl serum-free medium was
added to the upper chamber. Following a 48 h incubation at 37°C in
5% CO2, the chamber was washed with PBS and a cotton
swab was used to remove the cells that did not pass through the
basal membrane of the invasion chamber. Cells that had adhered to
the surface of the inferior chamber were fixed with 100% methanol
at room temperature for 30 min and the chamber was immersed in
Giemsa (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) staining
solution at room temperature for 20 min. Excess dye was removed
with PBS and the chambers were dried in air. Various fields of view
(magnification, ×100) were randomly captured by light microscope
(Olympus Corporation) and cells were counted. The number of
assessed fields of view was 200.
Statistical analyses
The Pearson's χ2 test, Fisher's exact
test, unpaired Student's t-tests, Wilcoxon signed-rank tests,
Mann-Whitney U tests and Kruskal-Wallis one-way analysis of
variance tests were performed using SPSS version 18.0 (SPSS, Inc.,
Chicago, IL, USA). The Bonferroni post hoc test used for the
χ2 tests and the Kruskal Wallis tests. Results were
expressed as the mean ± standard deviation. All P-values were
two-tailed, and P<0.05 was considered to indicate a
statistically significant difference.
Results
SPAG9 is highly expressed in HCC
tissues
In order to detect the mRNA expression levels of
SPAG9 in HCC and the adjacent non-cancerous tissues, 16 samples of
each were selected to perform RT-qPCR of the SPAG9 gene. The data
were analyzed using the 2−ΔΔCq method and the fold
change in the expression levels of these genes relative to the
internal control gene, GAPDH, were analyzed. The expression level
of the SPAG9 gene was higher in the HCC samples compared with in
the adjacent non-cancerous tissues, and the normalized SPAG9 gene
expression level in HCC was upregulated by 3.35-fold (P=0.003;
Fig. 1A).
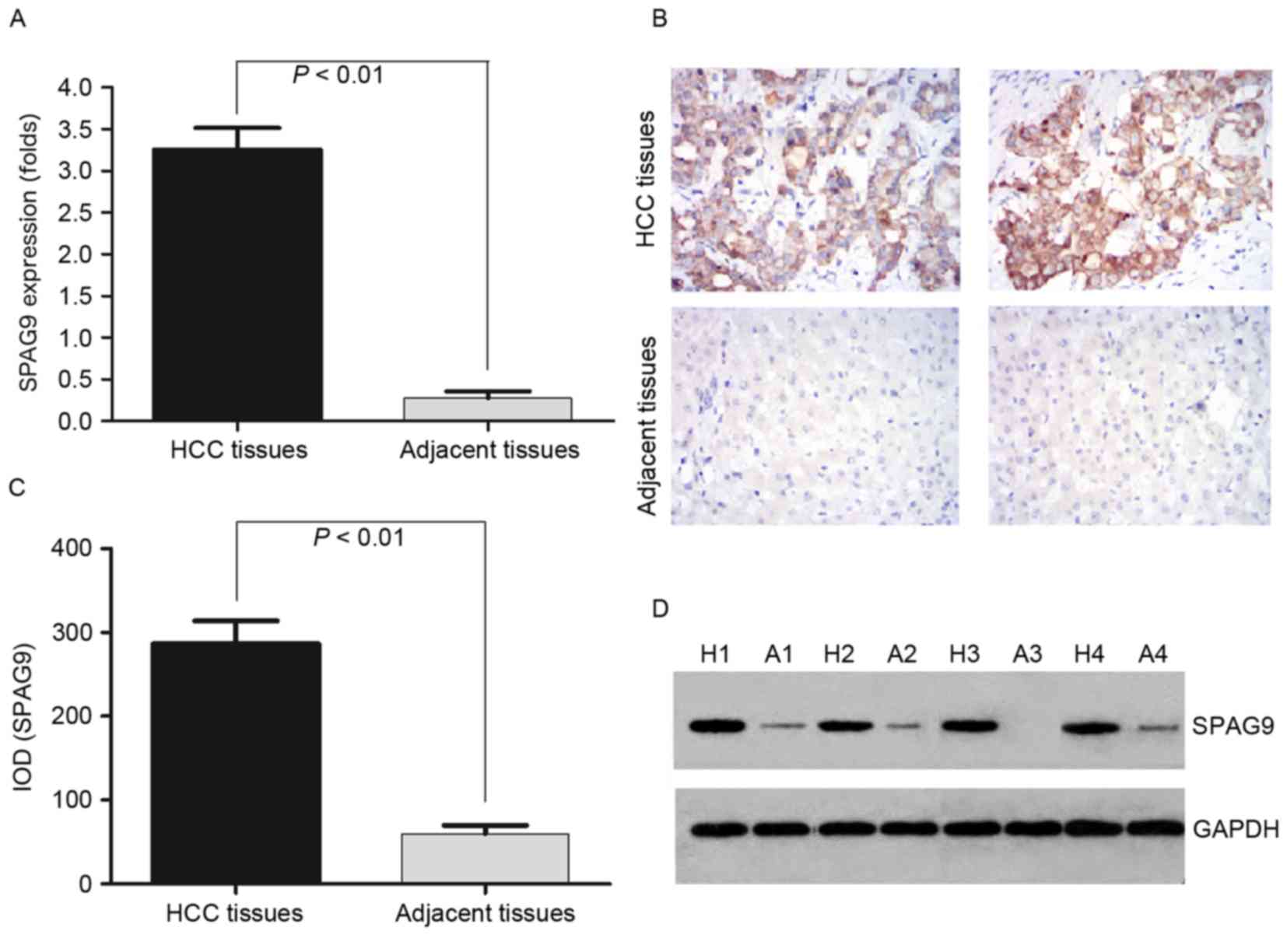 | Figure 1.Detection of SPAG9 expression levels
in the HCC tissues and the adjacent non-cancerous tissues by
RT-qPCR, IHC and western blotting. (A) RT-qPCR was performed to
validate the expression level of SPAG9 in HCC tissues and the
adjacent non-cancerous tissues. GAPDH was used as an internal
control and for normalization of the data. (B) IHC analysis of the
expression level of SPAG9 protein in HCC and adjacent non-cancerous
tissues. Brown grains indicate a positive signal. Original
magnification, ×400. (C) The protein expression levels of SPAG9
were significantly higher in the HCC tissues compared with in
adjacent non-cancerous tissues, as assessed by western blot
analysis. (D) SPAG9 protein expression levels in 4 tissues used in
the detection of mRNA expression levels by RT-qPCR. SPAG9,
sperm-associated antigen 9; HCC, hepatocellular carcinoma; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction; IHC,
immunohistochemistry; H, HCC tissues; A, adjacent non-cancerous
tissues; IOD, integrated optical density. |
To confirm the pattern of SPAG9 expression in HCC,
IHC was performed with antibodies against SPAG9 protein in HCC and
adjacent non-cancerous tissues. SPAG9 was identified as
differentially expressed between HCC tissues vs. the adjacent
non-cancerous tissues. IHC demonstrated a similar pattern in
protein expression level to the western blotting results. There was
a 75.0% (12/16) high score of SPAG9 expression level in HCC tissues
and 0% (0/16) in the adjacent non-cancerous tissues. The
distribution of a low score was 0% (0/16) and 62.5% (10/16) in HCC
and the adjacent non-cancerous tissues, respectively (P=0.0001;
Fig. 1B; Table II). This corresponded with the
RT-qPCR results.
 | Table II.Expression levels of SPAG9 in HCC
tissues compared with adjacent non-cancerous tissues, as assessed
by immunohistochemistry. |
Table II.
Expression levels of SPAG9 in HCC
tissues compared with adjacent non-cancerous tissues, as assessed
by immunohistochemistry.
|
|
| Score |
|
|
|---|
|
|
|
|
|
|
|---|
| Tissue | No. patients | Low (%) (0–2) | Moderate (%)
(3–4) | High (%) (5–6) | χ2 | P-value |
|---|
| HCC | 16 | 0 (0) | 4 (25.0) | 12
(75.0) | 22.40 | 0.0001 |
| Adjacent
tissues | 16 | 10
(62.5) | 6 (37.5) | 0 (0) |
|
|
To determine whether the SPAG9 protein was expressed
at a higher level in HCC compared with adjacent non-cancerous
tissues, the protein expression levels of SPAG9 were further
examined by western blotting in 1 to 4 samples (Fig. 1C and D). SPAG9 protein high expression
level was detected in cancerous tissue (IOD 286.84±75.91) and at
lower levels in the adjacent noncancerous tissues (IOD 29.86±34.91;
P<0.01; Fig. 1C), which
corresponded with the RT-qPCR results.
SPAG9 effects the proliferation of HCC
cells
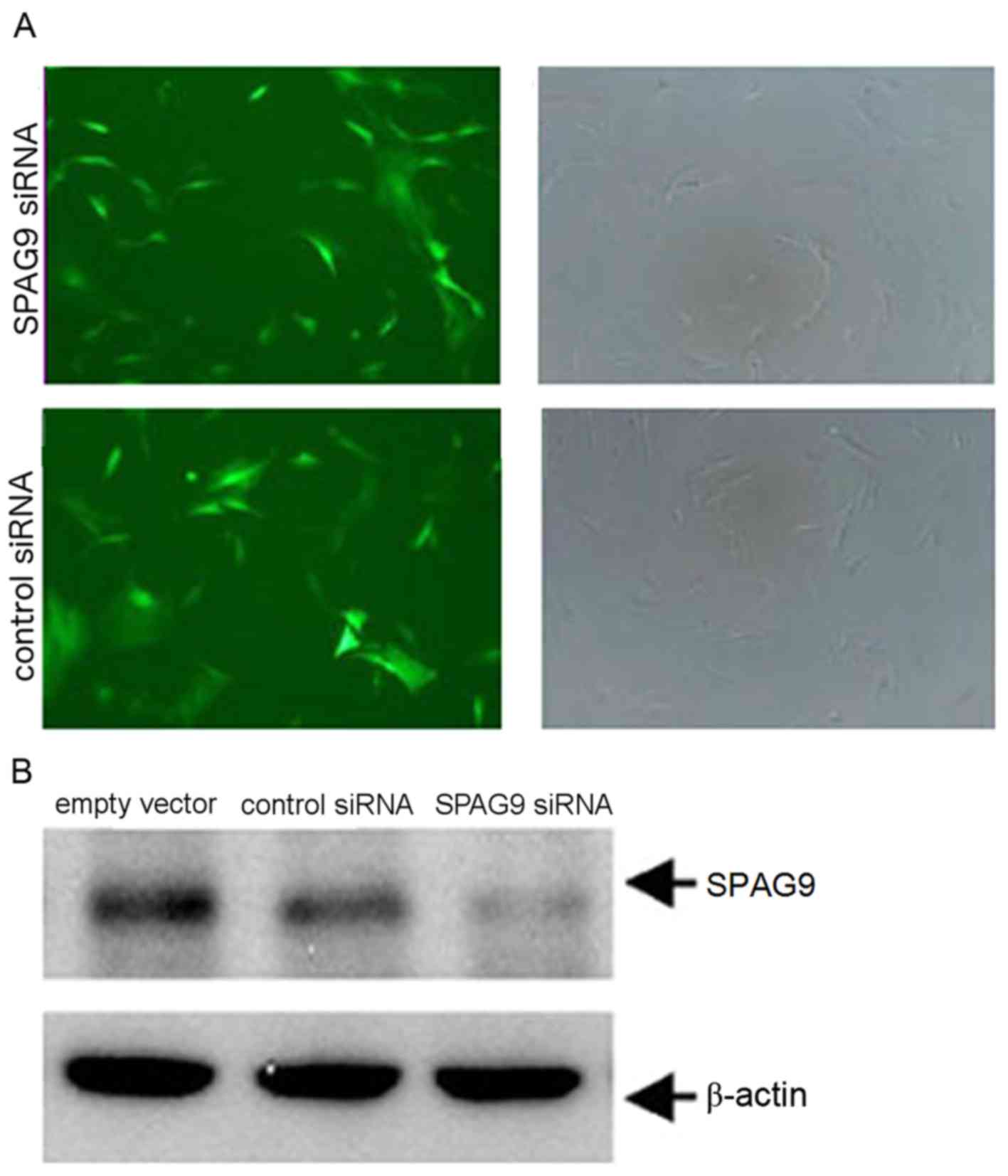
QGY human HCC cells were infected with lentiviral
vectors engineered to express siRNA targeting SPAG9 or a control
siRNA with an empty vector. Green fluorescence due to a virally
expressed GFP was observed in cells following infection for 12 h
using an inverted fluorescence microscope. Fluorescence increased
with infection time. The same vision field was randomly selected
and observed under fluorescence (73 perspectives) and white light
(76 perspectives) to determine infection efficiency. At 48 h, the
infection efficiency was ~92% (Fig.
2A). The expression level of SPAG9 protein in the SPAG9 siRNA
cells was significantly lower compared with in cells in the control
siRNA group (Fig. 2B). These results
revealed that the silencing of SPAG9 expression was successful in
QGY cells.
To investigate the effect of silencing of SPAG9, the
present study evaluated the proliferation of QGC cells between the
siRNA group and control group by MTT assay. The results
demonstrated that SPAG9-deficient cells did not proliferate as well
as cells treated with empty vector or cells that expressed the
control siRNA (Table III). Compared
with the control siRNA-expressing cells, proliferation of cells
that expressed SPAG9 siRNA were inhibited by 32.6% at 96 h.
 | Table III.Proliferation of QGY cells. |
Table III.
Proliferation of QGY cells.
|
| Incubation period
(h) |
|---|
|
|
|
|---|
| Group | 24 | 48 | 72 | 96 |
|---|
| SPAG9 siRNA |
0.294±0.007 |
0.376±0.021a |
0.584±0.063a |
0.662±0.127a |
| Control siRNA |
0.316±0.031 |
0.506±0.063 |
0.812±0.064 |
0.946±0.127 |
| Empty vector |
0.294±0.035 |
0.518±0.042 |
0.825±0.076 |
0.982±0.135 |
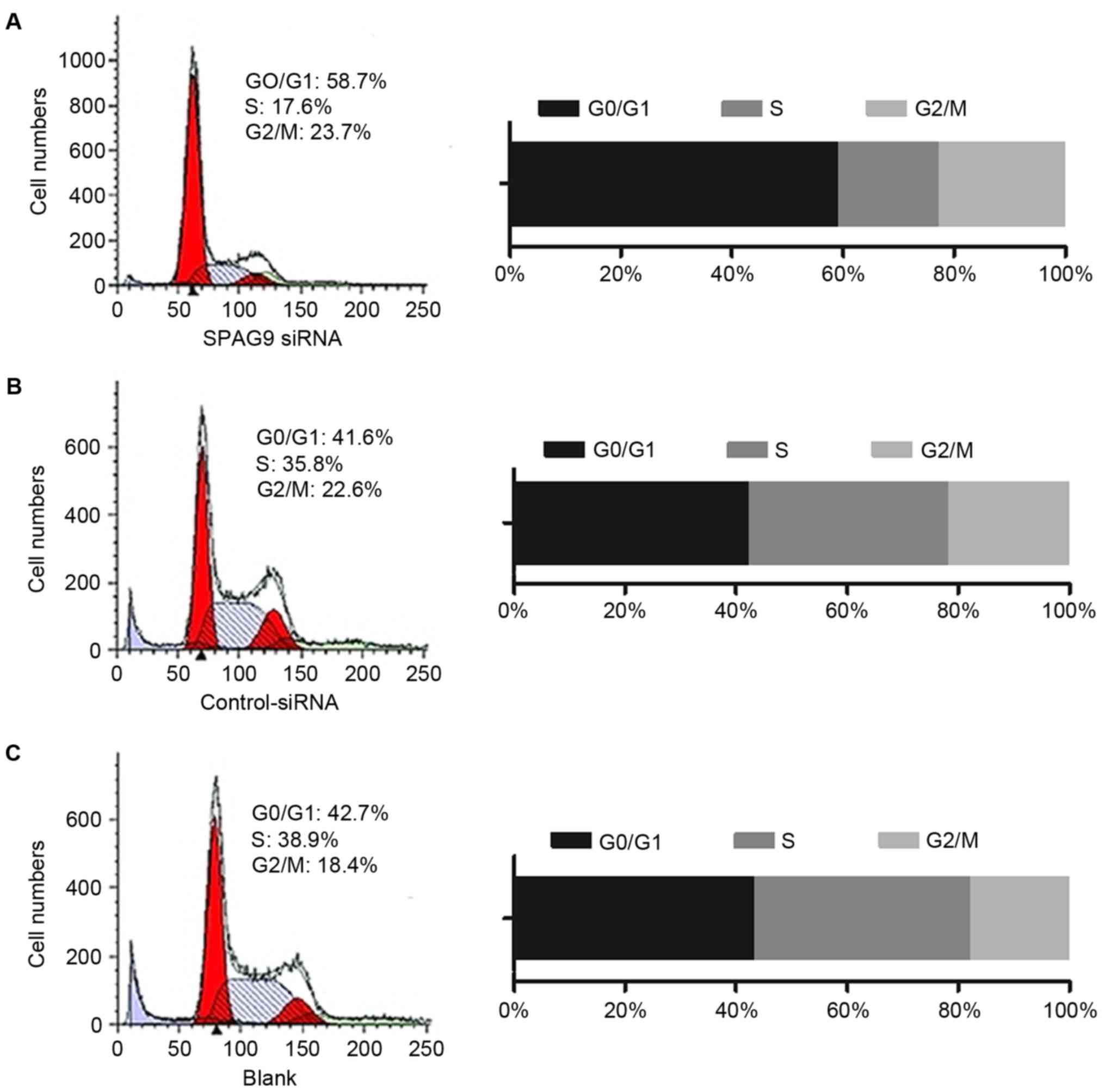
SPAG9 promotes cell cycle
progression
Flow cytometry was performed to detect the cell
cycle distributions of QGY cells (Fig.
3). In cells that expressed control siRNA, 41.6% were in the
G0/G1 cell cycle phase, whereas 58.7% of
cells that expressed the SPAG9 siRNA were in the
G0/G1 cell cycle phase. Compared with the
control siRNA-treated cells, the percentage of cells in
G0/G1 cell cycle phase were increased by
17.1% in SPAG9 siRNA-expressing cells and the percentage in the S
phase was reduced by 18.2% (Fig. 3).
There was no significant difference between cells that expressed
control siRNA and cells infected with the empty vector, suggesting
that inhibition of SPAG9 expression may inhibit the cell cycle in
the G1 and G2 phases.
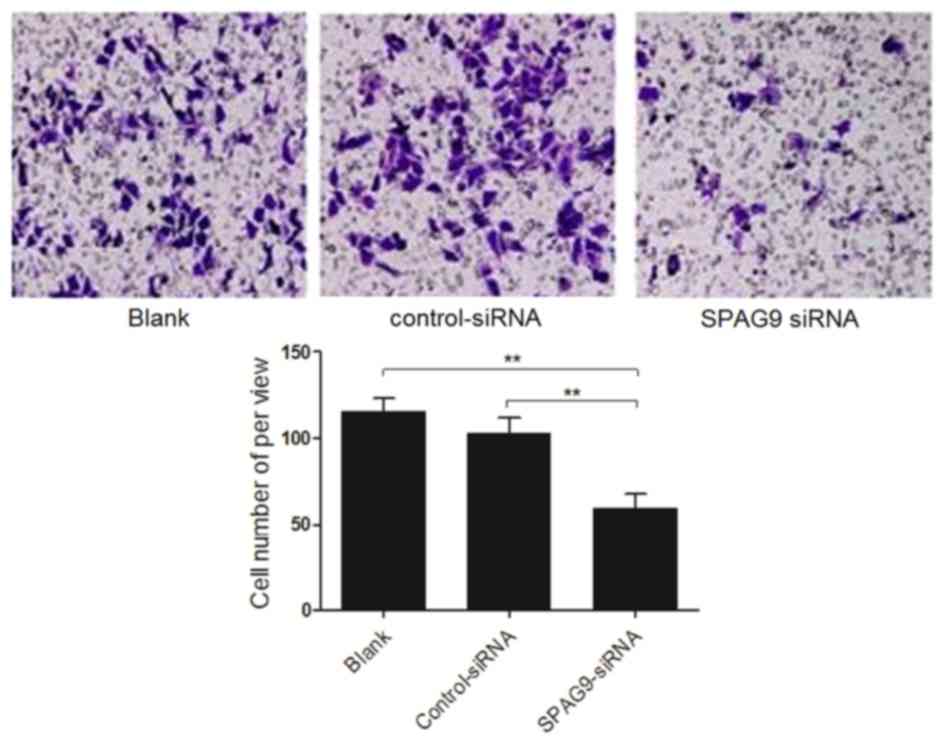
SPAG9 enhances cell migration
Finally, the migration of cells deficient in SPAG9
was compared with that of control cells using a Transwell chamber
assay. Compared with cells that expressed control siRNA, the
percentage of cells that expressed the SPAG9 siRNA that migrated
toward rich medium was significantly decreased (P<0.01). There
was no difference in migration of cells that expressed control
siRNA and cells infected with an empty vector (P>0.05; Fig. 4). The results of the present study
indicated that inhibiting SPAG9 expression levels reduced cell
migration.
Discussion
The occurrence and development of HCC are complex
processes associated with a variety of oncogenes, tumor suppressor
genes and certain cell cycle regulation factors (24,25).
Therefore, further investigation is required to fully understand
and improve treatments for HCC. Attempts to diagnose HCC early in
order to decrease the mortality rate and prolong the survival time
and quality of life have been made (26); however, there have been no major
breakthroughs at present. Cancer/testis antigens expressed in a
variety of human malignant tissues provide a novel direction for
the study of HCC, and SPAG9 is one of these cancer/testis antigens
(27). A previous study concerning
ovarian cancer suggested that SPAG9 regulates the MAPK signaling
pathway (28). The MAPK signaling
pathway is mediated by various stimuli, and the selectivity and
specificity of these reactions depends on scaffold proteins
(29). SPAG9 interacts with JNK in
the A549 lung cancer cell line and SPAG9 contributes to invasive
abilities via a mechanism mediated by matrix metalloproteinase 9
and the activation of JNK (14).
The present study revealed that the expression level
of the SPAG9 gene and protein was upregulated in the HCC tumor
tissues compared with in adjacent non-cancerous tissues
(P<0.01). The analysis of HCC tissues from patients in the
present study was consistent with previous studies that evaluated
SPAG9 expression levels in the QGY hepatoma cell line (11,27). RNAi
technology was used to inhibit the expression level of SPAG9 in
cultured cells and the results revealed that SPAG9 deficiency was
able to affect the proliferation and progression of HCC QGY cells.
The present study performed an MTT assay and demonstrated that,
compared with the control siRNA-expressing cells, proliferation of
cells that expressed SPAG9 siRNA was significantly inhibited from
48 h and the rate of inhibition achieved was 32.6% at 96 h. The
number of QGY cells in the G0 phase increased and the
number in the S phase decreased following SPAG9 gene silencing. A
Transwell chamber assay was performed and revealed that inhibiting
the expression level of SPAG9 reduced the migratory ability of QGY
cells; however, the specific mechanism underlying this effect
requires further study. Inhibiting SPAG9 expression may effectively
decrease the differentiation and proliferation of OGY cells,
indicating that SPAG9 may serve a specific function in accelerating
the cell cycle, promoting proliferation and migration of HCC. This
revealed that downregulation of scaffold proteins may induce
inactivation of numerous signaling pathways. Therefore, blocking
the tumor signaling pathway by knocking out the tumor scaffold
proteins is a novel concept for cancer treatment.
The present study evaluated a limited number of
patients and only one immortalized cell line. Due to the
heterogeneities of HCC, the expression levels of SPAG9 may differ.
SPAG9 protein levels were increased in the adjacent tissues of
patients 4 and 7 relative to the adjacent tissues, as assessed by
western blotting and IHC, but in the other patients the expression
level was low relative to that of the tumor tissues. The present
study observed an increased expression level of SPAG9 in the tumor
samples evaluated compared with adjacent non-cancerous samples and
SPAG9 expression was decreased in adjacent non-cancerous samples
compared with tumor samples. The results of the present study
suggested that SPAG9 was upregulated in HCC and may serve an
important function in cancer cell proliferation, differentiation
and invasion. Furthermore, that downregulation of SPAG9 expression
levels may inhibit the proliferation and invasion of HCC. Whether
SPAG9 is a potential diagnostic marker and therapeutic target of
human HCC requires additional investigation.
Acknowledgements
The present study was supported by the Hunan
Provincial Health Department Foundation (grant no. B2011-099) and
the Key Project of Hunan Provincial Second People's Hospital Fund
Foundation (grant no. 2013-1).
References
|
1
|
Li H, Peng Y, Niu H, Wu B, Zhang Y, Zhang
Y, Bai X and He P: SPAG9 is overexpressed in human prostate cancer
and promotes cancer cell proliferation. Tumour Biol. 35:6949–6954.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanojia D, Garg M, Gupta S, Gupta A and
Suri A: Sperm-associated antigen 9 is a novel biomarker for
colorectal cancer and is involved in tumor growth and
tumorigenicity. Am J Pathol. 178:1009–1020. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peng JR, Chen HS, Mou DC, Cao J, Cong X,
Qin LL, Wei L, Leng XS, Wang Y and Chen WF: Expression of
cancer/testis (CT) antigens in Chinese hepatocellular carcinoma and
its correlation with clinical parameters. Cancer Lett. 219:223–232.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang P, Huo Z, Liao H and Zhou Q:
Cancer/testis antigens trigger epithelial-mesenchymal transition
and genesis of cancer stem-like cells. Curr Pharm Des.
21:1292–1300. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garg M, Chaurasiya D, Rana R, Jagadish N,
Kanojia D, Dudha N, Kamran N, Salhan S, Bhatnagar A, Suri S, et al:
Sperm-associated antigen 9, a novel cancer testis antigen, is a
potential target for immunotherapy in epithelial ovarian cancer.
Clin Cancer Res. 13:1421–1428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garg M, Kanojia D, Khosla A, Dudha N, Sati
S, Chaurasiya D, Jagadish N, Seth A, Kumar R, Gupta S, et al:
Sperm-associated antigen 9 is associated with tumor growth,
migration, and invasion in renal cell carcinoma. Cancer Res.
68:8240–8248. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garg M, Kanojia D, Salhan S, Suri S, Gupta
A, Lohiya NK and Suri A: Sperm-associated antigen 9 is a biomarker
for early cervical carcinoma. Cancer. 115:2671–2683. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garg M, Kanojia D, Suri S, Gupta S, Gupta
A and Suri A: Sperm-associated antigen 9: A novel diagnostic marker
for thyroid cancer. J Clin Endocrinol Metab. 94:4613–4618. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kanojia D, Garg M, Gupta S, Gupta A and
Suri A: Sperm-associated antigen 9, a novel biomarker for early
detection of breast cancer. Cancer Epidemiol Biomarkers Prev.
18:630–639. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu P, Yan L, Zhang H, Lin X and Zhao X:
Expression and clinical significance of sperm-associated antigen 9
in patients with endometrial carcinoma. Int J Gynecol Cancer.
22:87–93. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garg M, Kanojia D, Suri S and Suri A:
Small interfering RNA-mediated down-regulation of SPAG9 inhibits
cervical tumor growth. Cancer. 115:5688–5699. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sinha A, Agarwal S, Parashar D, Verma A,
Saini S, Jagadish N, Ansari AS, Lohiya NK and Suri A: Down
regulation of SPAG9 reduces growth and invasive potential of
triple-negative breast cancer cells: Possible implications in
targeted therapy. J Exp Clin Cancer Res. 32:692013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yi F, Ni W, Liu W, Pan X, Han X, Yang L,
Kong X, Ma R and Chang R: SPAG9 is overexpressed in human
astrocytoma and promotes cell proliferation and invasion. Tumour
Biol. 34:2849–2855. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Dong Q, Miao Y, Fu L, Lin X and
Wang E: Clinical significance and biological roles of SPAG9
overexpression in non-small cell lung cancer. Lung Cancer.
81:266–272. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanojia D, Garg M, Saini S, Agarwal S,
Parashar D, Jagadish N, Seth A, Bhatnagar A, Gupta A, Kumar R, et
al: Sperm associated antigen 9 plays an important role in bladder
transitional cell carcinoma. PLoS One. 8:e813482013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Agarwal S, Parashar D, Gupta N, Jagadish
N, Thakar A, Suri V, Kumar R, Gupta A, Ansari AS, Lohiya NK and
Suri A: Sperm associated antigen 9 (SPAG9) expression and humoral
response in benign and malignant salivary gland tumors.
Oncoimmunology. 3:e9743822014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nordenstedt H, White DL and El-Serag HB:
The changing pattern of epidemiology in hepatocellular carcinoma.
Dig Liver Dis. 42 Suppl 3:S206–S214. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li P, Cao Y, Li Y, Zhou L, Liu X and Geng
M: Expression of Wnt-5a and β-catenin in primary hepatocellular
carcinoma. Int J Clin Exp Pathol. 7:3190–3195. 2014.PubMed/NCBI
|
|
20
|
Gauglhofer C, Paur J, Schrottmaier WC,
Wingelhofer B, Huber D, Naegelen I, Pirker C, Mohr T, Heinzle C,
Holzmann K, et al: Fibroblast growth factor receptor 4: A putative
key driver for the aggressive phenotype of hepatocellular
carcinoma. Carcinogenesis. 35:2331–2338. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chu R, Mo G, Duan Z, Huang M, Chang J, Li
X and Liu P: miRNAs affect the development of hepatocellular
carcinoma via dysregulation of their biogenesis and expression.
Cell Commun Signal. 12:452014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu WH, Zhang JB, Dang Z, Li X, Zhou T, Liu
J, Wang DS, Song WJ and Dou KF: Long non-coding RNA URHC regulates
cell proliferation and apoptosis via ZAK through the ERK/MAPK
signaling pathway in hepatocellular carcinoma. Int J Biol Sci.
10:664–676. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanojia D, Garg M, Gupta S, Gupta A and
Suri A: Sperm-associated antigen 9, a novel biomarker for early
detection of breast cancer. Cancer Epidemiol Biomarkers Prev.
18:630–639. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang H, Xiong FX, Lin M, Yang Y, Nie X and
Zhou RL: LAPTM4B-35 overexpression is a risk factor for tumor
recurrence and poor prognosis in hepatocellular carcinoma. J Cancer
Res Clin Oncol. 136:275–281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao ZK, Dong P, Gu J, Chen L, Zhuang M,
Lu WJ, Wang DR and Liu YB: Overexpression of LSD1 in hepatocellular
carcinoma: A latent target for the diagnosis and therapy of
hepatoma. Tumour Biol. 34:173–180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marquardt JU, Galle PR and Teufel A:
Molecular diagnosis and therapy of hepatocellular carcinoma (HCC):
An emerging field for advanced technologies. J Hepatol. 56:267–275.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xie C, Fu L, Liu N and Li Q:
Overexpression of SPAG9 correlates with poor prognosis and tumor
progression in hepatocellular carcinoma. Tumour Biol. 35:7685–7691.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ha JH, Yan M, Gomathinayagam R, Jayaraman
M, Husain S, Liu J, Mukherjee P, Reddy EP, Song YS and Dhanasekaran
DN: Aberrant expression of JNK-associated leucine-zipper protein,
JLP, promotes accelerated growth of ovarian cancer. Oncotarget.
7:72845–72859. 2016.PubMed/NCBI
|
|
29
|
Jagadish N, Rana R, Mishra D, Kumar M and
Suri A: Sperm associated antigen 9 (SPAG9): A new member of c-Jun
NH2 -terminal kinase (JNK) interacting protein exclusively
expressed in testis. Keio J Med. 54:66–71. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Edmondson HA and Steiner PE: Primary
carcinoma of the liver: A study of 100 cases among 48,900
necropsies. Cancer. 7:462–503. 1954. View Article : Google Scholar : PubMed/NCBI
|