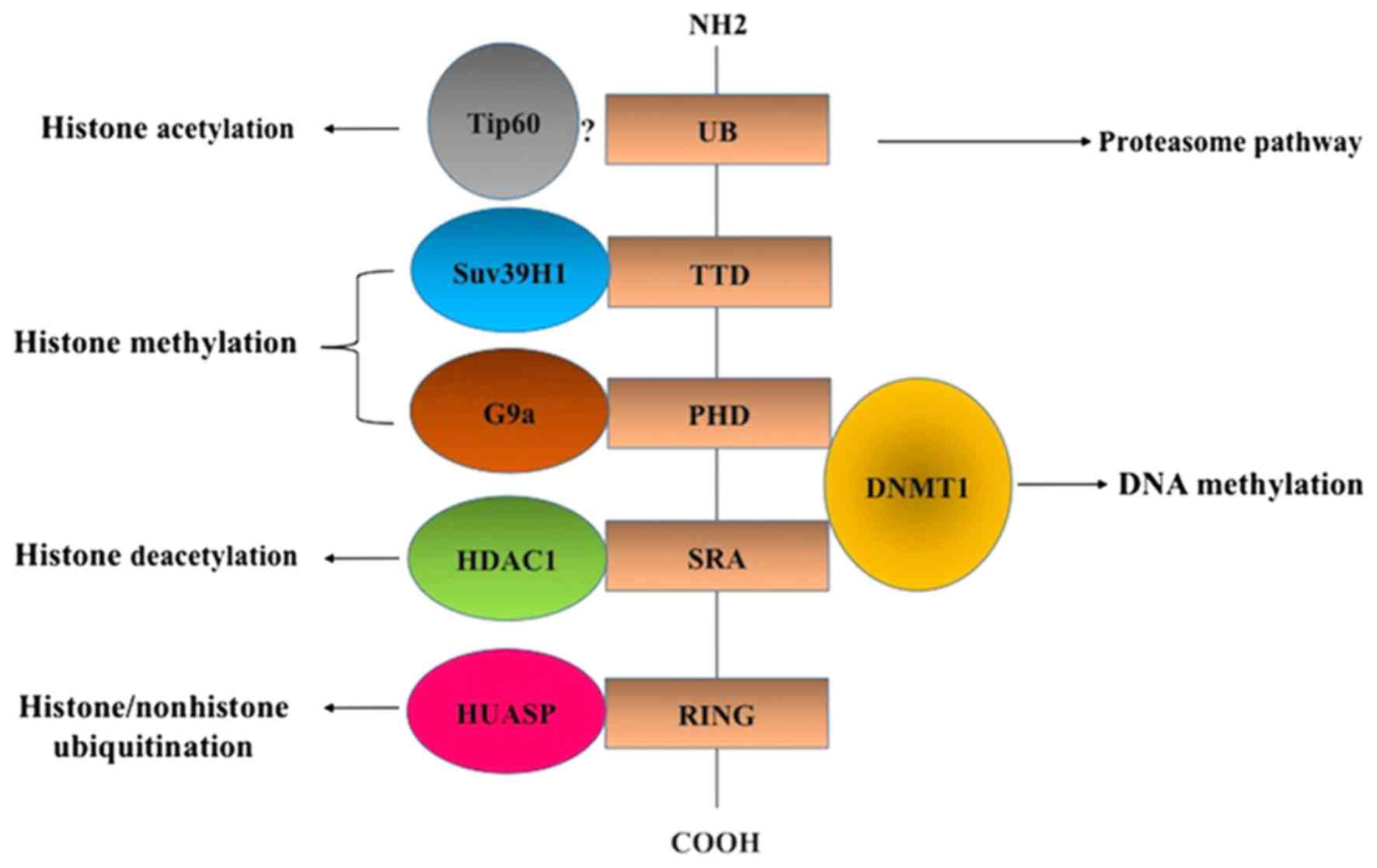
Epigenetic silencing of tumor suppressor genes
(TSGs) including breast cancer susceptibility gene 1
(BRCA1), human MutL homolog 1 (hMLH1),
p16INK4A and p14ARF involves
DNA methylation maintained by DNA methyltransferase 1 (DNMT1),
histone deacetylation and methylation through histone deacetylase 1
(HDAC1) and histone H3K9 methyltransferase G9a respectively
(1–3).
Ensuring a coordinated crosstalk between DNA methylation and
histone deacetylation and methylation, ubiquitin-like containing
plant homeodomain (PHD) and really interesting new gene domain
(RING) finger domains 1 (UHRF1) overexpressed in various human
cancer cells induces epigenetic silencing in several TSGs (4). UHRF1-mediated epigenetic silencing of
TSG is primarily due to the presence of the SET- and
RING-associated (SRA) domain (4).
Through the SRA domain, UHRF1 interacts with HDAC1 and DNMT1
(Fig. 1), leading to the inhibition
of several TSGs including p16INK4A,
p14ARF and retinoic acid receptor α (1,3,5,6).
Furthermore, the UHRF1 structure includes other functional domains
which contribute to its inhibitory activity on TSGs including the
ubiquitin-like domain, tandem Tudor domain (TTD), PHD and RING
domain (Fig. 1). A large
macromolecular protein complex termed epigenetic code replication
machinery (ECREM) is formed through interactions between the
different UHRF1 domains and several epigenetic coordinators
including HDAC1, DNMT1, histone acetyltransferase, Tat-interacting
protein 60 (Tip60), herpesvirus-associated ubiquitin specific
protease (HAUSP) and histone methyltransferase G9a and Suv39H1
(Fig. 1) (7–9).
The ECREM complex is considered to be orchestrated
by UHRF1 to ensure a coordinated transmission of silenced TSGs to
daughter cells during cell division (4,7,9). UHRF1 binds to H3K9me2, a repressive
chromatin mark, thus providing additional evidence of
UHRF1-mediated crosstalk between DNA methylation and histone
modification (10). Furthermore,
UHRF1 was also demonstrated to bind to H3K9me3 through the TTD
domain, an interaction involved in the regulation of
p16INK4A expression (11).
Although overexpression of wild-type UHRF1 induced
p16INK4A downregulation, such an effect was not
demonstrated when the TTD-mutated UHRF1 variant was overexpressed,
thus indicating that UHRF1 binding to H3K9me3 through the TTD
domain is involved in the silencing of p16INK4A
(11). In the same context, UHRF1 was
demonstrated to use its PHD domain to specifically bind to H3K9me3
(12) and cause large-scale
modifications of chromocenters which assisted in the recruitment of
HDAC1 and DNMT1, and led to the formation of pericentromeric
heterochromatin (13). The UHRF1,
through its RING domain, was demonstrated to possess E3 ubiquitin
ligase activity for histone 3, and was also involved in tumor
proliferation; however, the role of this domain remains unclear
(14,15). UHRF1 interacts with HAUSP (Fig. 1), a deubiquitinating enzyme involved
in the regulation of several TSGs (16). HAUSP protects UHRF1 from its own E3
ligase activity (autoubiquitination), suggesting that UHRF1 uses
its RING domain to target itself for degradation via
autoubiquitination in response to the downregulation of HAUSP
(17–19).
Collectively, these previous studies demonstrate
that UHRF1 through its several functional domains negatively
regulates the expression of TSGs via its interaction with numerous
proteins. Thus, the inhibition of the expression and/or activity of
UHRF1 may enable cancer cells to undergo apoptosis by reactivating
TSGs. As UHRF1 belongs to a large macromolecular complex, in which
it serves a function of a hub protein for the integration of
epigenetic information, microRNAs (miRNAs) which target UHRF1 may
have marked effects on cellular functions. In the present review,
the role of these specific miRNAs in the regulation of UHRF1
(Table I), and the associated
downstream events, as well as the importance of targeting
miRNA/UHRF1 pathways as a novel strategy in cancer therapy, are
discussed.
Several previous studies indicate that UHRF1 may be
a key regulator of the human epigenome through interactions with
domains in several types of coordinator (7,20–23). These interactions indicate that UHRF1
is involved in carcinogenesis through two key mechanisms. The first
is regarded as a participation in the onset phase of cancer,
whereas the second is with regard to the maintenance of the cancer
phenotype. Concerning the first, the driving of DNA hypomethylation
is a result of defective DNMT1-UHRF1 interaction (24,25). The
second is associated with the maintenance of DNA methylation
patterns, particularly the hypermethylation of the TSG promoters
and genome-wide hypomethylation (6).
Although the role of UHRF1 as a potent oncogene is
well-documented in several solid tumors and hematological
malignancies, its downregulation has been demonstrated to increase
the malignancy of carcinoma cells through the activation of
epithelial-mesenchymal transition (EMT) (3,4,26–35).
Numerous types of human cancer including leukemia, breast, bladder,
gastric, colorectal and astrocytoma express increased levels of
UHRF1, causing an increase in cell proliferation, migration,
metastasis and inhibition of apoptosis (7,36–39). The UHRF1 serves a crucial function in
the progression of the cell cycle at G1/S phase through
the p16INK4A-dependent pathway, and its
downregulation allows cancer cells to undergo apoptosis through DNA
demethylation and histone deacetylation-dependent reactivation of
several TSGs including p16INK4A, BRCA1, homeobox
protein CDX-2 (CDX2), runt-related transcription factor 3
(RUNX3), forkhead box protein O4,
peroxisome-proliferator-activated receptor γ and promyelocytic
leukemia (PML) (2,32,40). UHRF1
overexpression in cancer cells, compared with matched normal tissue
was suggested to be a potential biomarker for the prognosis and
diagnosis of several types of cancer including bladder and
colorectal carcinoma (23,36,39).
Considering that cancer cells overexpress UHRF1, deciphering the
upstream pathways involved in UHRF1 may shed light on the
underlying molecular mechanisms involved in the silencing of TSGs
in tumorigenesis.
High-throughput transcriptome analysis demonstrated
that the majority of transcriptional outcome is non-coding RNAs
(41). These non-coding RNAs are
classified, based on their length, into small non-coding RNAs
(<200 nucleotides), including miRNA, and long non-coding RNA
(>200 nucleotides), including nuclear paraspeckle assembly
transcript 1 (42,43). A number of non-coding RNAs have
potential transcriptional, post-transcriptional and epigenetic
regulatory functions, and are often dysregulated in many types of
disease including cancer (44–47).
miRNAs (18–25 nucleotides) are the most studied class of non-coding
RNAs and post-transcriptionally regulate mRNA stability and
translation (48). The biogenesis of
miRNA and the mechanism by which they degrade mRNA and inhibit RNA
translation is complex (49). Altered
miRNA expression has been observed in many types of cancer cell
line, xenograft, blood and clinical tissue (50,51).
Aberrant levels of miRNAs contribute to cancer formation and
progression by regulating expression levels of key genes involved
in tumorigenesis pathways which are responsible for cell
proliferation, tumor migration, invasion, integrin-mediated
adhesion, EMT and resistance to cancer therapy (52). Similar to protein-coding genes, miRNAs
are also subject to epigenetic regulatory modifications in cancer
(53,54). The majority of miRNA loci are
associated with CpG islands suggesting marked dependence on DNA
methylation.
In cancer, miRNAs are able to function as being
either oncogenic or a tumor suppressor, depending on the target
gene (55). For example, miR-21 was
the first miRNA to be identified as being oncogenic, and was
demonstrated to be overexpressed in numerous types of cancer
(56). Mechanistically, miR-21 was
observed to suppress the expression of many TSGs, including
phosphatase and tensin homolog, programmed cell death protein 4 and
sprout 1 (SPRY1) (57,58). However, miR-34b, miR-199b and miR-218
are examples of tumor suppressor miRNAs that were observed to be
downregulated in several types of tumor (59,60).
Advances in genomic technologies may lead to identification of
novel miRNAs involved in cancer, therefore the increase in the
understanding of their biological functions and target genes is
expected to enhance our knowledge on the role of miRNA in cancer
progression and permit the development of miRNA-associated cancer
biomarkers and consequently the formation of effective therapy.
Considering the fact that numerous types of human
cancer express increased levels of the oncogene UHRF1 in
association with decreased expression levels of several tumor
suppressor miRNAs, UHRF1 overexpression in cancer as a result of
altered miRNA expression is a subject worth investigating (36). Nevertheless, it has previously been
indicated that UHRF1 overexpression may also be a result of
increased stability, and of the inhibitory effects of several
miRNAs on its expression (36).
However, it should be noted that UHRF1 has been demonstrated to be
regulated by several other pathways including the cluster of
differentiation 47/nuclear factor κB axis, TSG p53 and p73, and the
thyroid hormone receptor α1/specificity protein 1 pathway (61–64)
Several previous studies have demonstrated that
UHRF1 is overexpressed in gastric cancer (GC) and therefore promote
the invasion and metastasis of this type of cancer; however, the
upstream regulatory mechanisms involved in UHRF1 overexpression are
currently unknown (32,65,66).
Increased levels of UHRF1 expression were identified in tissue
samples from patients with GC compared with normal controls, and
its overexpression was associated with patient age and lymph node
metastasis (66). The levels of UHRF1
were also significantly increased in tissues isolated from patients
with GC compared with corresponding normal tissues. Furthermore,
increased expression levels of UHRF1 were identified in GC tissues
in association with GC stage and grade (65). Similarly, UHRF1 was demonstrated to be
overexpressed in GC, and its downregulation induced upregulation of
several TSGs including RUNX3, BRCA1 and PML through a
promoter demethylation-dependent mechanism, inhibiting GC cell
proliferation and metastasis (32).
Furthermore, another study demonstrated that UHRF1 was
overexpressed in tissues from patients with GC compared with
matched normal tissues, and its increased expression levels were
associated with GC metastases (67).
Of note, UHRF1 depletion decreased GC migration and metastasis;
however, overexpression significantly promoted these effects
(67). Collectively, the results of
these previous studies demonstrate that UHRF1 is a primary factor
in GC development, and suggest that understanding the molecular
mechanisms underlying UHRF1 overexpression in GC may assist in
discovering the underlying molecular mechanisms involved in GC
tumorigenesis.
miR-146a and miR-146b are known to act as tumor
suppressors in several types of tumor including GC and lung cancer
(67–69). It has been demonstrated that UHRF1 is
regulated by miR-146a/b in GC (67).
Furthermore, miR-146a/b overexpression significantly downregulated
UHRF1 expression through directly targeting its 3′-untranslated
region (UTR) (67). The
downregulation was associated with DNA demethylation-dependent
reactivation of a number of TSGs including Slit guidance ligand 3,
cadherin 4 and RUNX3, and a decrease in GC cell migration
and metastasis (67). Furthermore,
the downregulation of miR-146a/b led to an increase in the
expression of UHRF1, indicating that UHRF1 is negatively regulated
by miR-146a/b in normal cells (67).
A previous study also demonstrated that miR-146a binds to the
3′-UTR of UHRF1, providing additional evidence of a direct
regulation of UHRF1 by miR-146a (65). Collectively, these results that
support the hypothesis that UHRF1 mRNA is a direct target of
miR-146a/b in GC, and miR-146a/b overexpression may be a promising
strategy to downregulate UHRF1 in order to achieve GC metastasis
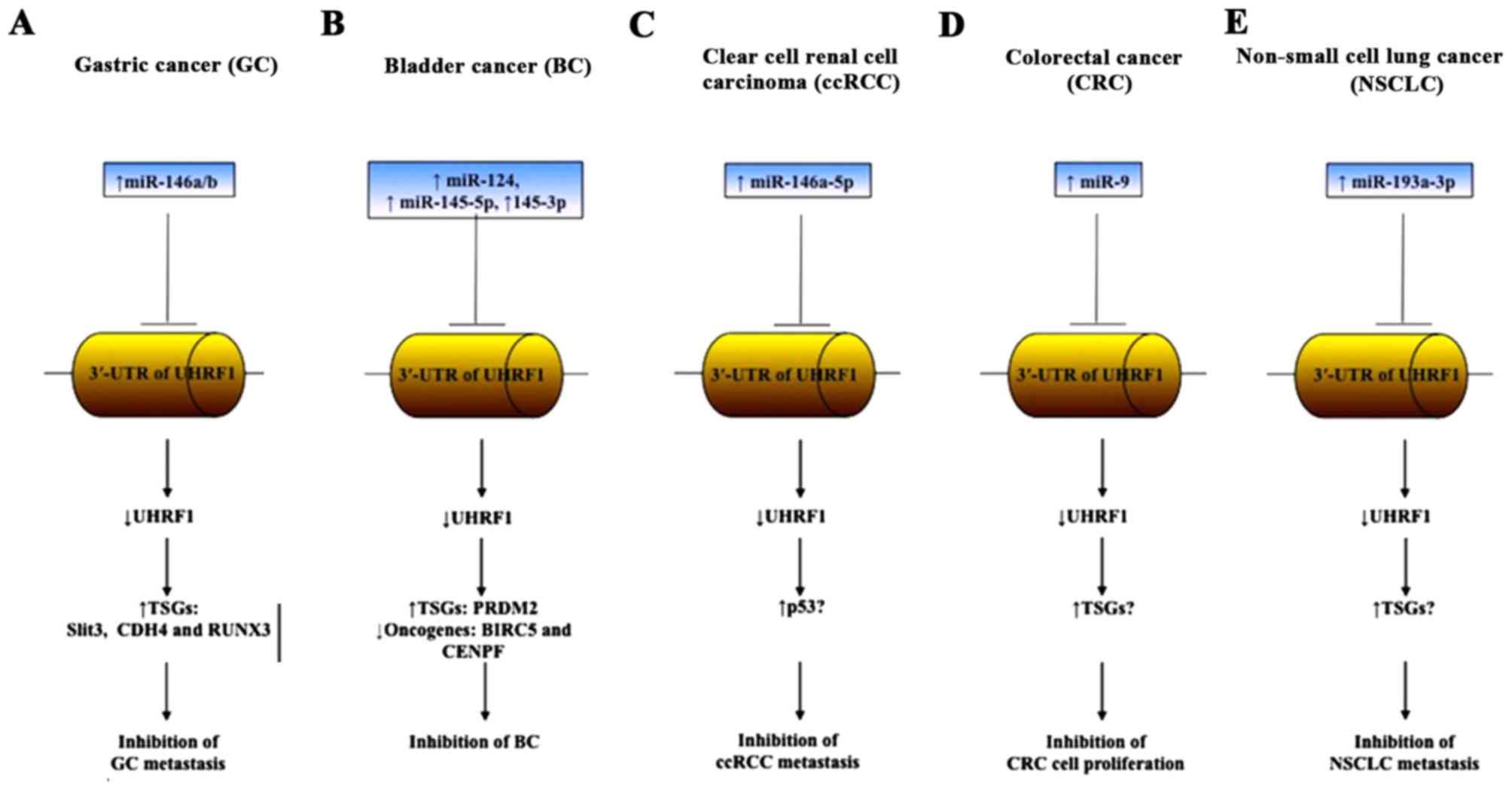
inhibition through the reactivation of TSGs (Fig. 2A).
Several previous studies have revealed the important
role of UHRF1 in human bladder cancer (BC) invasion (1,70–72). UHRF1 was demonstrated to be
overexpressed in bladder and kidney cancer, and its increased
expression levels were associated with the stage and grading of BC
(73). UHRF1 expression was increased
in BC compared with matched normal tissues, and its overexpression
was associated with tumor grade, relapse and survival rate
(70). Several underlying molecular
mechanisms have been suggested to explain the contribution of UHRF1
in the pathology of BC; however, the molecular mechanisms
underlying UHRF1 regulation in this type of tumor are largely
unknown. UHRF1 was demonstrated to promote the invasion of BC
through epigenetic silencing of the tumor suppressor kisspeptin
(KiSS1) and the regulator of G-protein signaling 2 (71,72). A
recent study also reported that the overexpression of UHRF1
detected in BC tissues was accompanied by decreased levels of
miR-124, acting as a tumor suppressor. Furthermore, miR-124
overexpression in human BC cells induced a decrease in UHRF1
expression, which led to the inhibition of cell proliferation,
metastasis and invasion (74).
Notably, a luciferase assay confirmed that miR-124 regulates UHRF1
expression by binding to the 3′-UTR of UHRF1, suggesting that UHRF1
overexpression observed in BC may be attributed at least in part to
the loss of the expression of the tumor suppressor miR-124. The
UHRF1 was recently demonstrated to be targeted in normal bladder
cells by other tumor suppressor miRNAs including miR-145-5p and
miR-145-3p (75). The downregulation
of these miRNAs in BC causes overexpression of UHRF1 (75). Furthermore, this study demonstrated
that BC expresses increased levels of UHRF1 compared with normal
tissue, and its overexpression is associated with decreased levels
of miR-145-5p and miR-145-3p. UHRF1 downregulation in BC cells
induced a reactivation of a number of TSGs including PR domain zinc
finger protein 2 (PRDM2), which acts as a histone
methyltransferase on H3K9, and also led to the inhibition of
several oncogenes including survivin (BIRC5) and centromere
protein F (CENPF) (75–77).
Notably, transfection of BC cells by miR-145-5p or miR-145-3p
induced UHRF1 downregulation and consequently cell cycle arrest and
apoptosis (75). Furthermore,
luciferase reporter assays confirmed that miR-145-5p and miR-145-3p
bind directly to different sites on 3′-UTR of UHRF1 (75). These results demonstrate that
overexpression of miR-124, miR-145-5p and miR-145-3p in BC are able
to decrease the expression levels of UHRF1, leading to BC
metastasis inhibition by downregulating the anti-apoptotic proteins
BIRC5 and CENPF (Fig. 2B).
Collectively, these results support that miRNAs regulate TSG
expression through the control of UHRF1, which dictates the level
of DNA methylation and histone H3 methylation levels via PRDM2
and/or G9a.
UHRF1 was also revealed to be overexpressed in clear
cell renal cell carcinoma (ccRCC), which represents ~70% of all
renal cell carcinomas (RCCs) (73,78,79). UHRF1
overexpression at the mRNA and protein levels in ccRCC was
associated with the downregulation of the TSG TP53 (79). This study demonstrated that UHRF1
directly binds to p53 protein causing ubiquitination-dependent
degradation of p53, leading to apoptosis inhibition and ccRCC
promotion (79). Recent studies have
demonstrated that UHRF1 is overexpressed in primary and metastatic
ccRCC tumors associated with decreased expression levels of
miR-146a-5p, which has also been demonstrated to act as a tumor
suppressor in non-small cell lung cancer (NSCLC) and prostate
cancer (78,80,81).
Notably, overexpression of miR-146a-5p in two different human
kidney cancer cell lines (786-O and ACHN) significantly decreased
the expression of UHRF1 (78). These
results indicate that miR-146a-5p negatively regulates the
expression of UHRF1 in ccRCC, and miR-146a-5p upregulation is
sufficient to induce UHRF1 degradation and p53 reactivation leading
to the inhibition of ccRCC progression (Fig. 2C). The overexpression of miR-101, a
tumor suppressor miRNA, was recently demonstrated to inhibit the
expression of UHRF1 in RCC (82–84).
Currently, it is not yet known whether UHRF1 is regulated by a
unique miRNA in a cell-specific manner, or by several miRNAs within
a same cell, and therefore further investigation is required.
Several previous studies have demonstrated that
UHRF1 is overexpressed in colorectal cancer (CRC) and induces
epigenetic silencing of several TSGs including
p16INK4A leading to cell growth and metastasis
(39,85,86). CRC
tissues and cell lines have exhibited overexpression of UHRF1 with
decreased levels of p16INK4A expression and increased
metastasis (39). Conversely, UHRF1
downregulation induced an upregulation of p16INK4A, cell
proliferation, migration inhibition, cell cycle arrest and
apoptosis (39). In the same context,
increased levels of UHRF1 were detected in specimens of CRC, and
in vitro UHRF1 downregulation resulted in inhibition of CRC
proliferation (86). Collectively,
these studies indicate that UHRF1 overexpression may be a primary
event in CRC development, and therefore its targeting may represent
a novel approach to CRC therapy. However, the upstream factors
which regulate UHRF1 expression in CRC remain unclear.
UHRF1 overexpression was associated with decreased
survival rates of patients with CRC, and a decrease in the
expression of the tumor suppressor miR-9 (87–90). The
luciferase assay demonstrated that UHRF1 is directly regulated by
miR-9, indicating that UHRF1 overexpression in CRC results from a
decrease in miR-9 expression (87).
Notably, miR-9 overexpression in CRC cells was able to
significantly decrease UHRF1 expression and cell proliferation, and
induce apoptosis (Fig. 2D). These
results demonstrate that another miRNA is involved in CRC compared
with the aforementioned types of cancer. Thus, these results
suggest a cell-specific dependence of UHRF1 regulation towards
miRNAs.
UHRF1 was also identified as being overexpressed in
several other types of NSCLC, and may serve as a diagnostic and
therapeutic marker for this type of cancer (91–93).
Increased expression levels of UHRF1 were identified in primary
NSCLC accompanied with increased levels of the three DNA
methyltransferases DNMT1, DNMT3A and DNMT3B concomitantly with
hypermethylation of several TSG promoters including
cyclin-dependent kinase inhibitor 2 (CDKN2A) and
Ras-associated domain-containing protein 1 (RASSF1)
(94). Notably, UHRF1 depletion
induced a promoter demethylation-dependent reactivation of
CDKN2A and RASSF1 with subsequent inhibition of cell
proliferation and metastasis (94).
These results suggest that UHRF1 overexpression is involved in the
molecular pathogenesis of NSCLC and that the upstream regulatory
mechanisms begin to be elucidated. Accordingly, miR-193a-3p serves
as a tumor suppressor in cancer, and its overexpression was
demonstrated to repress NSCLC progression (95). Recently, it has been identified that
miR-193a-3p inhibits NSCLC metastasis by downregulating several
oncogenes including UHRF1 (Fig. 2E),
suggesting that UHRF1 expression is inversely associated with
miR-193a-3p in NSCLC (93).
Furthermore, these studies collectively support the hypothesis that
each type of cancer has a different type of UHRF1 in terms of
miRNA.
The present review provides an insight into the
molecular mechanism underlying how miRNAs may function as tumor
suppressors by directly inhibiting the expression of the oncogene
UHRF1 which is considered to be a master of the epigenetic
silencing of several TSGs in cancer. It is also indicated that the
combined expression of regulator miRNA and UHRF1 may be a potential
diagnostic and prognostic marker in cancer. UHRF1 is overexpressed
in several types of human cancer, which contributes to the increase
in cell proliferation, metastasis and the inhibition of apoptosis.
Considering the fact that abnormalities in miRNA expression may be
a potent cause of cancer development, exploring the direct
association between UHRF1 and miRNAs will increase our
understanding of tumor pathology, and may also allow the
development of novel therapeutic strategies based on specific
targeting of the miRNA/UHRF1 pathways in several types of tumor
(Fig. 2). However, future
investigations are required to understand the downregulation of
miRNAs in the pathogenesis of cancer. Furthermore, the assessment
of the cell-specific miRNA-dependent regulation of UHRF1 as well as
the origin of the deregulation of these types of miRNA may also be
required.
The present review was supported by a French State
fund managed by the Agence Nationale de la Recherche under the
framework program Investissements d'Avenir ANR-10-IDEX-0002-02
(grant no. ANR-10-LABX-0030-INRT).
|
1
|
Unoki M, Nishidate T and Nakamura Y:
ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG
through its SRA domain. Oncogene. 23:7601–7610. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jin W, Chen L, Chen Y, Xu SG, Di GH, Yin
WJ, Wu J and Shao ZM: UHRF1 is associated with epigenetic silencing
of BRCA1 in sporadic breast cancer. Breast Cancer Res Treat.
123:359–373. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Achour M, Jacq X, Rondé P, Alhosin M,
Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A,
Hughes AD, et al: The interaction of the SRA domain of ICBP90 with
a novel domain of DNMT1 is involved in the regulation of VEGF gene
expression. Oncogene. 27:2187–2197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bronner C, Achour M, Arima Y, Chataigneau
T, Saya H and Schini-Kerth VB: The UHRF family: Oncogenes that are
drugable targets for cancer therapy in the near future? Pharmacol
Ther. 115:419–434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Berkyurek AC, Suetake I, Arita K,
Takeshita K, Nakagawa A, Shirakawa M and Tajima S: The DNA
methyltransferase Dnmt1 directly interacts with the SET and RING
finger-associated (SRA) domain of the multifunctional protein Uhrf1
to facilitate accession of the catalytic center to hemi-methylated
DNA. J Biol Chemistry. 289:379–386. 2014. View Article : Google Scholar
|
|
6
|
Alhosin M, Omran Z, Zamzami MA, Al-Malki
AL, Choudhry H, Mousli M and Bronner C: Signalling pathways in
UHRF1-dependent regulation of tumor suppressor genes in cancer. J
Exp Clin Cancer Res. 35:1742016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bronner C, Krifa M and Mousli M:
Increasing role of UHRF1 in the reading and inheritance of the
epigenetic code as well as in tumorogenesis. Biochem Pharmacol.
86:1643–1649. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alhosin M, Sharif T, Mousli M,
Etienne-Selloum N, Fuhrmann G, Schini-Kerth VB and Bronner C:
Down-regulation of UHRF1, associated with re-expression of tumor
suppressor genes, is a common feature of natural compounds
exhibiting anti-cancer properties. J Exp Clin Cancer Res.
30:412011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bronner C, Chataigneau T, Schini-Kerth VB
and Landry Y: The ‘Epigenetic Code Replication Machinery’, ECREM: A
promising drugable target of the epigenetic cell memory. Curr Med
Chemistry. 14:2629–2641. 2007. View Article : Google Scholar
|
|
10
|
Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J,
Koseki H and Wong J: UHRF1 targets DNMT1 for DNA methylation
through cooperative binding of hemi-methylated DNA and methylated
H3K9. Nat Commun. 4:15632013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nady N, Lemak A, Walker JR, Avvakumov GV,
Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X, et al:
Recognition of multivalent histone states associated with
heterochromatin by UHRF1 protein. J Biol Chem. 286:24300–24311.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Karagianni P, Amazit L, Qin J and Wong J:
ICBP90, a novel methyl K9 H3 binding protein linking protein
ubiquitination with heterochromatin formation. Mol Cell Biol.
28:705–717. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Papait R, Pistore C, Grazini U, Babbio F,
Cogliati S, Pecoraro D, Brino L, Morand AL, Dechampesme AM, Spada
F, et al: The PHD domain of Np95 (mUHRF1) is involved in
large-scale reorganization of pericentromeric heterochromatin. Mol
Biol Cell. 19:3554–3563. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jenkins Y, Markovtsov V, Lang W, Sharma P,
Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, et al:
Critical role of the ubiquitin ligase activity of UHRF1, a nuclear
RING finger protein, in tumor cell growth. Mol Biol Cell.
16:5621–5629. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Citterio E, Papait R, Nicassio F, Vecchi
M, Gomiero P, Mantovani R, Di Fiore PP and Bonapace IM: Np95 is a
histone-binding protein endowed with ubiquitin ligase activity. Mol
Cell Biol. 24:2526–2535. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li M, Chen D, Shiloh A, Luo J, Nikolaev
AY, Qin J and Gu W: Deubiquitination of p53 by HAUSP is an
important pathway for p53 stabilization. Nature. 416:648–653. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qin W, Leonhardt H and Spada F: Usp7 and
Uhrf1 control ubiquitination and stability of the maintenance DNA
methyltransferase Dnmt1. J Cell Biochem. 112:439–444. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Felle M, Joppien S, Németh A, Diermeier S,
Thalhammer V, Dobner T, Kremmer E, Kappler R and Längst G: The
USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1
and regulates the stability of UHRF1. Nucleic Acids Res.
39:8355–8365. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma H, Chen H, Guo X, Wang Z, Sowa ME,
Zheng L, Hu S, Zeng P, Guo R, Diao J, et al: M phase
phosphorylation of the epigenetic regulator UHRF1 regulates its
physical association with the deubiquitylase USP7 and stability.
Proc Natl Acad Sci USA. 109:pp. 4828–4833. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arita K, Ariyoshi M, Tochio H, Nakamura Y
and Shirakawa M: Recognition of hemi-methylated DNA by the SRA
protein UHRF1 by a base-flipping mechanism. Nature. 455:818–821.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Avvakumov GV, Walker JR, Xue S, Li Y, Duan
S, Bronner C, Arrowsmith CH and Dhe-Paganon S: Structural basis for
recognition of hemi-methylated DNA by the SRA domain of human
UHRF1. Nature. 455:822–825. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bostick M, Kim JK, Estève PO, Clark A,
Pradhan S and Jacobsen SE: UHRF1 plays a role in maintaining DNA
methylation in mammalian cells. Science. 317:1760–1764. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Unoki M, Brunet J and Mousli M: Drug
discovery targeting epigenetic codes: The great potential of UHRF1,
which links DNA methylation and histone modifications, as a drug
target in cancers and toxoplasmosis. Biochem Pharmacol.
78:1279–1288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hervouet E, Lalier L, Debien E, Cheray M,
Geairon A, Rogniaux H, Loussouarn D, Martin SA, Vallette FM and
Cartron PF: Disruption of Dnmt1/PCNA/UHRF1 interactions promotes
tumorigenesis from human and mice glial cells. PLoS One.
5:e113332010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pacaud R, Brocard E, Lalier L, Hervouet E,
Vallette FM and Cartron PF: The DNMT1/PCNA/UHRF1 disruption induces
tumorigenesis characterized by similar genetic and epigenetic
signatures. Sci Reports. 4:42302014. View Article : Google Scholar
|
|
26
|
Ge TT, Yang M, Chen Z, Lou G and Gu T:
UHRF1 gene silencing inhibits cell proliferation and promotes cell
apoptosis in human cervical squamous cell carcinoma CaSki cells. J
Ovarian Res. 9:422016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wan X, Yang S, Huang W, Wu D, Chen H, Wu
M, Li J, Li T and Li Y: UHRF1 overexpression is involved in cell
proliferation and biochemical recurrence in prostate cancer after
radical prostatectomy. J Exp Clin Cancer Res. 35:342016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abu-Alainin W, Gana T, Liloglou T,
Olayanju A, Barrera LN, Ferguson R, Campbell F, Andrews T, Goldring
C, Kitteringham N, et al: UHRF1 regulation of the Keap1-Nrf2
pathway in pancreatic cancer contributes to oncogenesis. J
Pathology. 238:423–433. 2016. View Article : Google Scholar
|
|
29
|
UHRF1 is an oncogene that promotes DNA
hypomethylation. Cancer Discov. 4:OF92014. View Article : Google Scholar
|
|
30
|
Guan D, Factor D, Liu Y, Wang Z and Kao
HY: The epigenetic regulator UHRF1 promotes ubiquitination-mediated
degradation of the tumor-suppressor protein promyelocytic leukemia
protein. Oncogene. 32:3819–3828. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Babbio F, Pistore C, Curti L, Castiglioni
I, Kunderfranco P, Brino L, Oudet P, Seiler R, Thalman GN, Roggero
E, et al: The SRA protein UHRF1 promotes epigenetic crosstalks and
is involved in prostate cancer progression. Oncogene. 31:4878–4887.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou L, Shang Y, Jin Z, Zhang W, Lv C,
Zhao X, Liu Y, Li N and Liang J: UHRF1 promotes proliferation of
gastric cancer via mediating tumor suppressor gene
hypermethylation. Cancer Biol Ther. 16:1241–1251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qu X, Davison J, Du L, Storer B, Stirewalt
DL, Heimfeld S, Estey E, Appelbaum FR and Fang M: Identification of
differentially methylated markers among cytogenetic risk groups of
acute myeloid leukemia. Epigenetics. 10:526–535. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim JH, Shim JW, Eum DY, Kim SD, Choi SH,
Yang K, Heo K and Park MT: Downregulation of UHRF1 increases tumor
malignancy by activating the CXCR4/AKT-JNK/IL-6/Snail signaling
axis in hepatocellular carcinoma cells. Sci Rep. 7:27982017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jung YD, Shim JW, Park SJ, Choi SH, Yang
K, Heo K and Park MT: Downregulation of UHRF1 promotes EMT via
inducing CXCR4 in human cancer cells. Int J Oncol. 46:1232–1242.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ashraf W, Ibrahim A, Alhosin M, Zaayter L,
Ouararhni K, Papin C, Ahmad T, Hamiche A, Mély Y, Bronner C and
Mousli M: The epigenetic integrator UHRF1: On the road to become a
universal biomarker for cancer. Oncotarget. 8:51946–51962.
2017.PubMed/NCBI
|
|
37
|
Cui L, Chen J, Zhang Q, Wang X, Qu J,
Zhang J and Dang S: Up-regulation of UHRF1 by oncogenic Ras
promoted the growth, migration, and metastasis of pancreatic cancer
cells. Mol Cell Biochem. 400:223–232. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qin L, Dong Z and Zhang JT: Reversible
epigenetic regulation of 14-3-3sigma expression in acquired
gemcitabine resistance by uhrf1 and DNA methyltransferase 1. Mol
Pharmacol. 86:561–569. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang F, Yang YZ, Shi CZ, Zhang P, Moyer
MP, Zhang HZ, Zou Y and Qin HL: UHRF1 promotes cell growth and
metastasis through repression of p16(ink(4)a) in colorectal cancer.
Ann Surg Oncol. 19:2753–2762. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Achour M, Mousli M, Alhosin M, Ibrahim A,
Peluso J, Muller CD, Schini-Kerth VB, Hamiche A, Dhe-Paganon S and
Bronner C: Epigallocatechin-3-gallate up-regulates tumor suppressor
gene expression via a reactive oxygen species-dependent
down-regulation of UHRF1. Biochem Biophys Res Commun. 430:208–212.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Choudhry H, Harris AL and McIntyre A: The
tumour hypoxia induced non-coding transcriptome. Mol Aspects Med.
47–48. 1–53. 2016.PubMed/NCBI
|
|
43
|
Choudhry H and Mole DR: Hypoxic regulation
of the noncoding genome and NEAT1. Brief Funct Genomics.
15:174–185. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Josse C and Bours V: MicroRNAs and
inflammation in colorectal cancer. Adv Exp Med Biol. 937:53–69.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Vicente R, Noël D, Pers YM, Apparailly F
and Jorgensen C: Deregulation and therapeutic potential of
microRNAs in arthritic diseases. Nat Rev Rheumatol. 12:4962016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Guedes JR, Santana I, Cunha C, Duro D,
Almeida MR, Cardoso AM, de Lima MC and Cardoso AL: MicroRNA
deregulation and chemotaxis and phagocytosis impairment in
Alzheimer's disease. Alzheimers Dement (Amst). 3:7–17.
2015.PubMed/NCBI
|
|
47
|
Irmak-Yazicioglu MB: Mechanisms of
MicroRNA Deregulation and MicroRNA Targets in Gastric Cancer. Oncol
Res Treat. 39:136–139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ruan X, Zuo Q, Jia H, Chau J, Lin J, Ao J,
Xia X, Liu H, Habib SL, Fu C and Li B: P53 deficiency-induced Smad1
upregulation suppresses tumorigenesis and causes chemoresistance in
colorectal cancers. J Mol Cell Biol. 7:105–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shah MY, Ferrajoli A, Sood AK,
Lopez-Berestein G and Calin GA: microRNA therapeutics in cancer-an
emerging concept. EBioMedicine. 12:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li G, Yang F, Gu S, Li Z and Xue M:
MicroRNA-101 induces apoptosis in cisplatin-resistant gastric
cancer cells by targeting VEGF-C. Mol Med Rep. 13:572–578. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xu J, Wang Z, Li S, Chen J, Zhang J, Jiang
C, Zhao Z, Li J, Li Y and Li X: Combinatorial epigenetic regulation
of non-coding RNAs has profound effects on oncogenic pathways in
breast cancer subtypes. Brief Bioinform. Oct 14–2016.(Epub ahead of
print). View Article : Google Scholar
|
|
54
|
Kaur S, Lotsari-Salomaa JE,
Seppänen-Kaijansinkko R and Peltomäki P: MicroRNA Methylation in
Colorectal Cancer. Adv Exp Med Biol. 937:109–122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Voorhoeve PM: MicroRNAs: Oncogenes, tumor
suppressors or master regulators of cancer heterogeneity? Biochim
Biophys Acta. 1805:72–86. 2010.PubMed/NCBI
|
|
56
|
Zhou X, Wang X, Huang Z, Wang J, Zhu W,
Shu Y and Liu P: Prognostic value of miR-21 in various cancers: An
updating meta-analysis. PLoS One. 9:e1024132014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Medina PP, Nolde M and Slack FJ: OncomiR
addiction in an in vivo model of microRNA-21-induced pre-B-cell
lymphoma. Nature. 467:86–90. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Selcuklu SD, Donoghue MT and Spillane C:
miR-21 as a key regulator of oncogenic processes. Biochem Soc
Transac. 37:918–925. 2009. View Article : Google Scholar
|
|
59
|
Favreau AJ, McGlauflin RE, Duarte CW and
Sathyanarayana P: miR-199b, a novel tumor suppressor miRNA in acute
myeloid leukemia with prognostic implications. Exp Hematol Oncol.
5:42016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Venkataraman S, Birks DK, Balakrishnan I,
Alimova I, Harris PS, Patel PR, Handler MH, Dubuc A, Taylor MD,
Foreman NK and Vibhakar R: MicroRNA 218 acts as a tumor suppressor
by targeting multiple cancer phenotype-associated genes in
medulloblastoma. J Biol Chem. 288:1918–1928. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Boukhari A, Alhosin M, Bronner C, Sagini
K, Truchot C, Sick E, Schini-Kerth VB, André P, Mély Y, Mousli M
and Gies JP: CD47 activation-induced UHRF1 over-expression is
associated with silencing of tumor suppressor gene p16INK4A in
glioblastoma cells. Anticancer Res. 35:149–157. 2015.PubMed/NCBI
|
|
62
|
Arima Y, Hirota T, Bronner C, Mousli M,
Fujiwara T, Niwa S, Ishikawa H and Saya H: Down-regulation of
nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage
checkpoint signals contributes to cell cycle arrest at G1/S
transition. Genes Cells. 9:131–142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Alhosin M, Abusnina A, Achour M, Sharif T,
Muller C, Peluso J, Chataigneau T, Lugnier C, Schini-Kerth VB,
Bronner C and Fuhrmann G: Induction of apoptosis by thymoquinone in
lymphoblastic leukemia Jurkat cells is mediated by a p73-dependent
pathway which targets the epigenetic integrator UHRF1. Biochem
Pharmacol. 79:1251–1260. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wu SM, Cheng WL, Liao CJ, Chi HC, Lin YH,
Tseng YH, Tsai CY, Chen CY, Lin SL, Chen WJ, et al: Negative
modulation of the epigenetic regulator, UHRF1, by thyroid hormone
receptors suppresses liver cancer cell growth. Int J Cancer.
137:37–49. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Soleimani A, Ghanadi K, Noormohammadi Z
and Irani S: The correlation between miR-146a C/G polymorphism and
UHRF1gene expression level in gastric tumor. J Dig Dis. 17:169–174.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ge M, Gui Z, Wang X and Yan F: Analysis of
the UHRF1 expression in serum and tissue for gastric cancer
detection. Biomarkers. 20:183–188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhou L, Zhao X, Han Y, Lu Y, Shang Y, Liu
C, Li T, Jin Z, Fan D and Wu K: Regulation of UHRF1 by miR-146a/b
modulates gastric cancer invasion and metastasis. FASEB J.
27:4929–4939. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Patnaik SK, Kannisto E, Mallick R and
Yendamuri S: Overexpression of the lung cancer-prognostic miR-146b
microRNAs has a minimal and negative effect on the malignant
phenotype of A549 lung cancer cells. PLoS One. 6:e223792011.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Xiao B, Zhu ED, Li N, Lu DS, Li W, Li BS,
Zhao YL, Mao XH, Guo G, Yu PW and Zou QM: Increased miR-146a in
gastric cancer directly targets SMAD4 and is involved in modulating
cell proliferation and apoptosis. Oncol Rep. 27:559–566.
2012.PubMed/NCBI
|
|
70
|
Yang GL, Zhang LH, Bo JJ, Chen HG, Cao M,
Liu DM and Huang YR: UHRF1 is associated with tumor recurrence in
non-muscle-invasive bladder cancer. Med Oncol. 29:842–847. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang Y, Huang Z, Zhu Z, Zheng X, Liu J,
Han Z, Ma X and Zhang Y: Upregulated UHRF1 promotes bladder cancer
cell invasion by epigenetic silencing of KiSS1. PLoS One.
9:e1042522014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ying L, Lin J, Qiu F, Cao M, Chen H, Liu Z
and Huang Y: Epigenetic repression of regulator of G-protein
signaling 2 by ubiquitin-like with PHD and ring-finger domain 1
promotes bladder cancer progression. FEBS J. 282:174–182. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Unoki M, Kelly JD, Neal DE, Ponder BA,
Nakamura Y and Hamamoto R: UHRF1 is a novel molecular marker for
diagnosis and the prognosis of bladder cancer. Br J Cancer.
101:98–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wang X, Wu Q, Xu B, Wang P, Fan W, Cai Y,
Gu X and Meng F: miR-124 exerts tumor suppressive functions on the
cell proliferation, motility and angiogenesis of bladder cancer by
fine-tuning UHRF1. FEBS J. 282:4376–4388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Matsushita R, Yoshino H, Enokida H, Goto
Y, Miyamoto K, Yonemori M, Inoguchi S, Nakagawa M and Seki N:
Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145
(miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell
aggressiveness. Oncotarget. 7:28460–28487. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Xie W, Li X, Chen X and Huang S and Huang
S: Decreased expression of PRDM2 (RIZ1) and its correlation with
risk stratification in patients with myelodysplastic syndrome. Br J
Haematol. 150:242–244. 2010.PubMed/NCBI
|
|
77
|
Kim KC, Geng L and Huang S: Inactivation
of a histone methyltransferase by mutations in human cancers.
Cancer Res. 63:7619–7623. 2003.PubMed/NCBI
|
|
78
|
Wotschofsky Z, Gummlich L, Liep J, Stephan
C, Kilic E, Jung K, Billaud JN and Meyer HA: Integrated microRNA
and mRNA signature associated with the transition from the locally
confined to the metastasized clear cell renal cell carcinoma
exemplified by miR-146-5p. PLoS One. 11:e01487462016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ma J, Peng J, Mo R, Ma S, Wang J, Zang L,
Li W and Fan J: Ubiquitin E3 ligase UHRF1 regulates p53
ubiquitination and p53-dependent cell apoptosis in clear cell renal
cell carcinoma. Biochem Biophys Res Commun. 464:147–153. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li YL, Wang J, Zhang CY, Shen YQ, Wang HM,
Ding L, Gu YC, Lou JT, Zhao XT, Ma Z and Jin YX: MiR-146a-5p
inhibits cell proliferation and cell cycle progression in NSCLC
cell lines by targeting CCND1 and CCND2. Oncotarget. 7:59287–59298.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sun Q, Zhao X, Liu X, Wang Y, Huang J,
Jiang B, Chen Q and Yu J: miR-146a functions as a tumor suppressor
in prostate cancer by targeting Rac1. Prostate. 74:1613–1621. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Riquelme I, Tapia O, Leal P, Sandoval A,
Varga MG, Letelier P, Buchegger K, Bizama C, Espinoza JA, Peek RM,
et al: miR-101-2, miR-125b-2 and miR-451a act as potential tumor
suppressors in gastric cancer through regulation of the
PI3K/AKT/mTOR pathway. Cell Oncology (Dordr). 39:23–33. 2016.
View Article : Google Scholar
|
|
83
|
Farhadi E, Zaker F, Safa M and Rezvani MR:
miR-101 sensitizes K562 cell line to imatinib through Jak2
downregulation and inhibition of NF-κB target genes. Tumour Biol.
37:14117–14128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Goto Y, Kurozumi A, Nohata N, Kojima S,
Matsushita R, Yoshino H, Yamazaki K, Ishida Y, Ichikawa T, Naya Y
and Seki N: The microRNA signature of patients with sunitinib
failure: Regulation of UHRF1 pathways by microRNA-101 in renal cell
carcinoma. Oncotarget. 7:59070–59086. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Walter A, Etienne-Selloum N, Brasse D,
Khallouf H, Bronner C, Rio MC, Beretz A and Schini-Kerth VB: Intake
of grape-derived polyphenols reduces C26 tumor growth by inhibiting
angiogenesis and inducing apoptosis. FASEB J. 24:3360–3369. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Kofunato Y, Kumamoto K, Saitou K, Hayase
S, Okayama H, Miyamoto K, Sato Y, Katakura K, Nakamura I, Ohki S,
et al: UHRF1 expression is upregulated and associated with cellular
proliferation in colorectal cancer. Oncol Rep. 28:1997–2002. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhu M, Xu Y, Ge M, Gui Z and Yan F:
Regulation of UHRF1 by microRNA-9 modulates colorectal cancer cell
proliferation and apoptosis. Cancer Sci. 106:833–839. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Bandres E, Agirre X, Bitarte N, Ramirez N,
Zarate R, Roman-Gomez J, Prosper F and Garcia-Foncillas J:
Epigenetic regulation of microRNA expression in colorectal cancer.
Int J Cancer. 125:2737–2743. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Cekaite L, Rantala JK, Bruun J, Guriby M,
Agesen TH, Danielsen SA, Lind GE, Nesbakken A, Kallioniemi O, Lothe
RA and Skotheim RI: MiR-9, −31, and −182 deregulation promote
proliferation and tumor cell survival in colon cancer. Neoplasia.
14:868–879. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lu MH, Huang CC, Pan MR, Chen HH and Hung
WC: Prospero homeobox 1 promotes epithelial-mesenchymal transition
in colon cancer cells by inhibiting E-cadherin via miR-9. Clin
Cancer Res. 18:6416–6425. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Unoki M, Daigo Y, Koinuma J, Tsuchiya E,
Hamamoto R and Nakamura Y: UHRF1 is a novel diagnostic marker of
lung cancer. Br J Cancer. 103:217–222. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wu H, Meng S, Xu Q, Wang X, Wang J, Gong
R, Song Y, Duan Y and Zhang Y: Gene expression profiling of lung
adenocarcinoma in Xuanwei, China. Eur J Cancer Prev. 25:508–517.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Deng W, Yan M, Yu T, Ge H, Lin H, Li J,
Liu Y, Geng Q, Zhu M, Liu L, et al: Quantitative proteomic analysis
of the metastasis-inhibitory mechanism of miR-193a-3p in non-small
cell lung cancer. Cell Physiol Biochem. 35:1677–1688. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Daskalos A, Oleksiewicz U, Filia A,
Nikolaidis G, Xinarianos G, Gosney JR, Malliri A, Field JK and
Liloglou T: UHRF1-mediated tumor suppressor gene inactivation in
nonsmall cell lung cancer. Cancer. 117:1027–1037. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F,
Sun L, Zhang Y, Cui Y, Zhang F, et al: MicroRNA-193a-3p and −5p
suppress the metastasis of human non-small-cell lung cancer by
downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Oncogene. 34:413–423. 2015. View Article : Google Scholar : PubMed/NCBI
|