Introduction
Osteosarcoma is a highly malignant bone tumor with
widespread histological heterogeneity, a lack of biomarkers, high
local aggressiveness and a rapid metastasizing potential (1). Osteosarcoma is the most common primary
bone malignancy and the third most common cancer in adolescents and
young adults (2). Chemotherapy
combined with surgery is the principal mode of treatment for
osteosarcoma. However, tumor resistance to chemotherapy and
molecularly targeted therapies limits their effectiveness (1,3). Tumor
cells may become cross-resistant to a broad spectrum of
chemotherapeutic agents with different mechanisms and structures
following a single-drug treatment; this is known as multidrug
resistance (MDR) (4). Within tumor
cells, various MDR mechanisms can operate, including increased drug
efflux, mutations of the drug target, enhanced repair of DNA
damage, activation of alternative signaling pathways and evasion of
cell death (3). Although the precise
mechanisms of MDR remain unclear, several cell membrane transporter
proteins are linked to the resistance observed with commonly used
chemotherapeutic agents. For example, multidrug resistance protein
1 (MDR1), also known as P-glycoprotein (P-gp), belongs to the
ATP-binding cassette (ABC) transporter family, and regulates the
efflux of multiple structurally and mechanistically unrelated
chemotherapeutic agents across the plasma membrane (3).
LIM kinase 1 (LIMK1) is a member of the
serine/threonine protein kinase family. It is activated when the
Thr508 residue is phosphorylated by Rho-GTPases such as Rho, Rac
and cell division cycle 42 (5,6). LIMK1
functions through regulating actin cytoskeleton remodeling by
phosphorylating and inactivating cofilin on the Ser3 residue
(7,8).
Increasing evidence suggests that LIMK1 serves an important role in
tumor metastasis and invasion (9).
However, few studies have addressed the role of LIMK1 in MDR in
tumor cells (10). In a previous
study, an osteosarcoma MDR model was established by using the human
osteosarcoma cell line MG63 and vincristine (VCR), a drug commonly
used in the treatment of osteosarcoma (11). Notably, the development of MDR was
associated with increased expression of LIMK1. Therefore, the
current study was designed to determine whether the overexpression
of LIMK1 contributes to MDR and to identify its potential
mechanism.
Materials and methods
Cells and cell culture
The human osteosarcoma cell lines U2OS and MG63
(purchased from the Institute of Biochemistry and Cell Biology,
Chinese Academy of Sciences, Shanghai, China) were cultured in high
glucose-Dulbecco's modified Eagle's medium (H-DMEM; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal
bovine serum (FBS; Hyclone, Logan, UT, USA). Human fetal
osteoblastic (hFOB) 1.19 cells (donated by the Pathology Laboratory
of Jilin University, Changchun, China) were cultured in DMEM-F12
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FCS.
All three cell types were incubated at 37°C in a humidified
atmosphere containing 5% CO2. Tumor samples from 6
patients with osteosarcoma were collected intra-operatively and
stored at −80°C (March 2015 form the China-Japan Union Hospital of
Jilan University, Changchun, China). Written consent was obtained
from all patients or their families, and the study was conducted in
accordance with the Declaration of Helsinki. The protocol was
approved by the Institutional Review Board of Jilin University.
Establishment of VCR-resistant cell
sublines
To generate VCR-resistant sublines, MG63 cells were
exposed to increasing concentrations of VCR as described previously
(11). Briefly, parental MG63 cells
were cultured initially in H-DMEM containing 10 ng/ml VCR (Shanghai
New Hualian Pharmaceutical Co., Ltd., Shanghai, China) for 72 h.
Surviving cells were collected and cultured in VCR-free medium for
1–2 weeks, and then further cultured with VCR-containing H-DMEM.
This procedure was repeated ≥5 times until the majority of cells
survived in the drug-containing medium. Cells were then exposed to
increasing concentrations (10–500 ng/ml) of VCR following the same
procedure. This process required 4–6 weeks to establish adequate
growth at each VCR concentration. Eventually, a subline resistant
to 500 ng/ml VCR was obtained, which was named MG63/VCR. Prior to
each experiment, MG63 and MG63/VCR cells were maintained in
drug-free media and subcultured ≥3 times.
Drug sensitivity assays
The Cell Counting kit-8 (CCK-8) (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) assay was used to determine
the sensitivities of cells to VCR, pirarubicin (THP), methotrexate
(MTX), doxorubicin (DOX) (all from Hualian Pharmaceutical Co.,
Shanghai, China) and paclitaxel (PTX) (Laboratories Pierre Fabre,
Castres, Cedex, France). Cells in the logarithmic growth phase were
trypsinized and plated onto 96-well plates at 8×103
cells/well, and cultured for 24 h in 100 µl H-DMEM supplemented
with 10% FBS (Hyclone). The culture medium was then replaced with
medium containing serial dilutions of each drug (125, 25, 5, 1,
0.2, 0.04, 0.008 and 0.0016 µg/ml). Drug-free medium was added to
the control and blank wells. After 48 h, the medium was replaced
with 10% CCK-8. After incubation at 37°C for 2 h, the plates were
analyzed in a plate reader at 450 nm. The percentage of inhibition
for each treatment was calculated using the following formula:
Inhibition=(ODcontrol-OD experimental
group)/ODcontrolx100. Based on this information,
the 50% inhibitory concentration (IC50) was calculated
using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla,
CA, USA). The resistance index (RI) was calculated as IC50
MG63/VCR/IC50 MG63.
Immunohistochemical staining
Immunohistochemical staining was performed as
described previously (12). Briefly,
tumor tissue sections were incubated with 3%
H2O2 to inactivate endogenous peroxidases,
followed by deparaffinization and rehydration. The antigen was
released with 3% proteinase K, followed by washing with PBS and
blocking with 10% goat serum (Santa Cruz Biotechnology, Inc.) for 1
h at room temperature. The sections were then incubated for 1 h at
room temperature with a mouse monoclonal antibody targeting β-actin
(sc-58673, dilution 1:500; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) or a rabbit polyclonal antibody targeting LIMK1 (sc-48346,
dilution 1:100; Santa Cruz Biotechnology, Inc.). Biotinylated
rabbit anti-mouse immunoglobulin (sc-2359, Santa Cruz
Biotechnology, Inc.) diluted 1:100 in 3% normal goat serum (Santa
Cruz Biotechnology, Inc.) was applied for 1 h at room temperature,
followed by incubation with a horseradish peroxidase
(HRP)-conjugated streptavidin-biotin complex (Santa Cruz
Biotechnology, Inc.) for 10 min at room temperature.
Paraffin-embedded sections were dehydrated, mounted with neutral
gum and observed under a light microscope (Olympus Corporation,
Tokyo, Japan).
Immunoprecipitation and western blot
analysis
U2OS, MG63 and hFOB1.19 cells were lysed with lysis
buffer [150 mM NaCl, 1% Nonidet P-40, 50 mM Tris-HCl (pH 8.0)
containing 20 µM phenylmethylsulfonyl fluoride] at 4°C for 30 min,
and then centrifuged at 12,000 × g at 4°C for 15 min. The
supernatant was stored at −70°C. Equal amounts of protein (100 ng),
15 µl of protein A agarose beads (Santa Cruz Biotechnology, Inc.)
and 1 µl of anti-LIMK1 antibody (sc-48346, Santa Cruz
Biotechnology, Inc.) were added to the tubes, which were rotated
overnight at 4°C. The agarose-antibody-antigen complexes were
collected by centrifugation for 20 sec at 12,000 × g and 4°C. The
supernatant was removed carefully, and the complexes were washed
twice with lysis buffer. The pellet was resuspended in gel-loading
buffer (Santa Cruz Biotechnology, Inc.), and the proteins were
denatured by boiling for 5 min. Protein A agarose beads were
removed by centrifugation at 12,000 × g for 20 sec at 4°C, and the
supernatant was transferred to a fresh tube. The proteins (20 µl
per lane) were separated by cellulose acetate membrane SDS-PAGE (5%
acrylamide in concentration gel and 12% in separation gel) and
analyzed by immunoblotting, as described previously (13,14), using
primary antibodies against LIMK1. Immunodetection was accomplished
using an HRP-conjugated goat anti-rabbit secondary antibody
(BS-13278, 1:1,000; Bioworld Technology, Inc., St. Louis Park, MN,
USA) rotated for 1 h at room temperature. The protein bands were
developed with a Novex ECL Chemiluminescent Substrate Reagent kit
(Invitrogen; Thermo Fisher Scientific, Inc.) and analyzed by ImageJ
software version 1.44 (National Institutes of Health, Bethesda, MD,
USA).
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA from the cell lines was isolated using
TRIzol reagent (Takara Bio, Inc., Otsu, Japan) and quantified using
a NanoDrop ND-2000 (NanoDrop; Thermo Fisher Scientific, Inc.,
Wilmington, DE, USA), and reversely transcribed into cDNA using the
EX Tag kit (TaKaRa, Japan) according to the manufacturer's
protocols. RT-PCR was performed utilizing TaKaRa EX Taq DNA
Polymerase (Takara Bio, Inc.) under the following conditions:
Denaturation at 98°C for 10 sec, annealing at 54–62°C for 30 sec
and elongation at 72°C for 1 min for 20–30 cycles. The PCR products
were visualized by electrophoresis on a 1.5% agarose gel stained
with GelRed™ (Biotium, Inc., Freemont, CA, USA). Images were
obtained using the Gel Doc XR+ system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) and analyzed using Image Lab™ software version
4.0 (Bio-Rad Laboratories, Inc.). The PCR primers used were: LIMK1,
forward 5′-TGAGACAGGTGAGGTGATGG-3′ and reverse
5′-AGGCTGAGTCTTCTCGTCCA-3′; MDR1, forward
5′-ATATCAGCAGCCCACATCAT-3′ and reverse 5′-GAAGCACTGGGATGTCCGGT-3′;
and β-actin, forward 5′-CTGGGACGACATGGAGAAAA-3′ and reverse
5′-AAGGAAGGCTGGAAGAGTGC-3′.
Plasmid transfections
To assess the effect of LIMK1 in the multidrug
resistance, four different plasmids were transfected into MG63/VCR
cells using Lipofectamine™ 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. An expression plasmid coding for wild-type
LIMK1-myc-enhanced cyan fluorescent protein (ECFP-LIMK1) and a
short hairpin RNA (shRNA)-targeting LIMK1 construct, namely
pSUPER-LIMK1-small interfering RNA (siRNA) (pSUPER-LIMK1), were
constructed as described previously (11,12). The
pSUPER-negative control-siRNA (pSUPER) vector containing a
scrambled non-target sequence and an empty vector containing
myc-ECFP, were used as negative controls. Transfected cells were
cultured for 24 h prior to being transferred into 60-mm dishes or
96-well plates for subsequent experiments.
DOX efflux assay
Exponentially growing cells were plated onto 60-mm
Petri dishes and incubated with 1.5 µM DOX for 3 h at 37°C.
Subsequently, cells were washed three times with ice-cold PBS, then
centrifuged and suspended in ice-cold PBS and the mono-dispersed
cells were analyzed by a flow cytometer (BD Biosciences, San Jose,
CA, USA) using an argon laser of 15 mW at 488 nm (15). Cell fluorescence was evaluated in
duplicate for each group, and all experiments were performed in
triplicate.
Measurement of apoptosis
MG63/VCR cells in the exponential growth phase were
plated onto 60-mm Petri dishes and incubated with 500 ng/ml VCR for
6 h at 37°C. Upon incubation, apoptosis was evaluated using
fluorescein isothiocyanate-Annexin V and PI (BD Biosciences)
according to the manufacturer's protocol. The analysis was
performed in a flow cytometer (BD Biosciences) using an argon laser
of 15 mW at 488 nm. Cells (≥10,000 cells/sample) were collected and
analyzed using Cell Quest software version 7.5.3 (BD
Biosciences).
Statistical analyses
All data are expressed as means ± standard
deviation. Statistical significance was assessed by the unpaired
Student's t-test when comparing means between two groups or one-way
analysis of variance with post-hoc Dunett's test to compare mean
values among three or more groups. Statistical analysis was
conducted with GraphPad Prism version 5.04 for Windows (GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
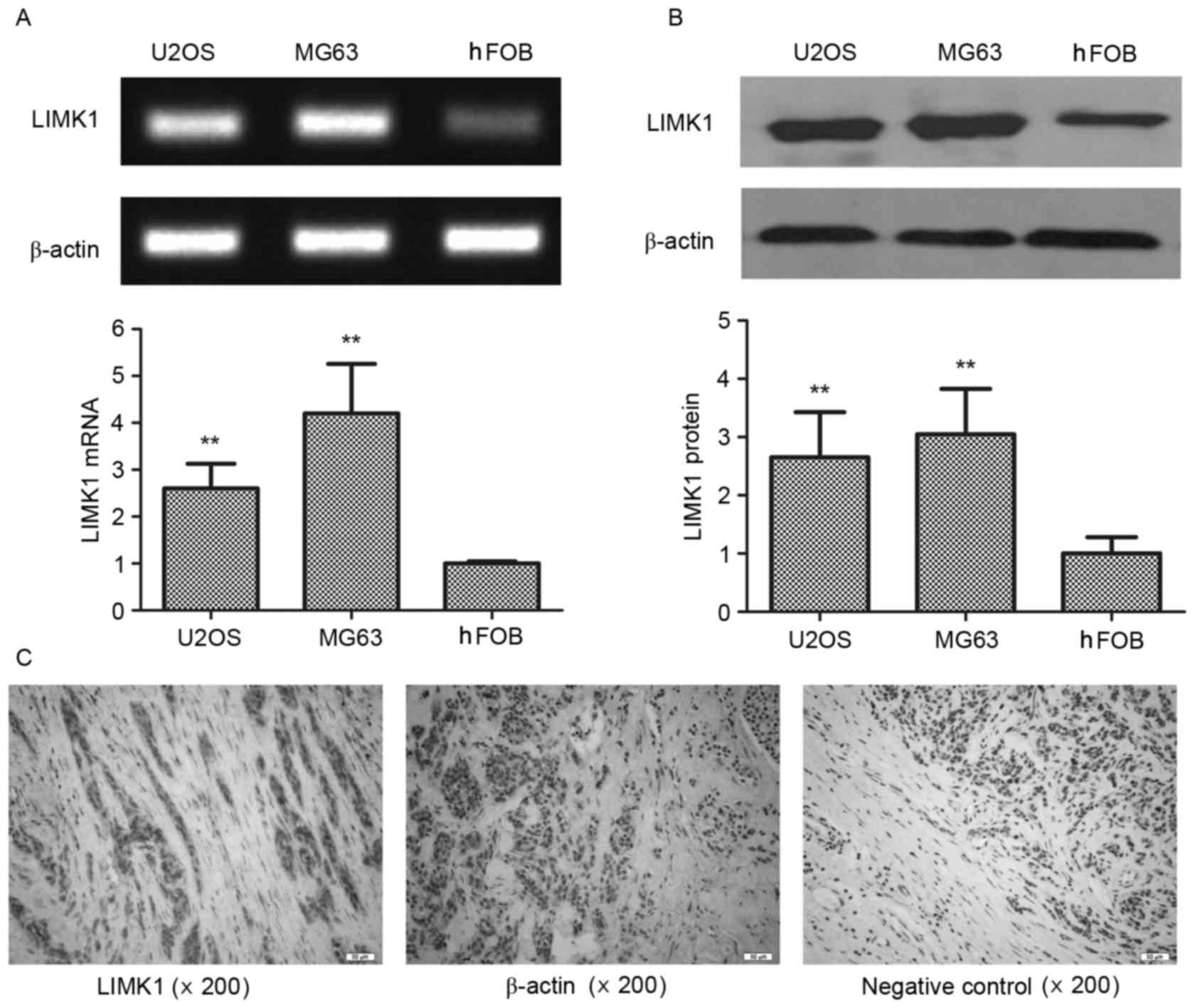
Elevated expression of LIMK1 in human
osteosarcoma cells
To explore the role of LIMK1 in the progression of
osteosarcoma, its expression in vitro was first assessed.
Two osteosarcoma cell lines, U2OS and MG63, were used and compared
with normal hFOB1.19 osteoblast cells. The expression of LIMK1
messenger RNA (mRNA) was significantly increased in both MG63 and
U2OS cells compared with that in hFOB1.19 cells (by 2.6- and
4.2-fold, respectively) (Fig. 1A). A
similar disparity was observed in western blot assays (P<0.01)
(Fig. 1B). The expression of LIMK1
was also analyzed in human osteosarcoma tissues by
immunohistochemistry. LIMK1 was highly expressed in the tumor
parenchyma compared with that in the mesenchyme (Fig. 1C) in 83.3% of the specimens derived
from 6 clinical patients. The expression of β-actin was unchanged
between the parenchyma and mesenchyme. These results indicated that
LIMK1 was overexpressed in osteosarcoma parenchyma, and suggested
that LIMK1 may affect the progression of this type of cancer.
LIMK1 and MDR1 are overexpressed in
multidrug resistant MG63/VCR cells
In previous study (11), a multidrug resistant MG63/VCR subline
was established by intermittent exposure of MG63 cells to gradually
increasing VCR concentrations, as specified in the Materials and
methods section. MG63/VCR cells were less sensitive to VCR-induced
cytotoxicity than parental cells (Fig.
2A). The IC50 values of MG63 and MG63/VCR cells were
1.0 and 453.4 ng/ml, respectively. MG63/VCR cells also exhibited
cross-resistance to other structurally and mechanistically
different drugs. The RI values of VCR, MTX, DOX, PTX and THP were
476.2, 4.7, 46.5, 379.0 and 23.7, respectively (Fig. 2B). To explore the mechanism by which
MG63/VCR cells exhibited increased drug resistance, the expression
of LIMK1 and MDR1/P-gp was examined by RT-PCR and western blot
analysis. The expression of LIMK1 and MDR1 mRNA was elevated in
MG63/VCR cells by 2.4- and 5.1-fold, respectively, compared with
that in MG63 cells (P<0.01) (Fig.
2C). A similar result was obtained in the western blot assay,
with increases by 4.7- and 6.6-fold in LIMK1 and MDR1 protein
expression, respectively (P<0.01) (Fig. 2D). These results indicated that LIMK1
and MDR1/P-gp may be important in the MDR of osteosarcoma.
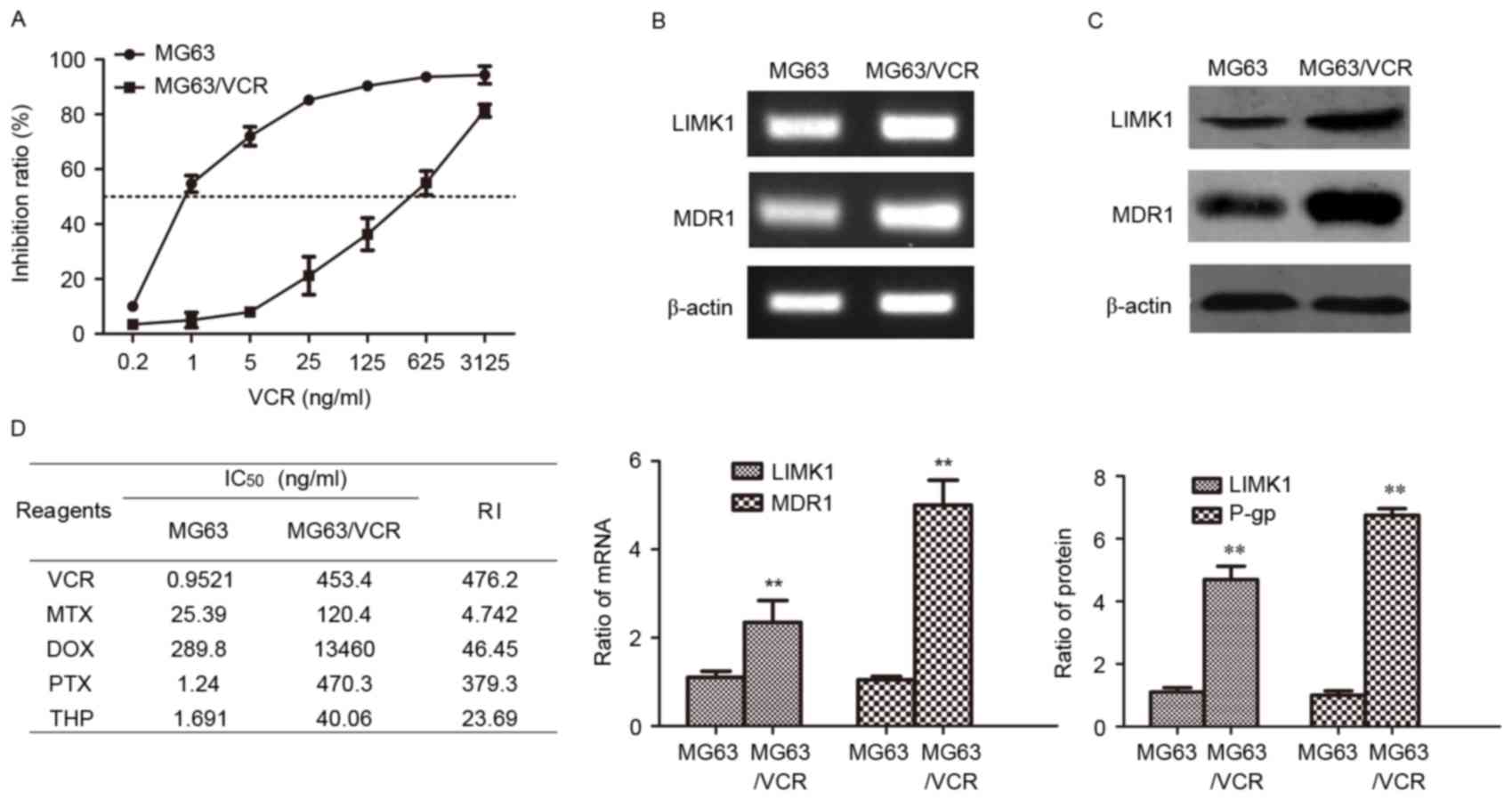 | Figure 2.Expression of LIMK1 and MDR1 in MG63
and MG63/VCR cells. (A) Dose-dependent cell viability curves of
cells treated with VCR. (B) Agarose gel electrophoresis of the
reverse transcription-polymerase chain reaction products for LIMK1
and MDR1. (C) Western blotting was performed to detect the protein
levels of LIMK1 and MDR1/P-gp proteins. (D) The concentration of
each drug that produced 50% inhibition of growth (IC50)
and the RI values of various commonly used anticancer drugs in MG63
and MG63/VCR cells are shown. **P<0.01 vs. MG63 using the
Student's t-test. All results are from three or four independent
experiments. Data are expressed as means ± standard deviation.
LIMK1, LIM kinase 1; MDR1, multidrug resistance protein 1; P-gp,
P-glycoprotein; mRNA, messenger RNA; IC50, 50%
inhibitory concentration; RI, resistance index; MG63/VCR, multidrug
resistant MG63 cells; VCR, vincristine; MTX, methotrexate; DOX,
doxorubicin; PTX, paclitaxel; THP, pirarubicin. |
LIMK1 serves a key role in the MDR of
MG63/VCR cells
To assess whether LIMK1 was involved in the MDR of
MG63/VCR cells, LIMK1 was knocked down by transfecting these cells
with pSUPER-LIMK1 plasmids coding LIMK1 shRNA target sequences. In
addition, LIMK1 expression was upregulated by transfecting the
cells with ECFP-LIMK1 plasmids coding wild-type LIMK1. The
viability of the transfected cells was evaluated upon treatment
with a series of diluted drugs. A representative result with VCR is
shown in Fig. 3A. The IC50
values for VCR, MTX, PTX and THP were calculated and shown in
Fig. 3B. Overall, cells expressing
high levels of LIMK1 exhibited strong resistance to all
chemotherapeutic drugs. DOX is a commonly used anticancer drug that
has autofluorescence at the same wavelength as PI (15). The concentration of DOX was evaluated
in cells transfected with different plasmids. The results revealed
that DOX accumulation was the highest in MG63/VCR cells transfected
with pSUPER-LIMK1 compared with in other groups, as evidenced by
the increased DOX fluorescence intensity in these cells. This
suggested that DOX efflux was downregulated along with LIMK1. By
contrast, cells transfected with the ECFP-LIMK1 plasmid exhibited
an increased efflux ability and reduced sensitivity to the drugs
tested (P<0.01) (Fig. 3C). The
results from the apoptosis assays were consistent with these
findings. Cytometric dot-plot images of MG63/VCR cells obtained
after 6 h of incubation with the IC50 of VCR were used
to evaluate the population of apoptotic cells in the lower right
quadrant of the graphs (which corresponds to Annexin V-positive and
PI-negative cells) (Fig. 3D). The
apoptosis rate in cells transfected with the pSUPER-LIMK1 plasmid
(25.6%) was higher than that in the empty vector group (6.2%) and
in the cells transfected with the ECFP-LIMK1 plasmid (1.3%)
(P<0.01) (Fig. 3D). These results
indicate that LIMK1 is important in the MDR of MG63/VCR cells.
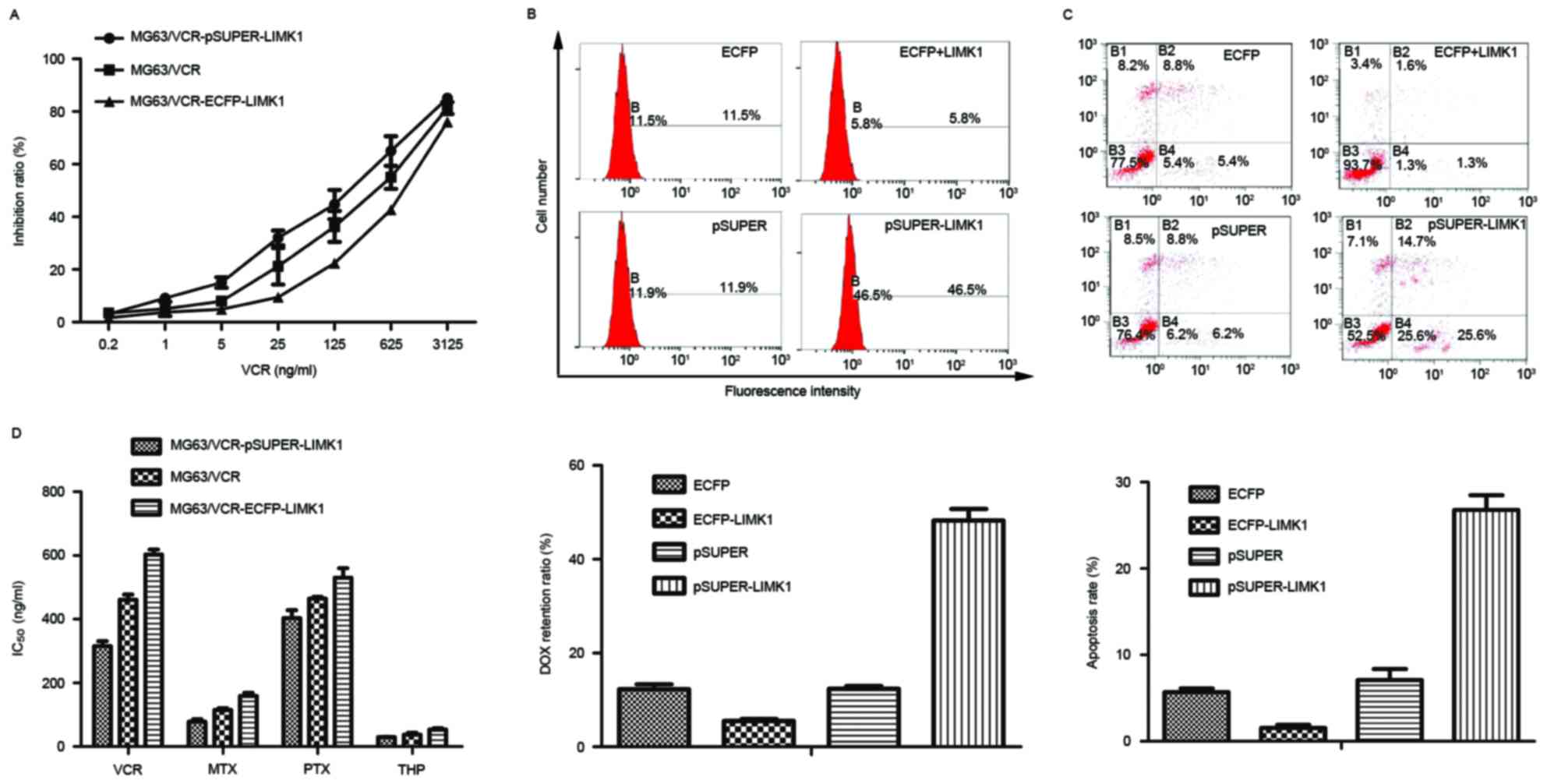 | Figure 3.LIMK1 is critical for drug resistance
in osteosarcoma cells. (A) MG63/VCR, MG63/VCR
pSUPER-LIMK1-transfected and MG63/VCR ECFP-LIMK1-transfected cells
were treated with different concentrations of VCR, and the cell
viability was analyzed using the Cell Counting kit-8 assay. (B) The
fluorescence intensity of DOX was analyzed quantitatively by flow
cytometry. The mean fluorescence intensity + SD is shown in the bar
graph. (C) Cell lines were incubated with VCR (IC50) and
assessed for early apoptosis by Annexin V/propidium iodide staining
and flow cytometry analysis. (D) The concentration at which each
drug produced 50% inhibition of growth (IC50) is shown
in the histogram. Representative data are shown. The histogram
represents means ± SD of five independent experiments. LIMK1, LIM
kinase 1; MG63/VCR, multidrug resistant MG63 cells; ECFP, enhanced
cyan fluorescent protein; IC50, 50% inhibitory
concentration; VCR, vincristine; MTX, methotrexate; DOX,
doxorubicin; PTX, paclitaxel; THP, pirarubicin; SD, standard
deviation. |
LIMK1 functions through regulating the
expression of MDR1/P-gp
LIMK1 and MDR1/P-gp were expressed at higher levels
in MG63/VCR cells than in MG63 cells (Fig. 2). To explore the connection between
the two genes, the expression of LIMK1 and MDR1/P-gp was assessed
in MG63/VCR cells following transfection with LIMK1 shRNA,
wild-type LIMK1 or negative control vector. The results indicated
that both LIMK1 mRNA and protein expression levels were increased
in MG63/VCR cells following transfection with wild-type LIMK1,
compared with those in cells transfected with the empty vector.
LIMK1 mRNA and protein expression levels were significantly reduced
in cells transfected with LIMK1 shRNA, compared with those in cells
transfected with the empty vector or wild-type LIMK1. Notably, the
change in MDR1 expression was completely consistent with that in
LIMK1 expression (P<0.05) (Fig. 4A and
B). Immunocytochemistry staining was performed to compare the
expression of MDR1 in the same cells. The results demonstrated that
MDR1/P-gp expression was increased significantly in MG63/VCR cells
transfected with ECFP-LIMK1 plasmid, while transfection with LIMK1
shRNA decreased MDR1/P-gp expression (Fig. 4C). These data suggest that LIMK1 may
affect MDR by regulating the expression of MDR1/P-gp.
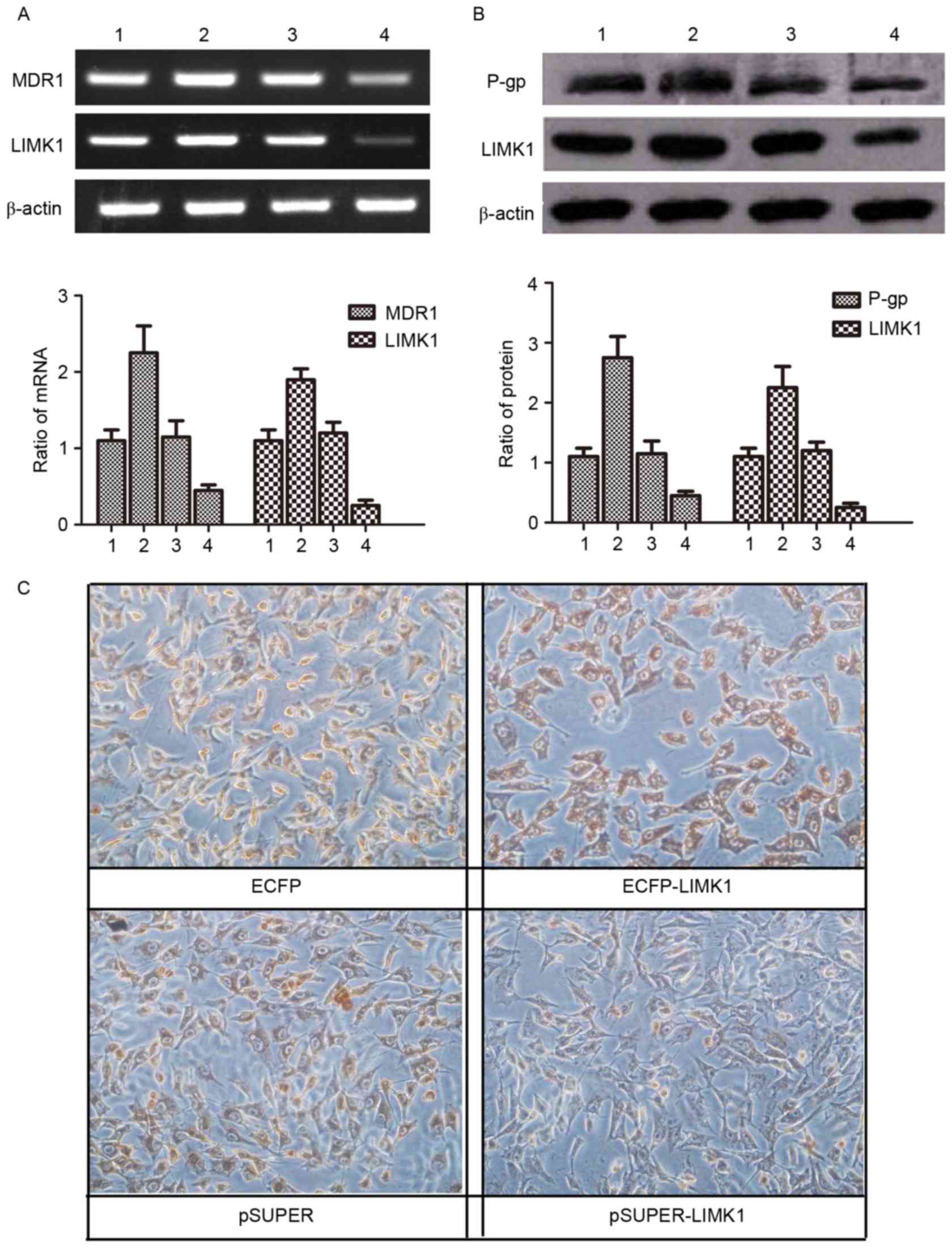 | Figure 4.LIMK1 functions through regulating the
expression of MDR1/P-gp. (A and B) Lanes 1, 2, 3 and 4 represent
MG63/VCR cells transfected with plasmids for ECFP, ECFP-LIMK1,
pSUPER and pSUPER-LIMK1 expression, respectively. (A) Agarose gel
electrophoresis of the reverse transcription-polymerase chain
reaction products for LIMK1 and MDR1 in MG63/VCR cells transfected
with different plasmids. (B) Western blot analysis of LIMK1 and
MDR1/P-gp proteins in MG63/VCR cells transfected with different
plasmids. (C) Immunohistochemistry staining using antibodies
against MDR1 in MG63 and MG63/VCR cells transfected with LIMK1
short hairpin RNA (pSUPER-LIMK1) or a control pSUPER-negative
control-small interfering RNA vector (magnification, ×200; scale
bar, 50 µm). LIMK1, LIM kinase 1; MG63/VCR, multidrug resistant
MG63 cells; mRNA, messenger RNA; MDR1, multidrug resistance protein
1; P-gp, P-glycoprotein; ECFP, enhanced cyan fluorescent
protein. |
Discussion
In the present study, a novel function of LIMK1 in
regulating MDR was identified. Previous studies demonstrated that
LIMK1 was overexpressed and highly active in cells and tissues of
certain malignant tumors, including prostate and breast cancer
(16,17). LIMK1 was considered to be a key
molecule that stimulated malignant tumor cell invasion and
metastasis, or possibly a new ‘oncogene’ (18,19).
However, few studies linked LIMK1 with MDR, which represents a
major challenge for managing the treatment of the majority cancers
(20). In the present study, the
expression of LIMK1 mRNA and protein in twohuman osteosarcoma cell
lines (MG63 and U2OS) was markedly higher than that in hFOB cells
(Fig. 1A and B). Furthermore, the
expression of LIMK1 in tumor parenchyma cells was significantly
higher compared with that in mesenchymal cells (Fig. 1C), suggesting that LIMK1 was
responsible for the genesis and development of osteosarcoma.
A multidrug resistant MG63/VCR subline was
established by intermittent exposure of MG63 cells to VCR. This
subline exhibited cross-resistance to other commonly used drugs to
treat osteosarcoma, but which are structurally and mechanistically
different to VCR (Fig. 2B) (1,21). These
drugs included an anti-microtubule agent (PTX), an antimetabolite
(MTX) and two topoisomerase II inhibitors (DOX and THP) (22). Notably, the mRNA and protein
expression levels of LIMK1 and MDR1/P-gp were higher in MG63/VCR
cells than in MG63 cells (Fig. 2C and
D). Therefore, it was hypothesized that there was a correlation
between LIMK1 and MDR in osteosarcoma.
In order to verify this conjecture, additional
experiments were conducted by upregulating and downregulating LIMK1
in MG63/VCR cells. Downregulation of LIMK1 inhibited the efflux of
DOX and decreased the MDR of these cells compared with that of the
controls. In addition, the sensitivity of these cells to VCR was
elevated (Fig. 3). Of note, the
expression of MDR1/P-gp, which was also increased in MG63/VCR cells
compared with that in MG63 cells, was changed in accordance with
the expression of LIMK1. MDR1 belongs to the ABC transporter family
that regulates the efflux across the plasma membrane of multiple
structurally and mechanistically unrelated chemotherapeutic agents
(3). MDR1/P-gp-mediated MDR has been
long considered a classical mechanism of MDR in malignant tumors,
including osteosarcoma (3,20). The expression of MDR1 in MG63/VCR
cells was increased significantly in cells overexpressing LIMK1
compared with that in cells transfected with empty vector. In
addition, downregulation of LIMK1 resulted in a significant
decrease in the expression of MDR1. These results indicate that
LIMK1 serves an important role in MDR through regulating the
expression of MDR1/P-gp. The RhoA/LIMK1/cofilin signaling pathway
may have certain connection with the ABC transporter family.
However, understanding the precise molecular mechanism requires
further investigation.
In summary, the overexpression of LIMK1 increases
MDR1/P-gp expression and has major effects on the MDR of human
osteosarcoma. The present study provides new insights into the
function of LIMK1 and suggests that altering LIMK1 is a potential
new therapeutic strategy for osteosarcoma.
Acknowledgements
The present study was supported by Projects of
International Cooperation of Jilin Provincial Science &
Technology Department (grant no. 20150101175JC) and the National
Natural Science Foundation of China (grant nos. 81172000 and
30772488).
Glossary
Abbreviations
Abbreviations:
|
CCK-8
|
Cell Counting kit-8
|
|
DOX
|
doxorubicin
|
|
H-DMEM
|
high glucose-Dulbecco's modified
Eagle's medium
|
|
hFOB
|
human fetal osteoblasts
|
|
LIMK1
|
LIM kinase 1
|
|
MDR
|
multidrug resistance
|
|
P-gp
|
P-glycoprotein
|
|
PI
|
propidium iodide
|
|
MTX
|
methotrexate
|
|
PTX
|
paclitaxel
|
|
THP
|
pirarubicin
|
|
VCR
|
vincristine
|
References
|
1
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Holohan C, Van Schaeybroeck S, Longley DB
and Johnston PG: Cancer drug resistance: An evolving paradigm. Nat
Rev Cancer. 13:714–726. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thomas H and Coley HM: Overcoming
multidrug resistance in cancer: An update on the clinical strategy
of inhibiting p-glycoprotein. Cancer Control. 10:159–165. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maekawa M, Ishizaki T, Boku S, Watanabe N,
Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K and Narumiya S:
Signaling from Rho to the actin cytoskeleton through protein
kinases ROCK and LIM-kinase. Science. 285:895–898. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohashi K, Nagata K, Maekawa M, Ishizaki T,
Narumiya S and Mizuno K: Rho-associated kinase ROCK activates
LIM-kinase 1 by phosphorylation at threonine 508 within the
activation loop. J Biol Chem. 275:3577–3582. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Arber S, Barbayannis FA, Hanser H,
Schneider C, Stanyon CA, Bernard O and Caroni P: Regulation of
actin dynamics through phosphorylation of cofilin by LIM-kinase.
Nature. 393:805–809. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang N, Higuchi O, Ohashi K, Nagata K,
Wada A, Kangawa K, Nishida E and Mizuno K: Cofilin phosphorylation
by LIM-kinase 1 and its role in Rac-mediated actin reorganization.
Nature. 393:809–812. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li R, Doherty J, Antonipillai J, Chen S,
Devlin M, Visser K, Baell J, Street I, Anderson RL and Bernard O:
LIM kinase inhibition reduces breast cancer growth and invasiveness
but systemic inhibition does not reduce metastasis in mice. Clin
Exp Metastasis. 30:483–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen Q, Jiao D, Hu H, Song J, Yan J, Wu L
and Xu LQ: Downregulation of LIMK1 level inhibits migration of lung
cancer cells and enhances sensitivity to chemotherapy drugs. Oncol
Res. 20:491–498. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang H, Wang Y, Xing F, Wang J, Wang Y,
Wang H, Yang Y and Gao Z: Overexpression of LIMK1 promotes
migration ability of multidrug-resistant osteosarcoma cells. Oncol
Res. 19:501–509. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang HS, Zhao JW, Wang H, Zhang HY, Ji
QY, Meng LJ, Xing FJ, Yang ST and Wang Y: LIM kinase 1 is required
for insulin-dependent cell growth of osteosarcoma cell lines. Mol
Med Rep. 9:103–108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takemura M, Mishima T, Wang Y, Kasahara J,
Fukunaga K, Ohashi K and Mizuno K:
Ca2+/calmodulin-dependent protein kinase IV-mediated LIM
kinase activation is critical for calcium signal-induced neurite
outgrowth. J Biol Chem. 284:28554–28562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao JW, Zhang MR, Ji QY, Xing FJ, Meng LJ
and Wang Y: The role of slingshot-1L (SSH1L) in the differentiation
of human bone marrow mesenchymal stem cells into cardiomyocyte-like
cells. Molecules. 17:14975–14994. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Peng X, Yu W, Hou S, Zhao Y,
Zhang Z, Huang X and Wu K: Alpha-tocopheryl succinate enhances
doxorubicin-induced apoptosis in human gastric cancer cells via
promotion of doxorubicin influx and suppression of doxorubicin
efflux. Cancer Lett. 307:174–181. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Davila M, Frost AR, Grizzle WE and
Chakrabarti R: LIM kinase 1 is essential for the invasive growth of
prostate epithelial cells: Implications in prostate cancer. J Biol
Chem. 278:36868–36875. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tapia T, Ottman R and Chakrabarti R: LIM
kinase1 modulates function of membrane type matrix
metalloproteinase 1: Implication in invasion of prostate cancer
cells. Mol Cancer. 10:62011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang TY, DerMardirossian C and Bokoch GM:
Cofilin phosphatases and regulation of actin dynamics. Curr Opin
Cell Biol. 18:26–31. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo H, Wu F, Wang Y, Yan C and Su W:
Overexpressed ubiquitin ligase Cullin7 in breast cancer promotes
cell proliferation and invasion via down-regulating p53. Biochem
Biophys Res Commun. 450:1370–1376. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kathawala RJ, Gupta P, Ashby CR Jr and
Chen ZS: The modulation of ABC transporter-mediated multidrug
resistance in cancer: A review of the past decade. Drug Resist
Updat. 18:1–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ta HT, Dass CR, Choong PF and Dunstan DE:
Osteosarcoma treatment: State of the art. Cancer Metastasis Rev.
28:247–263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang JZ, Ma SR, Rong XL, Zhu MJ, Ji QY,
Meng LJ, Gao YY, Yang YD and Wang Y: Characterization of
multidrug-resistant osteosarcoma sublines and the molecular
mechanisms of resistance. Mol Med Rep. 14:3269–3276. 2016.
View Article : Google Scholar : PubMed/NCBI
|