Introduction
The hypoxic microenvironment serves a function in
tumor growth, metastasis and recurrence, as tumors with extensive
low oxygen tension tend to lead to a poor prognosis and exhibit
resistance to conventional therapies (1). The underlying molecular mechanisms of
the cellular response to oxygen deprivation have been the subject
of numerous studies and are known to be complex. Hypoxia-inducible
factors (HIFs) regulate >100 genes in response to a decrease in
oxygen and are known to serve a function in the hypoxic condition
(2). MicroRNAs (miRNAs) have also
been demonstrated to be involved in the process of cellular
adaptation to the hypoxic microenvironment (3). miRNAs are non-coding single-stranded
RNAs of ~22 nucleotides that mediate sequence-dependent
post-transcriptional negative regulation of gene expression, and
are known to serve a function in fundamental processes including
cell proliferation (4), metabolism
(5) and cancer metastasis (6), in addition to canonical signaling
pathways including Notch, mitogen-activated protein kinase (MAPK)
(7) and the Toll-like receptor
signaling pathways (7,8). The study of the biological significance
and utility of miRNAs is an expanding field, and it is increasingly
evident that miRNAs serve complex functions and diverse functions
specific to individual miRNA sequence, cell type and tissue
environment. Although there are reports of hypoxia-regulated miRNAs
in several types of tumor, to the best of our knowledge,
cell-specific regulation and disease-specific alterations of miRNA
in hepatocellular carcinoma (HCC) have yet to be investigated. In
the present study, mRNA and miRNA expression profile changes were
analyzed using microarray technology to uncover the underlying
function of miRNA in the adaptation of HCC cells to the hypoxic
microenvironment.
Materials and methods
Cell culture
Huh7 cells, purchased from the Institute of
Biochemistry and Cell Biology (Shanghai, China), were maintained in
Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum. Cells
were incubated at 37°C with 5% CO2. To create hypoxic
conditions, cells 12 h post-seeding were placed in a hypoxia
workstation (In vivo 200, Ruskinn Technology, Ltd.,
Bridgend, UK) for 48 h at 1% O2, 5% CO2 and
37°C.
Extraction and labeling of sample
RNA
Total RNA was extracted and purified using
TRIzol® reagent (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. RNA integration was
verified using an Agilent Bioanalyzer 2100 (Agilent Technologies,
Inc., Santa Clara, CA, USA). miRNA in the total RNA was labeled
using the miRNA Complete Labeling and Hybkit (Agilent Technologies,
Inc.), according to the manufacturer's protocol.
Array hybridization, image scanning
and bioinformatic data processing
Each slide was hybridized with 100 ng cyanine
3-labeled RNA using the miRNA Complete Labeling and Hybkit in a
hybridization oven (Agilent Technologies, Inc.) at 55°C, 20
revolutions/min for 20 h, according to the manufacturer's protocol.
Following hybridization, slides were washed in staining dishes
(Thermo Fisher Scientific, Inc.) with Gene Expression Wash Buffer
kit (Agilent Technologies, Inc.). Slides were scanned using an
Agilent Microarray Scanner (Agilent Technologies, Inc.) and read
using Feature Extraction Software (version 10.7; Agilent
Technologies, Inc.) using default settings. Raw data were
normalized using the Quantile algorithm with GeneSpring software
(version 12.6; Agilent Technologies, Inc.).
Gene Ontology (GO) and Kyoto
encyclopedia of genes and genomes (KEGG) analysis
Using the GO and KEGG databases, functional
classifications and pathways of differentially expressed genes were
analyzed. TargetScan and miRanda databases were used to analyze
differentially expressed miRNAs and their associated negatively
regulated genes, and the GO and KEGG pathway labels were applied to
determine the significant functions of the differentially expressed
miRNAs (9,10). Fisher's exact test was applied to
identify the significant GO categories and the false discovery rate
was used to correct the P-values.
miRNA-target network
The association between the miRNAs and their gene
targets was analyzed on the basis of differential expression
values, and an miRNA-target network was built on the interactions
of miRNA and genes identified in the Sanger miRNA database
(miRBase.org). Circles represent genes and arrows
represent miRNAs, and their association is presented as an edge.
The size of the arrow changes on the basis of the degree of
contribution of the miRNA to gene expression, where key miRNAs in
the network are presented as larger arrows.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Certain genes and miRNAs identified in these
analyses were selected for verification using RT-qPCR.
cDNA was synthesized using random primers and PCR
was performed using a SYBR master mix (Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol; GAPDH was used as
a control. The primer sequences were used as follows: RAP2A;
forward: 5′-GACCCCACCATCGAGGACTTCTAC-3′, reverse,
5′-TGGCACTTTCTCATACCGCTTCAC-3′;. GPR4; forward,
5′-TCCCTAACTGCGCTGTGTCCTATC-3′; reverse,
5′-ATCGCTGGGCAATGGTGAGAA-3′; RASA4; forward,
5′-GGCAGGGCTTTTGTGGGTTATG-3′, reverse,
5′-GCTGGGAGGGAGGAGGCTTTAG-3′; RAPGEF2; forward,
5′-CAGTGGATTCCGAAGACGACGAC-3′, reverse,
5′-CCACCACTGCGAACACCATCAC-3′; METTL20; forward,
5′-AGAGGCTTAATCATTGGGCACTG-3′, reverse,
5′-CGAAAAGCATACAACACGCAAGAA-3′; RAPH1; forward,
5′-TCCCCCTACCCCTCCTGTTCC-3′, reverse,
5′-ACTGGCTGGCTATCTGCTTCACG-3′; CXCL-2; forward,
5′-TGCGCCCAAACCGAAGTCATA-3′, reverse,
5′-GTGGCCTCTGCAGCTGTGTCTCT-3′; FUBP-1; forward,
5′-ACACCCGAAAGGATAGCAC-3′, reverse, 5′-TTGCCTTGACCTCTACCTC-3′; YAP;
forward, 5′-AGGAGAGGCTGCGGCTGAAAC-3′, reverse,
5′-TGAGACATCCCGGGAGAAGACACT-3′; GAPDH; forward,
5′-GGGGCTCTCCAGAACATCATCC-3′, reverse,
5′-ACGCCTGCTTCACCACCTTCTT-3′. The miRNA-19a (assay ID 002424;
Thermo Fisher Scientific, Inc.), miR-34a (assay ID 000425; Thermo
Fisher Scientific, Inc.), miR-25 (assay ID 000403; Thermo Fisher
Scientific, Inc.) and miR-1207 (assay ID 241060; Thermo Fisher
Scientific, Inc.) expression levels were measured using TaqMan™
MicroRNA Assays (cat. no. 4427975; Thermo Fisher Scientific, Inc.).
Thermal cycling parameters were as follows: Polymerase activation
at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and
60°C for 1 min. The expression levels of miRNAs were analyzed using
the TaqMan miRNA assays and TaqMan miRNA RT kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) on an Applied
Biosystems 7500 RT-qPCR system (11).
Results
Differential gene expression under
normoxia and hypoxia
To investigate gene expression profile changes in
hypoxic cells, microarray analysis was conducted on Huh7 cells
after 48 h of culture under hypoxic conditions. Among the
transcripts tested, the expression levels of 4,146 genes were
identified to be upregulated and those of 7,362 genes were
identified to be downregulated (fold change >1.5; P<0.05).
These differentially expressed genes included HIFs,
platelet-derived growth factor subunit B, Krüppel-like factor 8,
C-X-C motif chemokine ligand 5 and ribonucleotide reductase
catalytic subunit M1, which are known to be involved in cell cycle
regulation, transcription and apoptosis (Fig. 1A). GO analysis was performed on
upregulated and downregulated genes. Downregulated genes were
primarily identified to be involved in the cell cycle, cell
division and transcription, whereas upregulated genes were
typically associated with G-protein-coupled receptor signaling
pathways, signal transduction, response to stimuli and synaptic
transmission (Fig. 1B). KEGG pathway
analysis revealed that downregulated genes were primarily
associated with cell cycle, ubiquitin-mediated proteolysis, RNA
transport and DNA replication, and the upregulated genes were
associated with neuroactive ligand-receptor interaction, steroid
biosynthesis, insulin secretion and the calcium signaling pathway
(Fig. 1C). Together, the GO and KEGG
pathway analysis results revealed that the annotated differentially
expressed genes were involved in a wide variety of signaling
pathways.
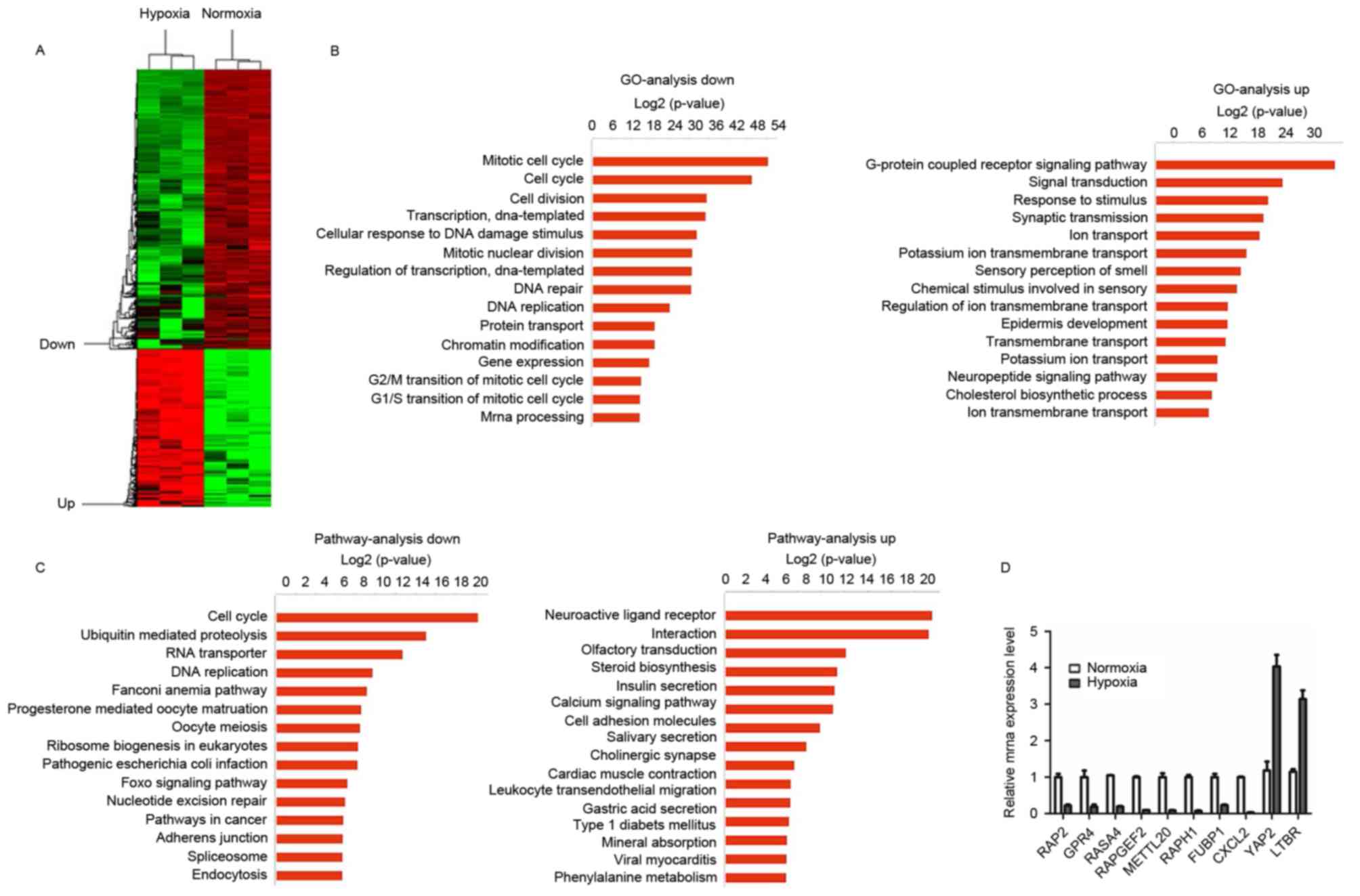 | Figure 1.Differentially expressed mRNAs in Huh7
cells after 48 h under hypoxia. (A) A heat map presents
differential gene expression patterns (fold change >2 or
<0.5). (B) GO analysis of downregulated and upregulated mRNAs,
respectively. (C) Pathways of downregulated and upregulated mRNAs,
respectively. (D) Reverse transcription-quantitative polymerase
chain reaction verification of the selected genes under normal and
hypoxic conditions. GO, Gene Ontology; RAP2, Ras-related protein
Rap-2a; GPR4, G-protein-coupled receptor 4; RASA4, Ras
GTPase-activating protein 4; RAPGEF2, Rap
guanine-nucleotide-exchange factor 2; METTL20,
methyltransferase-like 20; RAPH1, Ras-associated and pleckstrin
homology domain-containing protein 1; FUBP1, far-upstream
element-binding protein 1; CXCL2, C-X-C motif chemokine ligand 2;
YAP2, yes-associated protein 1; LTBR, lymphotoxin β receptor. |
Huh7 cells were identified to exhibit a number of
changes, including changes in the cell cycle, DNA replication and
transcription in response to the hypoxic conditions. RT-qPCR was
performed in order to confirm the gene expression changes observed
in the microarray analysis. The results revealed that Ras-related
protein Rap-2a, G-protein-coupled receptor 4, Rasp21 protein
activator 4, Rap guanine-nucleotide-exchange factor 2,
methyltransferase-like 20, Ras-associated and pleckstrin homology
domain-containing protein 1, far-upstream element-binding protein 1
and C-X-C motif chemokine ligand 2 expression were significantly
decreased, and the expression of yes-associated protein 1 and
lymphotoxin β receptor was significantly increased. These results
demonstrated a trend that was consistent with the results of
microarray analysis (Fig. 1D).
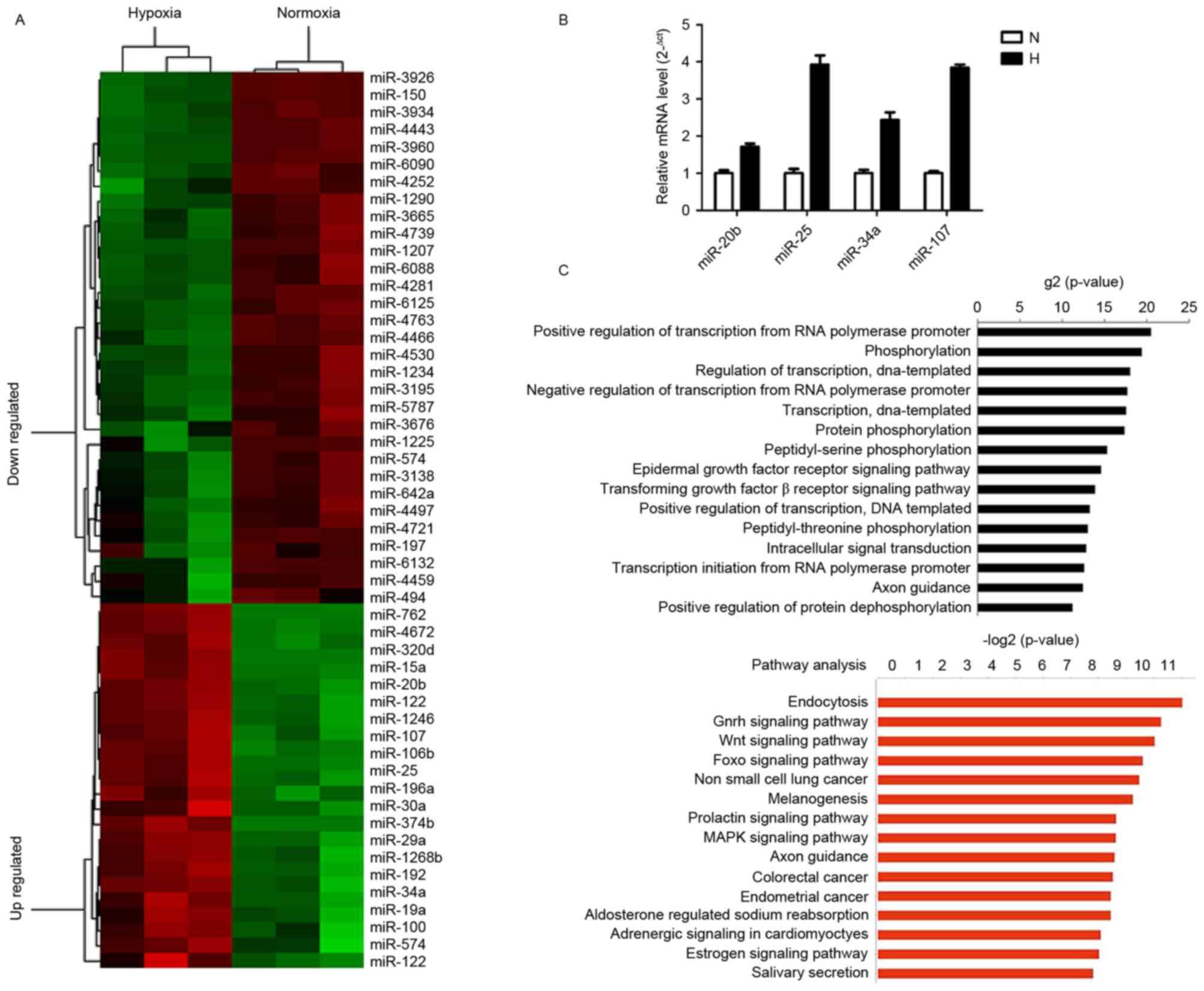
miRNAs are differentially expressed in
Huh7 cells under hypoxic and normoxic conditions
Microarray analysis demonstrated that 58 miRNAs were
differentially expressed under hypoxic conditions. Among these, 35
miRNAs were revealed to be downregulated and 23 were identified to
be upregulated (fold change >1.5; P<0.05; Fig. 2A). The 10 most upregulated miRNAs were
miR-574-3P, miR-320d, miR-151a-5p, miR-374b-5p, miR-19a-3p,
miR-1246, miR-122-3p, miR-122-5p, miR-3132 and miR-29b-3p, and the
10 most downregulated miRNAs were miR-3926, miR-3138, miR-150-3p,
miR-4252, miR-762, miR-6090, miR-3934-5p, miR-3960, miR-4430 and
miR-4443; all these miRNAs exhibited a fold change greater than 3
in hypoxic cells. Differentially expressed miRNAs were confirmed
using RT-qPCR. Among the validated miRNAs, 3 were demonstrated to
be significantly different between hypoxic and normoxic conditions,
of which one exhibited a similar expression change to that observed
using microarray analysis, although no significant difference was
observed (Fig. 2B).
Using the TargetScan and miRanda software packages,
differentially expressed miRNA targets were predicted. Predicted
genes, which exhibit negative co-expression with differentially
expressed miRNAs, were used to analyze the miRNA gene regulation
network. GO analysis revealed that the primary functions of the
differentially expressed miRNAs were as positive and negative
regulators of transcription from the RNA polymerase promoter,
phosphorylation and regulation of transcription. These results
demonstrate that the majority of differentially expressed miRNAs
regulate transcription-modifying and phosphorylation-associated
genes. KEGG pathway analysis revealed that genes associated with
endocytotic, gonadotropin-releasing hormone, wingless-related
integration site and forkhead box O (FOXO) signaling pathways were
involved in the 15 most altered pathways list (Fig. 2C). These associated signaling pathways
are known to exhibit aberrant activation in HCC and serve crucial
functions in the regulation of cellular proliferation, apoptosis,
stem and progenitor cell expansion, and drug resistance.
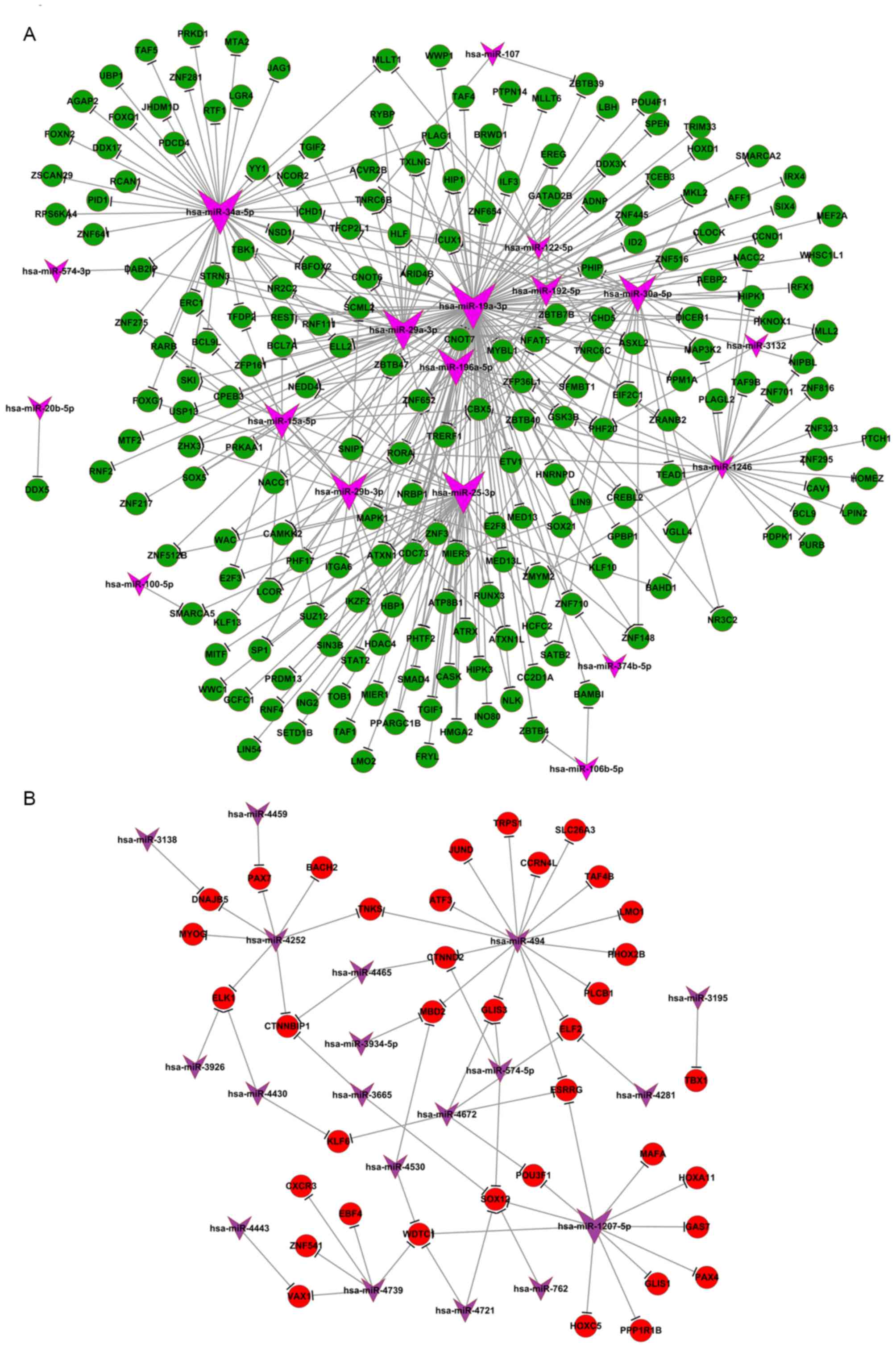
Significance of miRNA function on
transcription and phosphorylation
From the aforementioned results, it was identified
that the majority of differentially expressed miRNAs were involved
in the regulation of transcription and modification of
phosphorylation. Transcription is the initial step of gene
expression, and is performed in the nucleus by RNA polymerase,
which requires a promoter sequence and a set of DNA-binding
proteins to initiate the process. Potential target genes of the
differentially expressed miRNAs were revealed to be positive and
negative regulators of transcription from the RNA polymerase
promoter. It was identified that certain DNA-binding proteins
including transcriptional regulating factor 1, inhibitor of DNA
binding 2 and zinc-finger proteins 1–3, all of which are required
for gene transcription, had a distinct expression profile under
hypoxic conditions, and may be the potential targets of several of
the identified miRNAs (miR-19a, miR-196a, miR-192 and miR-34a).
Certain transcription factors, including mothers against
decapentaplegic homolog 4, E2 factor (E2F), specificity protein 1
and Krüppel-like factor 13, also exhibited marked expression fold
changes under hypoxic conditions and were revealed to control the
rate of transcription. Differentially expressed miRNAs identified
under hypoxic conditions (including miR-19a, miR-34a and miR-122)
are known to be involved in p53 (12,13), c-myc
(14) and E2F (15) signaling pathways. These miRNAs
associated with cell proliferation and epithelial-mesenchymal
transition (16,17) were identified in the miRNA-mRNA
network and had potential targets genes including RNA polymerase
elongation factor II, RNA-binding motif,
single-stranded-interacting protein 1, and signal transducer and
activator of transcription 3, all of which regulate gene
transcription (Fig. 3). The
miRNA-mRNA negative expression network also revealed that a large
portion of mRNA was downregulated, whereas the majority of the
associated miRNA was upregulated, compared with control (normoxic)
conditions (Fig. 3).
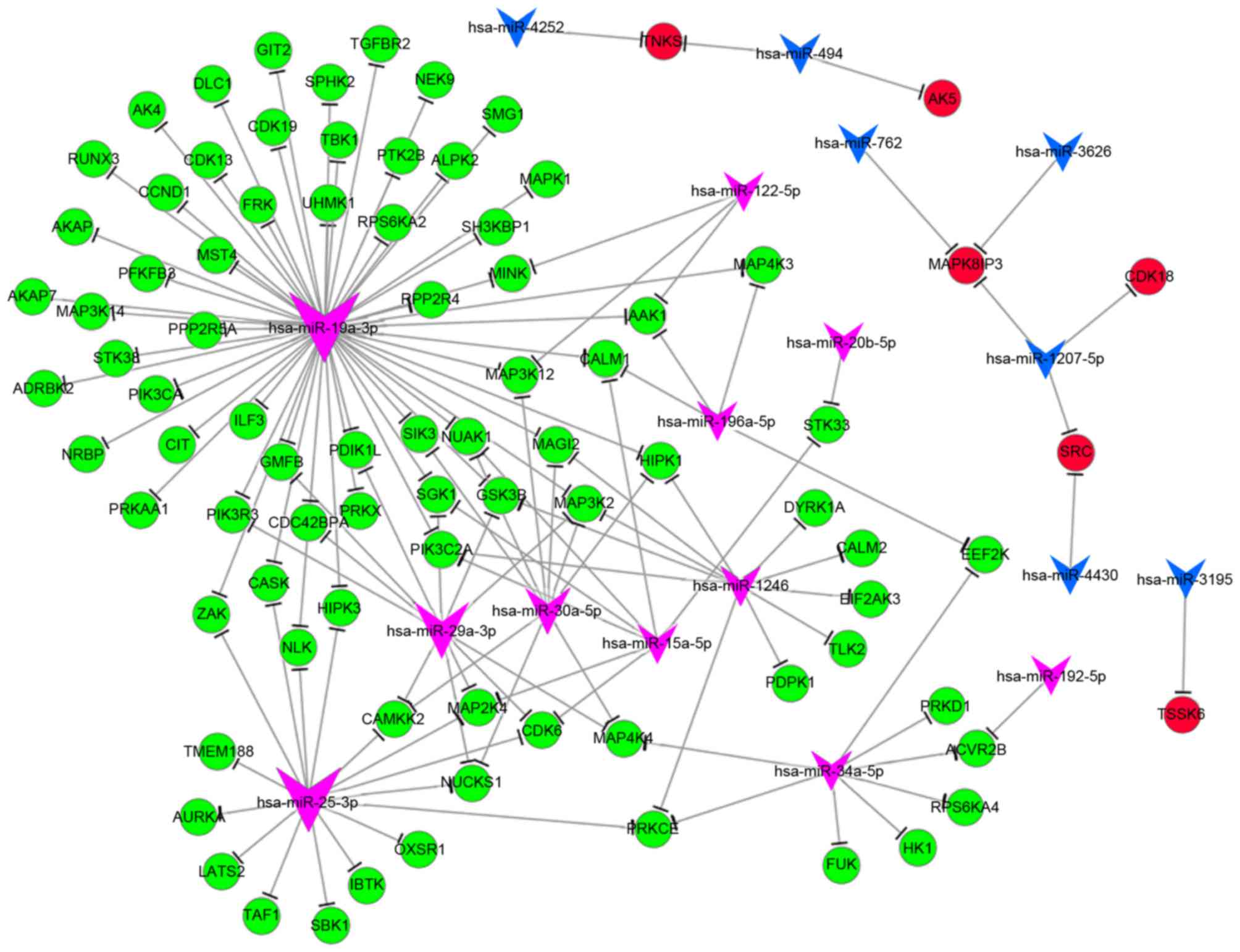
Phosphorylation is a post-translational protein
modification, which activates enzymes, alters their function and
activity and serves a function in a wide range of cellular
processes. In hypoxia, the majority of genes associated with
phosphorylation [including MAPK1, MAPK kinase kinase (MAP3K)4 and
phosphoinositide 3-kinase C2 domain-containing α polypeptide] were
downregulated, and a number of genes [including cyclin-dependent
kinase (CDK) 18, proto-oncogene tyrosine-protein kinase and
MAPK8-interacting protein 3] were upregulated. The GO analysis
revealed that differentially expressed miRNAs serve a function in
phosphorylation. Upon analyzing the miRNA-mRNA network of
phosphorylation, the key miRNAs were identified to be miR-19a,
miR-25, miR-29 and miR-34a, and their potential target genes were
MAP3K12, MAPK1, CDK1 and phosphoinositide3-kinase regulatory
subunit 3 (Fig. 4).
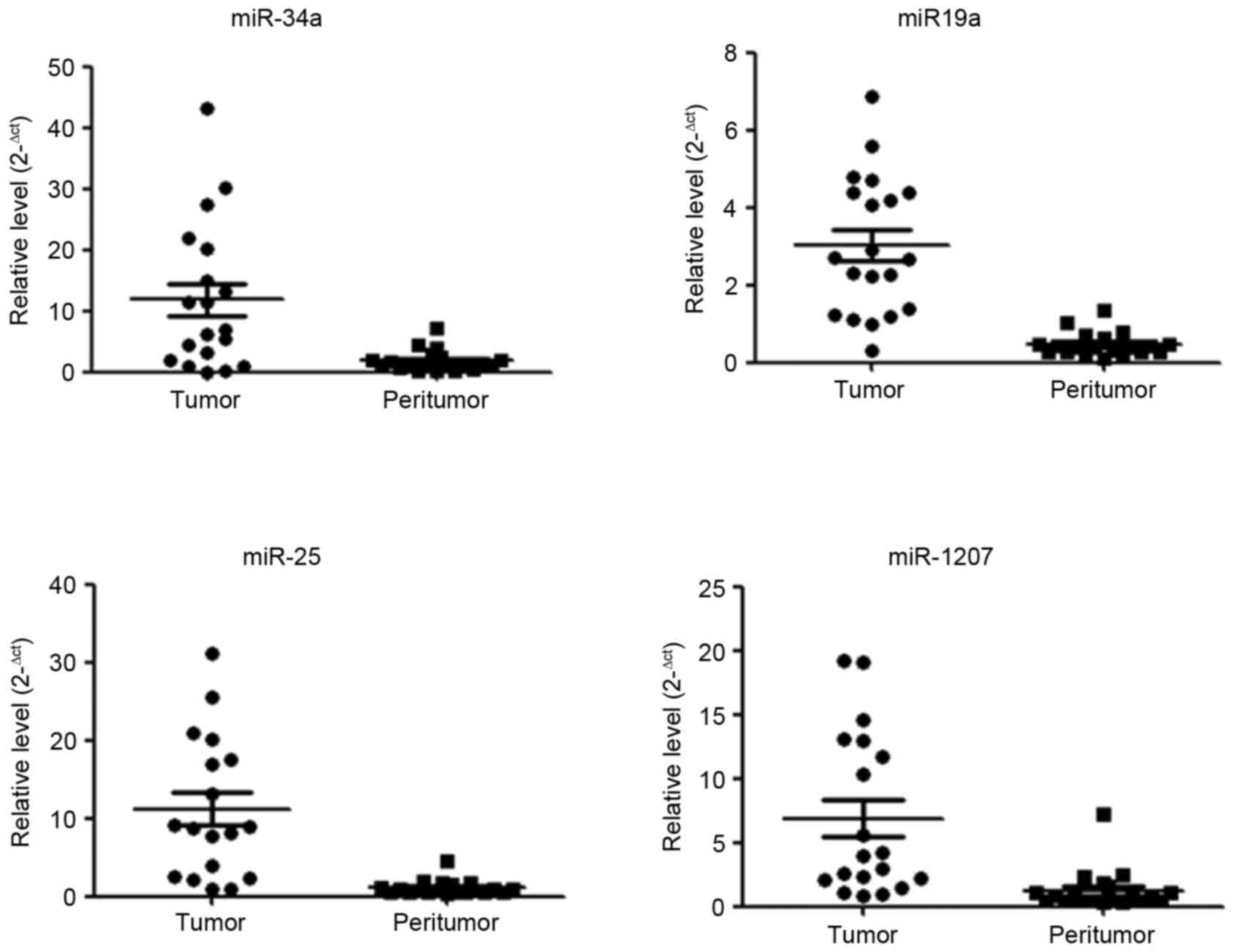
Key miRNA expression in HCC
The aforementioned results identified several miRNAs
have a function under hypoxic conditions, whose potential target
genes involve phosphorylation and transcription. It was also
revealed that key miRNAs miR-34a, miR-19a, miR-25 and miR-1207
exhibited increased expression levels in HCC tissue compared with
corresponding peritumor tissue (Fig.
5). As tumor growth and progression results in increasing
hypoxia in the tumor microenvironment and the results of the
present study demonstrate that the miRNAs which increased under
hypoxic conditions are increased in tumor tissue, it is possible
that these identified miRNAs are of clinical importance.
Discussion
miRNAs are regulators of tumorigenesis that regulate
gene expression on the basis of sequence complementarity,
particularly between the seed region and the 3′ untranslated
regions of the target. Dysregulation of miRNA expression has been
observed in almost all neoplasms, demonstrating the importance of
miRNAs in tumor biology. Notably, different types of cancer tend to
exhibit specific expression patterns and the underlying molecular
mechanisms of these shifts in the tumor profile remain unresolved
(18,19). A previous study has demonstrated that
miRNA levels differ in low oxygen and contribute to the regulation
of specific genes under hypoxic conditions (20). In the present study, the altered
expression of miRNAs under hypoxic pressure was examined in an HCC
cell line and it was revealed that 58 miRNAs were aberrantly
expressed in hypoxia, a number of which (including miR-106, miR-15a
and miR-29a) were identified to be associated with cell cycle
changes, drug resistance and abnormal expression in certain types
of cancer. Additionally, certain key miRNAs were identified in the
miRNA-mRNA network of HCC cells in response to hypoxic pressure,
including miR-29a, miR-34a, miR-19a and miR-25.
A feature in numerous types of cancer, including
HCC, hypoxia results in the expression of specific miRNAs, referred
to as hypoxamirs (21). A large
number of hypoxamirs have been identified in retinoblastoma,
glioblastoma (21,22) and other types of tumor. In the present
study, hypoxamirs from an HCC cell line were identified. Normal
expression of certain miRNAs in liver cells was revealed to be
altered. For example, miR-122 is the most highly abundant miRNA
expressed in hepatocytes, where it has been demonstrated to
regulate hepatic cholesterol and lipid metabolism, and aberrant
expression leads to the development of fibrosis and HCC (23,24). Under
hypoxic conditions miR-122-3p and miR-122-5p were upregulated by a
factor of >2. Additionally, the expression of certain key miRNAs
was tested in HCC tumor tissue compared with their corresponding
peritumor tissue and it was revealed that expression levels were
consistently changed as a result of the hypoxic conditions in the
tumor. Hypoxamirs in the HCC cell may be used as a library to
screen miRNAs as potential biomarkers and therapeutic targets.
Since each miRNA potentially regulates a large
number of targets, understanding the function of miRNAs in
tumorigenesis may provide insight into the key questions that
remain in cancer research. In the present study, mRNA and miRNA
expression changes induced by hypoxic conditions in Huh7 HCC cells
were evaluated. It was observed that mRNA and miRNA undergo changes
in expression profiles, and GO analysis revealed that the majority
of these changes are involved in transcriptional regulation and
phosphorylation, particularly the differentially expressed miRNA.
In total, 40% of the 15 most differentially expressed miRNAs (6/15)
are involved in regulating transcription. AnmiRNA-mRNA negative
association network analysis revealed that differentially expressed
microRNA target genes include zinc-finger proteins (ZNF445, ZNF64,
etc.), transcription factors (neural precursor cell expressed
developmentally downregulated protein 4, runt-related transcription
factor 3, FOXO1, transcription factor Dp-2, etc.) and others that
are known to regulate the rate of transcription. Key miRNAs
identified in the present study include miR-34a, miR-19a and
miR-25, which exhibited a significantly increased expression level
in HCC tumors compared with their surrounding tissue. Taken
together, the results of the present study demonstrate that miRNA
serves an important function in tumor biology by regulating a
number of transcription factors and targeting transcriptional
regulators in HCC cells under hypoxic conditions.
Acknowledgements
The present study was accomplished through combined
grant support from the National Key Sci-Tech Project (grant nos.
2013ZX10002011-004 and 2012ZX0930100-007), National Natural Science
Foundation of China (grant nos. 81302100, 81572884 and 81372317),
Zhongshan Hospital Outstanding Youth Fund (grant no. 2015ZSYXQN03),
the Specialized Research Fund for the Doctoral Program of Higher
Education (grant no. 20120071120068) and The Pujiang Scholars Fund
of Shanghai (grant no. 13PJD007).
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
HCC
|
hepatocellular carcinoma
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
HIFs
|
hypoxia-inducible factors
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
References
|
1
|
Zhou TY, Zhuang LH, Hu Y, Zhou YL, Lin WK,
Wang DD, Wan ZQ, Chang LL, Chen Y, Ying MD, et al: Inactivation of
hypoxia-induced YAP by statins overcomes hypoxic resistance
tosorafenib in hepatocellular carcinoma cells. Sci Rep.
6:304832016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kai AK, Chan LK, Lo RC, Lee JM, Wong CC,
Wong JC and Ng IO: Down-regulation of TIMP2 by
HIF-1α/miR-210/HIF-3α regulatory feedback circuit enhances cancer
metastasis in hepatocellular carcinoma. Hepatology. 64:473–487.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jia YY, Zhao JY, Li BL, Gao K, Song Y, Liu
MY, Yang XJ, Xue Y, Wen AD and Shi L: miR-592/WSB1/HIF-1α axis
inhibits glycolytic metabolism to decrease hepatocellular carcinoma
growth. Oncotarget. 7:35257–35269. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Candini O, Spano C, Murgia A, Grisendi G,
Veronesi E, Piccinno MS, Ferracin M, Negrini M, Giacobbi F, Bambi
F, et al: Mesenchymal progenitors aging highlights a miR-196 switch
targeting HOXB7 as master regulator of proliferation and
osteogenesis. Stem Cells. 33:939–950. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pogribny IP and Beland FA: Role of
microRNAs in the regulation of drug metabolism and disposition
genes in diabetes and liver disease. Expert Opin Drug Metab
Toxicol. 9:713–724. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krzeszinski JY, Wei W, Huynh H, Jin Z,
Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, et
al: miR-34a blocks osteoporosis and bone metastasis by inhibiting
osteoclastogenesis and Tgif2. Nature. 512:431–435. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu G, Zhang Y, Wei J, Jia W, Ge Z, Zhang Z
and Liu X: MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell
proliferation through repression of mitogen-activated protein
kinase-kinase 3. BMC Cancer. 13:4692013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Shi Y, McCaw L, Li YJ, Zhu F,
Gorczynski R, Duncan GS, Yang B, Ben-David Y and Spaner DE:
Microenvironmental interleukin-6 suppresses toll-like receptor
signaling in human leukemia cells through miR-17/19A. Blood.
126:766–778. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. Gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Draghici S, Khatri P, Tarca AL, Amin K,
Done A, Voichita C, Georgescu C and Romero R: A systems biology
approach for pathway level analysis. Genome Res. 17:1537–1545.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hamilton MP, Rajapakshe K, Hartig SM, Reva
B, McLellan MD, Kandoth C, Ding L, Zack TI, Gunaratne PH, Wheeler
DA, et al: Identification of a pan-cancer oncogenic microRNA
superfamily anchored by a central core seed motif. Nat Commun.
4:27302013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chevalier B, Adamiok A, Mercey O, Revinski
DR, Zaragosi LE, Pasini A, Kodjabachian L, Barbry P and Marcet B:
miR-34/449 control apical actin network formation during
multiciliogenesis through small GTPase pathways. Nat Commun.
6:83862015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Han YC, Vidigal JA, Mu P, Yao E, Singh I,
González AJ, Concepcion CP, Bonetti C, Ogrodowski P, Carver B, et
al: An allelic series of miR-17~92-mutant mice uncovers functional
specialization and cooperation among members of a microRNA
polycistron. Nat Genet. 47:766–775. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marzi MJ, Puggioni EM, Dall'Olio V, Bucci
G, Bernard L, Bianchi F, Crescenzi M, Di Fiore PP and Nicassio F:
Differentiation-associated microRNAs antagonize the Rb-E2F pathway
to restrict proliferation. J Cell Biol. 199:77–95. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: Corrigendum. IL-6R/STAT3/miR-34a feedback loop promotes
EMT-mediated colorectal cancer invasion and metastasis. J Clin
Invest. 125:13622015. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kulshreshtha R, Ferracin M, Wojcik SE,
Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM,
Negrini M, et al: A microRNA signature of hypoxia. Mol Cell Biol.
27:1859–1867. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Agrawal R, Pandey P, Jha P, Dwivedi V,
Sarkar C and Kulshreshtha R: Hypoxic signature of microRNAs in
glioblastoma: Insights from small RNA deep sequencing. BMC
Genomics. 15:6862014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu X, Jia R, Zhou Y, Song X, Wang J, Qian
G, Ge S and Fan X: Microarray-based analysis: Identification of
hypoxia-regulated microRNAs in retinoblastoma cells. Int J Oncol.
38:1385–1393. 2011.PubMed/NCBI
|
|
23
|
Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ,
Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, et al: MicroRNA-122
plays a critical role in liver homeostasis and
hepatocarcinogenesis. J Clin Invest. 122:2884–2897. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hsu SH, Wang B, Kota J, Yu J, Costinean S,
Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, et al: Essential
metabolic, anti-inflammatory, and anti-tumorigenic functions of
miR-122 in liver. J Clin Invest. 122:2871–2883. 2012. View Article : Google Scholar : PubMed/NCBI
|