Introduction
Hypoxia refers to a decrease in the concentration of
oxygen available and the decrease in the pressure of oxygen below
normal range. Hypoxia is able to limit and even halt the
physiological function of organs, organisms and cells (1). Tumor cells commonly induce hypoxic
conditions, owing to the rapid growth of tumor cells and the
relatively limited blood supply in tumors (2,3). Clinical
d experimental research indicates that the hypoxic tumor
environment may be associated with the development and metastasis
of solid tumors (4,5).
Breast cancer is the most common type of cancer in
women (6). The pathological grade and
prognosis of breast cancer are directly associated with tumor
hypoxia (7). Therefore, the study of
hypoxic microenvironments may have a crucial function in targeted
therapy adopted by clinics in the future. Hypoxia-inducible
factor-1α (HIF-1α) is an essential transcriptional regulatory
factor in hypoxia microenvironments and has 100 types of downstream
gene, including cell proliferation, angiogenesis and energy
metabolism (8,9). A previous study indicated that the
expression of HIF-1α, C-X-C motif chemokine receptor 4 (CXCR4) and
vascular endothelial growth factor (VEGF) is involved in tumor
progression, angiogenesis, metastasis and survival, and their
expression may be induced by hypoxia (10).
Understanding how to mimic a precise hypoxic
environment in vitro and establish a reliable
easy-to-operate model of hypoxia is the first step in studying the
hypoxic tumor microenvironment. Cobalt ions are substrates of the
iron-chelating enzymes; they can substitute for the iron ions of
the oxygen sensor hemoglobin and combine with oxygen at high
concentrations, leading to molecules entering the deoxidization
phase (11). As has been documented
in previous studies, treatment with CoCl2 is able to
mimic hypoxia (12). In the present
study, different concentrations of CoCl2 and MCR-7
breast cancer cells were cultured together in vitro to find
the optimal hypoxia model. The changes in the biological behavior
of breast cancer cells in a hypoxic microenvironment were examined
and the effects of CoCl2 on the MCR-7 cell proliferation
of breast cancer and the tumor angiogenesis factor
investigated.
Materials and methods
Cell culture
The breast cancer cell line MCF-7 was purchased from
Shanghai Bo Valley Biological Technology Co., Ltd. (Shanghai,
China), and incubated in Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing
10% fetal bovine serum, 100 U/ml penicillin and 0.1 mg/ml
streptomycin at 37°C with 5% CO2. CoCl2 was
purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany).
MCF-7 cells in the exponential phase were used for further
detection. Different CoCl2 concentrations were added to
DMEM. Cells were incubated with 50, 100, 150 and 200 µM
CoCl2 for different periods of time [0, 24, 48 and 72 h
for MTT assay; 24 h for reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay], with an equivalent
volume of PBS added to the control group. The morphological changes
of MCF-7 cells were observed using an inverted phase-contrast
microscope following treatment with CoCl2.
MTT assay
MCF-7 cells (1×103) were seeded in
96-well plates and cultured overnight. Subsequently, the culture
medium was removed and fresh DMEM containing the aforementioned
concentrations of CoCl2 was added. Cells were then
incubated for different periods of time, and the culture medium was
removed and replaced with fresh DMEM with different concentrations
of CoCl2 as previously used. MTT solution (5 mg/ml, 10
µl) was added to each well prior to incubation for 4 h. Next,
culture medium was removed and 100 µl dimethylsulfoxide was added
to dissolve the formazan crystals. The absorbance value was
determined at 490 nm.
RT-qPCR analysis
RT-qPCR was performed to quantitatively estimate the
changes in the expression of HIF-1α, CXCR4 and VEGF mRNA in MCF-7
cells treated with the aforementioned CoCl2
concentrations for 24 h. Total RNA was isolated from cells using an
RNeasy kit (Sigma-Aldrich; Merck KGaA). The RNA was
reverse-transcribed into cDNA using the PrimeScript®
First Strand cDNA Synthesis kit (Takara Bio, Inc., Otsu, Japan) at
42°C for 15 min, then at 85°C for 5 min. The Maxima®
SYBR Green qPCR Master Mix (2X) kit (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) was used to perform qPCR. Primer sequences
for β-actin, HIF-1α, CXCR4 and VEGF are presented in Table I (13,14). The
following PCR conditions were used: 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 60 sec (annealing and
extension, respectively). The experiment was performed in
triplicate and independently repeated at least twice. RT-qPCR data
were normalized and quantified using the 2−ΔΔCq method
(15). The relative expression level
of HIF-1α, CXCR4 and VEGF mRNA was calculated by determining the
ratio between the amount of the gene and β-actin.
 | Table I.Primer sequences used in reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Primer sequences used in reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene | Primer
sequence | Product size,
bp |
|---|
| β-actin (13) |
| 162 |
|
Forward |
5′-GACTTAGTTGCGTTACACCCTTTCT-3′ |
|
|
Reverse |
5′-GAACGGTGAAGGTGACAGCAGT-3′ |
|
| HIF-1α (14) |
| 150 |
|
Forward |
5′-TCTGGGTTGAAACTCAAGCAACTG-3′ |
|
|
Reverse |
5′-CAACCGGTTTAAGGACACATTCTG-3′ |
|
| CXCR4 (14) |
| 184 |
|
Forward |
5′-TCTGTGACCGCTTCTACC-3′ |
|
|
Reverse | 5′-
AGGATGAGGATGACTGTGG-3′ |
|
| VEGF (14) |
| 176 |
|
Forward |
5′-TGCTTCTGAGTTGCCCAGGA-3′ |
|
|
Reverse |
5′-TGGTTTCAATGGTGTGAGGACATAG-3′ |
|
Western blotting
The protein expression of HIF-1α, CXCR4 and VEGF was
assessed by western blotting. MCF-7 cells were treated with the
aforementioned CoCl2 concentrations for 24 h and cells
were lysed with ice-cold radioimmunoprecipitation assay buffer (150
mM NaCl, 1.0% NP-40, 0.1% Triton X-100, 0.5% sodium deoxycholate,
0.1% SDS and 50 mM Tris-HCl, pH 8.0) and protease inhibitors (AEBSF
at 2 mM, Aprotinin at 0.3 µM, Bestatin at 116 µM, E-64 at 14 µM,
Leupeptin at 1 µM and EDTA at 1 mM; Beijing Solarbio Science &
Technology Co., Ltd., Beijing, China). Protein concentrations were
quantified using a Bicinchoninic Acid Protein assay kit. Proteins
(30 µg/lane) were separated by SDS-PAGE (10% gels) and transferred
onto nitrocellulose membranes. Membranes were blocked for 1 h with
5% non-fat dried milk in Tris-buffered saline containing 20%
Tween-20 (TBST) and incubated overnight at 4°C with the primary
antibody Following washing with TBST, membranes were incubated for
1 h with secondary antibodies. Labeled protein bands were detected
using the enhanced chemiluminescence method (ProteinTech Group,
Inc., Chicago, IL, USA). Western blotting was performed three
times. The relative expression level of HIF-1α, CXCR4 and VEGF
proteins was quantified by densitometry analysis using ImageJ
(version 1.6.0; National Institutes of Health, Bethesda, MD, USA)
relative to β-actin. The antibodies were as follows: Anti-HIF-1α
(ab69836; 1:600), anti-CXCR4 (ab124824; 1:1,000) and anti-β-actin
(ab8226; 1:1,000) were purchased from Abcam (Cambridge, MA, USA);
anti-VEGF (AF5131, 1:1,000) was purchased from Affinity Biosciences
(Cincinnati, OH, USA), and the secondary antibodies were purchased
from ProteinTech Group, Inc.
Terminal deoxynucleotidyl
transferase-mediated dUTP nick-end labeling (TUNEL) assay
In situ detection of apoptosis was performed
on sides using the TUNEL technique using an In Situ Cell
Death Detection kit (Roche Applied Science, Penzberg, Germany).
MCF-7 cells were seeded onto sterile glass cover slips in a 6-well
plate, and incubated with 150 µM CoCl2 for 48 h. Cells
were then assessed by a TUNEL assay, according to the
manufacturer's protocol. Steps were as follows: Cells were fixed
with 4% paraformaldehyde (pH 7.4, Beijing Solarbio Science&
Technology Co., Ltd.) at room temperature for 1 h and washed with
PBS for 5 min; cells were incubated in Blocking solution (3%
H2O2 in methanol) for 10 min at room
temperature and washed with PBS for 5 min. Cells were then
incubated with penetrating fluid (0.1% Triton X-100 in 0.1% sodium
citrate solution) on ice for 2 min and then 50 µl TUNEL reaction
mixture (1:9) was added prior to incubation in a damp box at 37°C
for 1 h. Following washing with PBS for 5 min, a drop of PBS was
added to the slide. For staining of the nuclei, cells were washed
with PBS and incubated with 1.0 µg/ml DAPI (Sigma-Aldrich; Merck
KGaA). The samples were analyzed using a fluorescence microscope
(magnification, ×100). A total of 10 fields of view were
analyzed.
Statistical analysis
SPSS statistical software (version 19.0; IBM Corp.,
Armonk, NY, USA) to analyze the experimental data. Quantitative
data are presented as the mean ± standard deviation. One-way
analysis of variance with Least Significant Difference post-hoc
test was used to compare between groups. Student's t-test was used
to determine the significance for all pairwise comparisons of
interest. P<0.05 was considered to indicate a statistically
significant difference.
Results
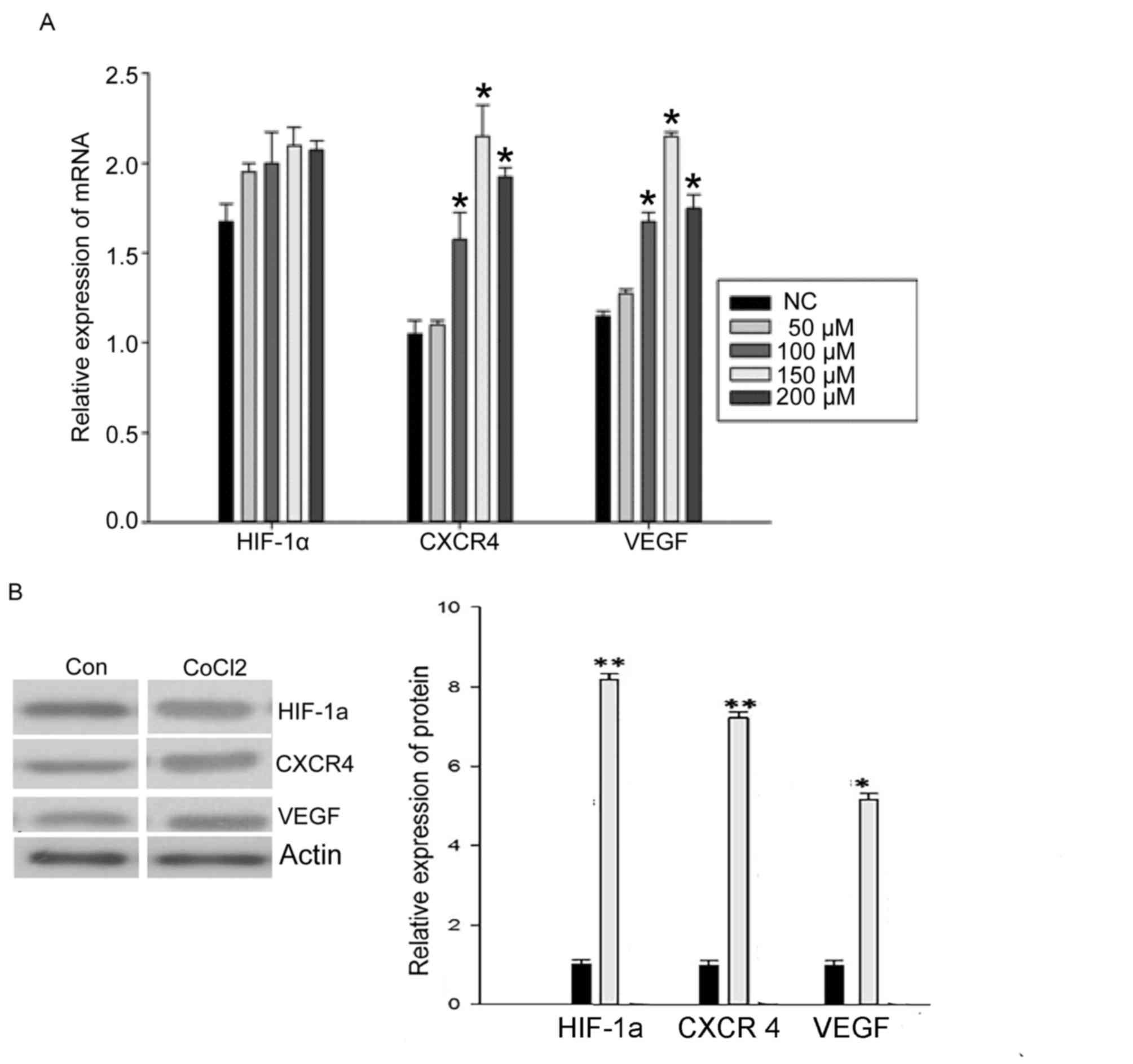
CoCl2 induces the expression of CXCR4
and VEGF mRNA, but not HIF-1α mRNA
To examine whether hypoxia affected the mRNA
expression level of HIF-1α, CXCR4 and VEGF, RT-qPCR was used to
quantify mRNA levels in MCF-7 cells cultured for 48 h with
different CoCl2 concentrations. There was no significant
change in the expression of HIF-1α mRNA following treatment with
different CoCl2 concentrations (Fig. 1A). However, the expression of CXCR4
mRNA and VEGF mRNA was significantly increased by treatment with
CoCl2 (50, 100, 150 and 200 µM). The 150 µM
concentration of CoCl2 induced the maximum effect
(P<0.05; Fig. 1A), which indicated
that hypoxia is able to promote the expression of CXCR4 and VEGF
mRNA.
CoCl2 induces the protein expression
of HIF-1α, CXCR4 and VEGF
To examine further whether hypoxia affects the
protein expression of HIF-1α, CXCR4 and VEGF, levels were examined
by western blotting in MCF-7 cells cultured for 48 h with different
CoCl2 concentrations. The expression of HIF-1α protein
significantly increased following treatment with CoCl2
(150 µM; P<0.05; Fig. 1B).
Expression of CXCR4 and VEGF protein was also significantly
increased by CoCl2 treatment (P<0.05; Fig. 1B). These results indicated that
hypoxia also promotes the expression of HIF-1α, CXCR4 and VEGF
protein. Owing to these results, a concentration of 150 µM
CoCl2 was used for further experiments.
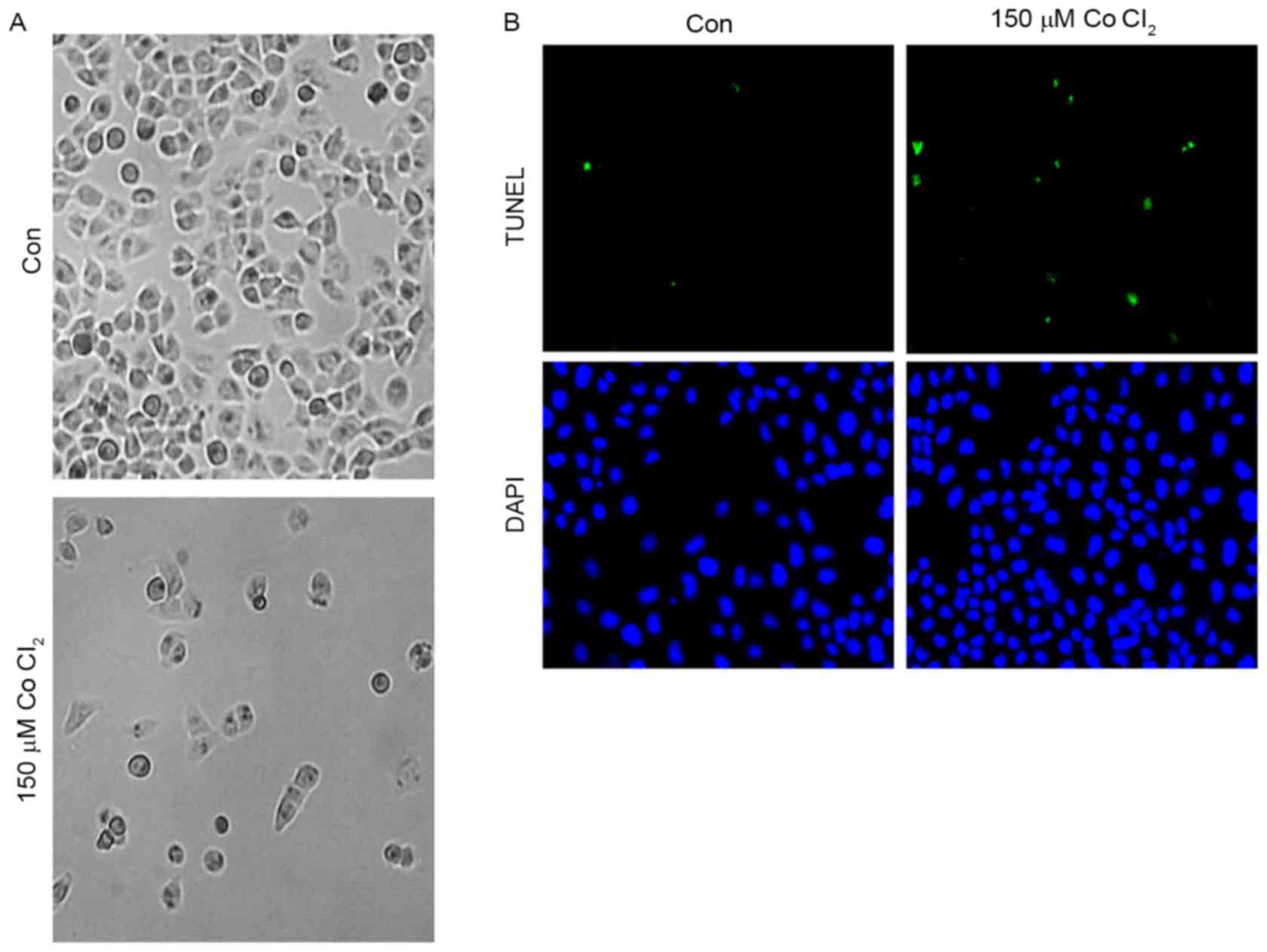
Effects of CoCl2 on MCF-7 cell
apoptosis
First, in order to determine the effect of
CoCl2 on MCF-7 cell morphology, 150 µM CoCl2
was added to MCF-7 cells for 48 h. As shown, the MCF-7 cell
morphology did not change significantly after treatment for 24 h.
However, 48 h later, an accumulation of cell metabolites was
observed in the culture dish, and a number of the cells exhibited
plasmatorrhexis (Fig. 2A). These
results indicated that the high intensity of the hypoxia
microenvironment may have an effect on cell morphology. A TUNEL
assay was performed to investigate whether CoCl2 was
able to trigger apoptosis of MCF-7 cells. As presented in Fig. 2B, the results of the TUNEL assay
demonstrated that incubation with 150 µM CoCl2 for 48 h
induced MCF-7 cells to exhibit a significant increase (5±2%
TUNEL-positive cells in the control group vs. 30±5% of
TUNEL-positive cells in the CoCl2-treatment group;
P<0.05) in TUNEL-positive cells, which indicated that
CoCl2 is involved in the regulation of apoptosis in
MCF-7 cells.
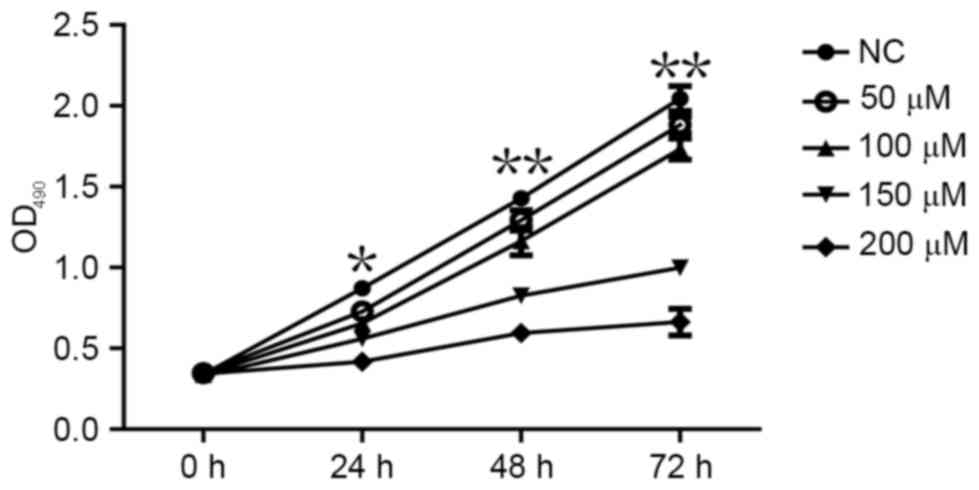
CoCl2 inhibits MCF-7 cell
proliferation
An MTT assay was performed to determine whether
CoCl2 affects the proliferation of MCF-7 cells. The MTT
assay demonstrated that MCF-7 cell proliferation significantly
decreased following treatment with CoCl2 compared with
the control group (P<0.05; Fig.
3). The effects of different CoCl2 concentrations on
MCF-7 cell proliferation were as follows: The higher the
concentration of CoCl2, the higher the inhibition
efficiency and the longer the treatment time, the higher the
inhibition efficiency (Fig. 3). The
inhibitory effect of CoCl2 on MCF-7 cell proliferation
was therefore dependent on the treatment time periods and on
CoCl2 concentration, which indicated that the time and
intensity of hypoxia was able to inhibit the proliferation of cells
to various extents.
Discussion
A hypoxic tumor microenvironment is a key feature in
a number of solid tumor tissues (16). This is due to the rapid proliferation
of tumor cells and the presence of tumor vascular structure and
functional abnormalities (17,18). The
presence of hypoxia may lead to a series of biological behaviors in
solid tumors, and these changes may possibly become the primary
reason for the development of resistance to radiotherapy and
chemotherapy (19). On the basis of
previous studies, tissue hypoxia is known to alter the oxygen
balance in tumor microenvironment (16). All of the aforementioned features may
elicit adverse effects in patients with breast cancer.
Breast cancer is the most frequently diagnosed
cancer in women and a major cause of mortality in women worldwide,
with high relapse rates (20–22). Breast cancer is sensitive to hypoxia;
25–40% of invasive breast cancer exhibits hypoxic microenvironments
(23,24). Hypoxia may promote the stemming of
breast cancer cells and epithelial-mesenchymal transition-mediated
breast cancer cell migration, with this intratumoral hypoxia having
a negative impact on the survival rate of patients with breast
cancer (25).
Hypoxia causes extensive responses in cells and
tissues; the expression of the transcription factor 1 HIF-1 is key
to allowing cells to adapt to the hypoxic environment (26,27). HIF-1
regulates the expression of a series of hypoxia-inducible genes,
resulting in a series of hypoxia adaptations. It has been
demonstrated that the expression of HIF-1α and its target genes are
increased in breast cancer (27).
HIF-1α expression may have a notable function early in breast
cancer progression (27). High levels
of HIF-1α expression at diagnosis may be used to predict early
recurrence and metastasis, and are also associated with poor
clinical outcomes in patients with breast cancer (28,29).
CXCR4 is a member of the C-X-C motif chemokine
receptor family associated with aggressive, proliferative and
motile breast cancer phenotypes (30–32). CXCR4
may represent a novel independent prognostic marker for patients
with lymph-node-positive breast cancer (33). Previous studies have demonstrated that
HIF-1α can markedly induce and regulate the expression of CXCR4 and
its ligand stromal cell-derived factor 1 (SDF-1) in breast cancer
tissues and cells, providing it with a vital function in the
migration of tumor cells (34–36). The
CXCR4-SDF-1 interaction potentially mediates the trafficking of
circulating tumor cells in primary breast cancer (36).
VEGF is able to regulate a number of cell functions,
including mitosis, permeability and vasoconstrictor tension
(37). VEGF expression is closely
associated with tumor angiogenesis and lymphatic formation in
breast cancer (38). VEGF is a target
gene of HIF-1 (39). The HIF-1
transcription complex is able to induce the expression of VEGF and
induce the corresponding biological effects (40,41).
HIF-1, CXCR4 and VEGF represent important targets in
the prevention and treatment of breast cancer under hypoxic
conditions. In recent years, HIFs have become the focus of a great
deal of research (42,43); however, only certain studies concern
HIF-1α, CXCR4 and VEGF and their association with the oxygen
homeostasis of microenvironments in breast tumors (44). For this reason, it is important to
investigate the association between the expression of these three
factors in breast cancer. Therefore, the present study established
an in vitro model to simulate the hypoxic microenvironment
present in human breast cancer cells. The present study revealed
that CoCl2 inhibited MCF-7 cell proliferation, and this
inhibitory effect was dependent on the length of time and
CoCl2 concentration. The results of RT-qPCR and western
blot analysis revealed that the expression of HIF-1α mRNA was not
significantly induced by CoCl2 (P>0.05); however, the
expression of CXCR4 and VEGF mRNA increased significantly upon
treatment with a range of different CoCl2 concentrations
(100, 150 and 200 µM). The results of western blotting revealed
that CoCl2 significantly induced the protein expression
of HIF-1α, CXCR4 and VEGF. Additionally, the
CoCl2-simulated hypoxic conditions generated
cytotoxicity and apoptosis in MCF-7 cells. The expression of
HIF-1α, CXCR4 and VEGF was associated with the CoCl2
treatment length and concentration. Thus, CoCl2
treatment was identified to induce the proliferation and metastasis
of tumors.
Further efforts to develop a suitable model of
hypoxia, or to discover an anticancer antioxidant to prevent damage
to cells may help to decrease tumor cell proliferation and decrease
the expression and transferal ability of HIF-1α, CXCR4 and VEGF.
This may provide a novel method for the prevention and treatment of
breast cancer.
Acknowledgements
The present study was supported by the Natural
Science Funds of Shandong Province Project: Mutation and Expression
of Parathyroid Carcinoma Susceptibility Gene HRPT2, MEN1, CyclinD1
and RET (grant no. ZR2012HL04); and the Science and Technology
Development Plan of Shandong Province Project: Clinical Application
of Selective ALND of CN+ Breast Cancer Patients (grant
no. 2012YD18062).
References
|
1
|
Hales CA: Physiological Function of
Hypoxic Pulmonary VasoconstrictionHypoxic Pulmonary
Vasoconstriction. Springer US; pp. 3–14. 2004, View Article : Google Scholar
|
|
2
|
K L Eales, Hollinshead KE and Tennant DA:
Hypoxia and metabolic adaptation of cancer cells. Oncogenesis.
5:e1902016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vaupel P and Harrison L: Tumor hypoxia:
Causative factors, compensatory mechanisms and cellular response.
Oncologist. 9 Suppl 5:S4–S9. 2004. View Article : Google Scholar
|
|
4
|
Yamada D, Kobayashi S, Yamamoto H,
Tomimaru Y, Noda T, Uemura M, Wada H, Marubashi S, Eguchi H,
Tanemura M, et al: Role of the hypoxia-related gene, JMJD1A, in
hepatocellular carcinoma: Clinical impact on recurrence after
hepatic resection. Ann Surg Oncol. 19 Suppl 3:S3552011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Andersen S, Donnem T, Al-Saad S, Al-Shibli
K, Stenvold H, Busund LT and Bremnes RM: Correlation and
coexpression of HIFs and NOTCH markers in NSCLC. Anticancer Res.
31:1603–1606. 2011.PubMed/NCBI
|
|
6
|
Coughlin SS and Ekwueme DU: Breast cancer
as a global health concern. Cancer Epidemiol. 33:315–318. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chakrabarti J, Turley H, Campo L, Han C,
Harris AL, Gatter KC and Fox SB: The transcription factor DEC1
(stra13, SHARP2) is associated with the hypoxic response and high
tumour grade in human breast cancers. Br J cancer. 91:954–958.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hänze J, Eul BG, Savai R, Krick S, Goyal
P, Grimminger F, Seeger W and Rose F: RNA interference for
HIF-1alpha inhibits its downstream signalling and affects cellular
proliferation. Biochem Biophys Res Commun. 312:571–577. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang H, Chen J, Liu F, Gao C, Wang X,
Zhao T, Liu J, Gao S, Zhao X, Ren H and Hao J: CypA, a gene
downstream of HIF-1α, promotes the development of PDAC. PLoS One.
9:e928242014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Teicher BA and Fricker SP: CXCL12
(SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 16:2927–2931.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang Y, Du KM, Xue ZH, Yan H, Li D, Liu
W, Chen Z, Zhao Q, Tong JH, Zhu YS and Chen GQ: Cobalt chloride and
low oxygen tension trigger differentiation of acute myeloid
leukemic cells: possible mediation of hypoxia-inducible
factor-1alpha. Leukemia. 17:2065–2073. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li S, Zhang J, Yang H, Wu C, Dang X and
Liu Y: Copper depletion inhibits CoCl2-induced aggressive phenotype
of MCF-7 cells via downregulation of HIF-1 and inhibition of
Snail/Twist-mediated epithelial-mesenchymal transition. Sci Rep.
5:124102015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shen B, Zheng MQ, Lu JW, Jiang Q, Wang TH
and Huang XE: CXCL12-CXCR4 promotes proliferation and invasion of
pancreatic cancer cells. Asian Pac J Cancer Prev. 14:5403–5408.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo HQ, Xu M, Zhong WT, Cui ZY, Liu FM,
Zhou KY and Li XY: EGCG decreases the expression of HIF-1α and VEGF
and cell growth in MCF-7 breast cancer cells. J BUON. 19:435–439.
2014.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data usingreal-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Semenza GL: The hypoxic tumor
microenvironment: A driving force for breast cancer progression.
Biochim Biophys Acta. 1863:382–391. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsutsui S, Matsuyama A, Yamamoto M,
Takeuchi H, Oshiro Y, Ishida T and Maehara Y: The Akt expression
correlates with the VEGF-A and -C expression as well as the
microvessel and lymphatic vessel density in breast cancer. Oncol
Rep. 23:621–630. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Z, Wu J, Liang B and Huangfu CS:
Correlationbetween hypoxia inducible factor (HIF-1α) and
epithelium-mesenchyma transform of ductal carcinoma with invasive
breast cancer. Shandong Med J. 52:90–92. 2012.
|
|
19
|
Moser C, Lang SA, Mori A, Hellerbrand C,
Schlitt HJ, Geissler EK, Fogler WE and Stoeltzing O: ENMD-1198, a
novel tubulin binding agent rdduces HIF-lalpha and STAT3 activity
in human hepatocellular carcinoma (HCC) cells and inhibits growth
and vascularization in vivo. BMC Cancer. 8:2062008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Marmot MG, Altman DG, Cameron DA, Dewar
JA, Thompson SG and Wilcox M: The benefits and harms of breast
cancer screening: An independent review. Br J Cancer.
108:2205–2240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lech R and Przemyslaw O: Epidemiological
models for breast cancer risk estimation. Ginekol Pol. 82:451–454.
2011.PubMed/NCBI
|
|
23
|
Liu ZJ, Semenza GL and Zhang HF:
Hypoxia-inducible factor 1 and breast cancer metastasis. J Zhejiang
Univ Sci B. 16:32–43. 2015.(In Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lundgren K, Holm C and Landberg G: Hypoxia
and breast cancer: Prognostic and therapeutic implications. Cell
Mol Life Sci. 64:3233–3247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park SJ, Kim JG, Kim ND, Yang K, Shim JW
and Heo K: Estradiol, TGF-β1 and hypoxia promote breast cancer
stemness and EMT-mediated breast cancer migration. Oncol Lett.
11:1895–1902. 2016.PubMed/NCBI
|
|
26
|
Bradbury J: Breathing hard to keep up with
HIF-1. Lancet. 358:17042001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Generali D, Berruti A, Brizzi MP, Campo L,
Bonardi S, Wigfield S, Bersiga A, Allevi G, Milani M, Aguggini S,
et al: Hypoxiainducible factor-1alpha expression predicts a poor
response to primary chemoendocrine therapy and disease-free
survival in primary human breast cancer. Clin Cancer Res.
12:4562–4568. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gruber G, Greiner RH, Hlushchuk R,
Aebersold DM, Altermatt HJ, Berclaz G and Djonov V:
Hypoxia-inducible factor 1alpha in high-risk breast cancer: An
independent prognostic parameter. Breast Cancer Res. 6:R191–R198.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Goswami S, Sahai E, Wyckoff JB, Cammer M,
Cox D, Pixley FJ, Stanley ER, Segall JE and Condeelis JS:
Macrophages promote the invasion of breast carcinoma cells via a
colony-stimulating factor-1/epidermal growth factor paracrine loop.
Cancer Res. 65:5278–5283. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rahimi M, Toth TA and Tang CK: CXCR4
suppression attenuates EGFRVIII mediated invasion and induces p38
MAPK-dependent protein trafficking and degradation of EGFRvIII in
breast cancer cells. Cancer Lett. 306:43–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Helbig G, Christopherson KW II,
Bhat-Nakshatfi P, Kumar S, Kishimoto H, Miller KD, Broxmeyer HE and
Nakshatri H: NF-kappaB promotes breast cancer cell migration and
metastasis by inducing the expression of the chemokine receptor
CXCR4. J Biol Chem. 278:21631–21638. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Parker CC, Kim RH, Li BD and Chu QD: The
chemokine receptor CXCR4 as a novel independent prognosic marker
for node-positive breast cancer patients. J Surg Oncol.
106:393–398. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dunn LK, Mohammad KS, Fournier PG, McKenna
CR, Davis HW, Niewolna M, Peng XH, Chirgwin JM and Guise TA:
Hypoxia and TGF-beta drive breast cancer bone metastases through
parallel signaling pathways in tumor cells and the bone
microenvironment. PLoS One. 4:e68962009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Citti A, Boldrini R, Inserra A, Alisi A,
Pessolano R, Mastronuzzi A, Zin A, De Sio L, Rosolen A, Locatelli F
and Fruci D: Expression of multidrug resistance-associated proteins
in paediatric soft tissue sarcomas before and after chemotherapy.
Int J Oncol. 41:117–124. 2012.PubMed/NCBI
|
|
36
|
Mego M, Cholujova D, Minarik G, Sedlackova
T, Gronesova P, Karaba M, Benca J, Cingelova S, Cierna Z, Manasova
D, et al: CXCR4-SDF-1 interaction potentially mediates trafficking
of circulating tumor cells in primary breast cancer. Bmc Cancer.
16:1272016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ferrara N: VEGF and the quest for tumour
angiogenesis factors. Nat Rev Cancer. 2:795–803. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Timoshenko AV, Chakraborty C, Wagner GF
and Lala PK: COX-2-mediated stimulation of the lymphangiogenic
factor VEGF-C in human breast cancer. Br J Cancer. 94:1154–1163.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Komatsu DE and Hadjiargyrou M: Activation
of the transcription factor HIF-1 and its target genes, VEGF, HO-1,
iNOS, during fracture repair. Bone. 34:680–688. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Semenza GL: Expression of
hypoxia-inducible factor 1: Mechanisms and consequences. Biochem
Pharmacol. 59:47–53. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li G, He L, Zhang E, Shi J, Zhang Q, Le
AD, Zhou K and Tang X: Overexpression of human papillomavirus (HPV)
type 16 oncoproteins promotes angiogenesis via enhancing HIF-1α and
VEGF expression in non-small cell lung cancer cells. Cancer Lett.
311:160–170. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dong X, Wang YS, Dou GR, Hou HY, Shi YY,
Zhang R, Ma K, Wu L, Yao LB, Cai Y and Zhang J: Influence of Dll4
via HIF-1α-VEGF signaling on the angiogenesis of choroidal
neovascularization under hypoxic conditions. PLoS One.
6:e184812011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ward C, Langdon SP, Mullen P, Harris AL,
Harrison DJ, Supuran CT and Kunkler IH: New strategies for
targeting the hypoxic tumour microenvironment in breast cancer.
Cancer Treat Rev. 39:171–179. 2013.(In Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lopez-Haber C, Barrio-Real L,
Casado-Medrano V and Kazanietz MG: Heregulin/ErbB3 signaling
enhances CXCR4-driven rac1 activation and breast cancer cell
motility via hypoxia-inducible factor 1α. Mol Cell Biol.
36:2011–2026. 2016. View Article : Google Scholar : PubMed/NCBI
|