Introduction
Genomic DNA is sensitive to a variety of exogenous
damage, including hydrolysis, oxidation, mismatch, and endogenous
damages such as UV radiation and chemicals. Endogenous damage can
also result in gene disruption and deletion, ultimately causing
apoptosis or tumorigenesis (1,2).
Abnormal DNA repair is closely associated with
tumorigenesis and tumor multi-drug resistance (3). Nucleotide excision repair (NER) is one
of the primary defensive barriers against tumorigenesis and a major
repair system for chemotherapy-induced DNA damage. Chemotherapy is
widely applied to induce apoptosis of tumor cells (1,4,5). Thus, NER reduces the efficacy of
chemotherapy to a certain degree.
NER is comprised of two pathways: Global genome
repair (GGR) and transcription-coupled repair (TCR). GGR is
involved in injury repair for any genomic sequence, which is
crucial to prevent carcinogenesis (6). The major role of TCR is to delay aging
by repairing the DNA damage present in activated transcriptional
chains (2). Generally, the NER
process is divided into the following three steps: i) Damage
recognition and shear complex assemble; ii) double-stranded DNA
separation and damage removal; and iii) DNA repair synthesis and
double-strand linkage. During the whole process, the recognition of
DNA damage is required to trigger and initiate the following repair
via certain signal transduction pathways (7).
Among these NER genes, xeroderma pigmentosum gene
group C (XPC) serves a key role in the process of GGR (1,2,8,9). Neither
GGR nor TCR can be initiated in the absence of XPA (10,11). XPF
combines with DNA excision repair protein ERCC-1 (ERCC1) to form a
dimer that functions as a 5′ DNA endonuclease, whereas XPG
functions as a DNA ligase and a 3′ DNA endonuclease (1,2,8).
Previous studies have demonstrated that the NER
genes are associated with the genesis and development of tumors
(12–15). Huang et al (14) revealed that haplotypes of XPC
polymorphisms containing XPC 499V modified the smoking-associated
risks of advanced colorectal adenoma. Previously, it has been
confirmed that there is no significant relation between XPD genetic
variation and non-Hodgkin's lymphoma (NHL) risk (12). However, the presence of the XPD 751Gln
allele was identified to be associated with a two-fold decreased
risk of developing diffuse large B-cell lymphoma (12).
In the present study the expression levels of the
NER genes XPC, XPA, XPG, XPF, ERCC1 and XPD were determined in
human colorectal carcinoma (CRC) and corresponding normal tissues.
The role of differential genes in chemotherapeutic resistance of
CRC was investigated. In view of this, the present study aimed to
clarify the role of these NER genes in the chemotherapeutic
sensitivity of CRC, and provide evidence of the efficacy of
targeting these genes in the treatment of CRC clinically in the
future.
Materials and methods
Clinic data and specimens
collection
A total of 46 samples of fresh CRC and 20 samples of
adjacent normal colorectal tissues were obtained from Department of
General Surgery, Xinhua Hospital (Shanghai, China) between January
2014 and May 2015. The patient cohort included 25 males and 21
females. The mean age of the patients was 58.4±14.8 years old. All
patients underwent surgical resection and cisplatin chemotherapy.
The specimens included 10 cases of mucinous adenocarcinoma, 22
cases of adenocarcinoma and 14 cases of mucinous adenocarcinoma
complicated with adenocarcinoma.
All patients were diagnosed as having CRC following
biopsy. The adjacent tissue that was 5-cm away from the CRC was
removed and selected as a normal control, which was also confirmed
by pathological examination. All patients provided written informed
consent. This study was approved by the Ethics Committee of Xinhua
Hospital.
Main reagents
TRIzol reagent and reverse transcriptase M-MLV were
purchased from Invitrogen; Thermo Fisher Scientific, Inc. (Waltham,
MA, USA). Quantitative PCR reagents IQ™ SYBR®-Green I
Supermix was obtained from Bio-Rad Laboratories, Inc. (Hercules,
CA, USA). An Annexin V-Fluorescein isothiocyanate (FITC) apoptosis
assay kit was provided by Beijing Baosai Biological Technology Co.,
Ltd. (Beijing, China). A Silencer T small interfering RNA (siRNA)
construction kit was obtained from Ambion; Thermo Fisher
Scientific, Inc. Cisplatin was provided by Sigma-Aldrich; Merck
KGaA (Darmstadt, Germany). The primers for XPC, XPA, XPG, XPF,
ERCC1, and XPD (Table I) were
synthesized by Takara Biotechnology Co., Ltd. (Dalian, China).
 | Table I.Reverse transcription-quantitative
polymerase chain reaction primer pairs for nucleotide excision
repair genes. |
Table I.
Reverse transcription-quantitative
polymerase chain reaction primer pairs for nucleotide excision
repair genes.
| Gene | Primer pairs | Product size,
bp |
|---|
| GAPDH | F:
5′-CTCTCTGCTCCTCCTGTTCGAC-3′ | 69 |
|
| R:
5′-TGAGCGATGTGGCTCGGCT-3′ |
|
| XPA | F:
5′-GGTCTCTTGAAGTTTGGGGTAGTC-3′ | 142 |
|
| R:
5′-TTCCACACGCTGCTTCTTACTG-3′ |
|
| XPC | F:
5′-ACACCTACTACCTCTCAAACC-3′ | 115 |
|
| R:
5′-ATGGACCAATTCCTCATCATCTCG-3′ |
|
| XPD | F:
5′-GCCTGAACGCTCTTCTAA-3′ | 324 |
|
| R:
5′-TTACAGGCGGTGGCGATAAT-3′ |
|
| XPF | F:
5′-TTTGTGAGGAAACTGTATCTGTGG-3′ | 125 |
|
| R:
5′-GTCTGTATAGCAAGCATGGTAGG-3′ |
|
| XPG | F:
5′-AGGTAGAGTCAAGGAGAGT-3′ | 97 |
|
| R:
5′-TGCTCCTGTCATTGTTGTA-3′ |
|
| ERCC1 | F:
5′-CTGCTGCGGGATGAGAAC-3′ | 193 |
|
| R:
5′-ATCGGAATAAGGGCTTGGC-3′ |
|
A plasmid, which carried an XPC gene cDNA, was
constructed in our laboratory according to protocols described
previously (16,17). Following SfiI digestion, the
XPC gene cDNA was further removed from the plasmid and then
inserted into the SfiI site of pcDNA3.1(+) (Invitrogen;
Thermo Fisher Scientific, Inc.) to prepare the pcDNA3-XPC
plasmid.
Cell line and culture condition
The CRC HCT116, HCT8, HT29, LS174T, LOVO, SW480,
SW620 cell lines and the normal human colorectal FHC cell line were
provided by the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China). All the cell lines were cultured in
RPMI-1640 medium supplemented with 10% fetal bovine serum and
cultured at 37°C in an atmosphere of 5% CO2. When the
cells reached a confluence of ~90% (every ~3 days), they were
passaged. Cells at passages 3–5 were used for experimental
analyses.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from normal and CRC tissues
or cancer cells using TRIzol. Total RNA preparation was performed
in accordance with the manufacturer's protocol. Following DNase I
(Takara Biotechnology Co., Ltd., Dalian, China) treatment, 2 µg of
RNA was reverse transcribed using a Takara RNA LA PCR kit (AMV)
(Takara Biotechnology Co., Ltd.).
The 25 µl standard reaction system included 12.5 µl
of Real-Time PCR Master Mix SYBR-Green I, 0.5 µl of primer forward
(10 µmol/l), 0.5 µl of primer reverse (10 µmol/l), 1 µl of cDNA and
10.5 µl of ddH2O. The sequences of all primers are
listed in Table I. The reaction
condition included initial denaturation at 95°C for 3 min, then
denaturation at 95°C for 4.5 min, annealing at 60°C for 40 sec and
extension at 72°C for 40 sec. The following reactions were
performed for 40 cycles. The data were analyzed using iQ5 Gene
expression software (Bio-Rad Laboratories, Inc.). The reactions
were performed and values were normalized to the housekeeping gene
GAPDH, Cq values were determined by using the 7500
System SDS software (version.1.2.3; Applied Biosystems; Thermo
Fisher Scientific, Inc.). Expression ratios were calculated using
the 2−ΔΔCq method (18).
Western blot assay
Approximately 100 mg of the cancer cells were lysed
with 1 ml of pre-cooled radioimmunoprecipitation assay buffer
containing 150 mM NaCl, 1.0% NP-40 or 0.1% Triton X-100, 0.5%
sodium deoxycholate, 0.1% SDS (sodium dodecyl sulphate), 50 mM
Tris-HCl (pH 8.0) and protease inhibitors (Abcam, Cambridge, UK)
for 15 sec and then in an ice bath for another 10 min. The lysate
was then centrifuged at 12,000 × g at 4°C for 10 min and the
supernatant was harvested. The concentration of the total protein
was quantified using the Bradford method.
A total of 50 µg protein per lane was separated by
12% SDS-PAGE and then the proteins were transferred onto a
polyvinylidene fluoride transfer membrane. The transfer membrane
was semidried at 20 V for 15 min. The membrane was then blocked
with 5% skim milk for 4 h at 4°C. The membranes were washed three
times with TBS for 5 min each. Subsequently, goat anti-human XPC
polyclonal antibody (Santa Cruz Biotechnology, Inc., Dallas, TX,
USA; 1:200) was added and the membranes were incubated at 4°C
overnight. Next, horseradish peroxidase-conjugated rabbit anti-goat
IgG (cat. no. TA130025; Origene Technologies, Inc., Beijing, China;
1:3,000) was added and incubated at room temperature for another 2
h. The membrane was stained with Enhanced Chemiluminescence reagent
(Pierce; Thermo Fisher Scientific, Inc.) and imaged on X-ray film
(Fujifilm Corporation, Tokyo, Japan) by autoradiography. Quantity
One® 1-D analysis software (Bio-Rad Laboratories, Inc.)
were used to quantitatively analyze the density of the bands.
β-actin (cat. no. Ab8227; Abcam, Cambridge, UK; 1:1,000) was
selected as an internal control. The relative protein level was
expressed as a ratio between the densities of XPC and β-actin.
Immunohistochemistry analysis
A total of 46 specimens (25 males and 21 females;
58.4±14.8 years old) were used for this experiment. Archived
samples from these 36 cases were retrieved from the surgical
pathology files. These CRC tissues, according to the Vienna
modified classification (2002) (19),
were assigned pathologically to poor differentiated (11 cases),
moderately differentiated (20 cases) and highly differentiated (15
cases).
The 5-µm tissue sections were deparaffinized with
xylene, rehydrated in graded alcohol, and processed using the
streptavidin immunoperoxidase method. Briefly, the sections were
submitted to antigen retrieval by heating to 95°C for 10 min in a
citrate buffer (0.01 mol/l, pH 6.0). Subsequently, the slides were
incubated in 10% goat normal serum (cat. no. C-0005; Shanghai
Haoran Biotechnology Co., Ltd., Shanghai, China) for 30 min at room
temperature, followed by overnight incubation at 4°C with goat
anti-human XPC polyclonal antibody (Santa Cruz Biotechnology, Inc.;
1:100). Following this, the samples were incubated with
biotinylated rabbit anti-goat immunoglobulin G (cat. no. ZDR-5308;
Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing,
China; 1:2,000) for 15 min at 37°C, followed by streptavidin
peroxidase complexes (cat. no. SP Kit-D1; Beijing Dingguo
Changsheng Biotechnology Co., Ltd., Beijing, China) for 15 min at
37°C. 3, 3′-diaminobenzidine was used as a chromogen, and
hematoxylin was used for nuclear counterstaining for 10 min at room
temperature. Following this, immunostaining was quantified using a
CM-2000B imaging analysis system (Beijing University of Aeronautics
and Astronautics, Beijing, China). Identification of
immunohistochemistry results was in accordance to the criteria
proposed by Maruyama et al (20). The differences between the two random
groups were analyzed using χ2 test.
Plasmid construction of siRNA
targeting XPC
An effective sequence targeting XPC
(5′-GGATGAAGCCCTCAGCGAT-3′) was screened using GenBank (no.
NM_004628; https://www.ncbi.nlm.nih.gov/nuccore/NM_004628.4).
As a template, the oligonucleotide chains were designed based on
the base pairing rule.
The following nucleotide sequences were used:
Forward,
5′-GATCCGGATGAAGCCCTCAGCGATTTCAAGAGAATCGCTGAGGGCTTCATCCTTTTTTGGAA-3′
and reverse,
5′-AGCTTTTCCAAAAAAGGATGAAGCCCTCAGCGATTCTCTTGAAATCGCTGAGGGCTTCATCCG-3′.
The control sequences forward,
5′-GATCCGGATGAAGCCCTCAGCGATTTCAAGAGAGTGCACCGAGTCCTTCTGTATTTTTGGAAA-3′
and reverse,
5′-AGCTTTTCCAAAAAATTACAGAAGGACTCGGTGCACTCTCTTGAAATCGCTGAGGGCTTCATCCG-3′
were also selected. The oligonucleotides were synthesized by
Invitrogen; Thermo Fisher Scientific, Inc.
A pSilencer™ 5.1-H1 Retro Vector (Ambion, No.
AM5784) was digested using HindIII and BamHI
restriction enzymes, followed by ligation with T4 DNA ligase. Next,
the recombinant plasmid was transformed into fresh competent E.
Coli DH5α cells. The recombinant clones were selected from a
Luria-Bertani agar plate containing 100 µg/ml ampicillin. The
positive clones were confirmed by PCR and then sent to Shanghai
GeneChem Co., Ltd. (Shanghai, China) for sequencing. The confirmed
vector was named pSilencer™ 5.1-XPC siRNA and the control vector
was named pSilencer™ 5.1-XPC control. Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used together
with pSilencer™ 5.1-XPC siRNA (20 µg/µl) or pSilencer™ 5.1-XPC
control (20 µg/µl) to transfect SW480 cells for 20 min. Additional
puromycin (0.4 µg/ml) was added to screen the positive clones 48 h
following transfection.
Stable transfection of CRC cells with
siRNA-XPC or pcDNA3-XPC plasmid
SW480 cells were seeded in 100-mm cell culture
dishes and cultured to reach a confluence of 70–80%. The cells were
then transfected with the siRNA-XPC (0.2 µg/µl) or the pcDNA3-XPC
plasmid DNA using a cationic lipid (0.2 µg/µl) (10 µg of plasmid
DNA/50 µl Lipofectamine 2000/100-mm dish) for 6 h. As a control,
cells were transfected with the pcDNA3.
Cell susceptibility assay
The treated SW480 cells (1×106/ml) were
inoculated in a 96-well plate (100 µl/well) and treated with
cisplatin (5 µmol/l) (cat. no. 479306-1G; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) for 4 h. Cell susceptibility was measured
at 24 h upon the addition of the tetrazolium salt XTT (0.12 mg/ml)
to the culture medium. The concentration of formazan dye formed was
measured by at 492 nm using a microplate reader (Bio-Rad
Laboratories, Inc.).
Cell apoptosis assay
The SW480 cells were treated with 5 µmol/l cisplatin
for 4 h. Subsequently, the cells were digested with 0.1% trypsin.
Next, the cell suspension was centrifuged at 150 × g for 5 min at
4°C and the cells were harvest. The supernatant was removed and the
precipitate was washed twice with PBS. The SW480 cells were
resuspended in PBS and adjusted to a concentration of
1×106/ml.
A total of 100 µl Annexin-V-FITC reagent was added
to the cells for 10–15 min at room temperature in the dark. Cells
were then centrifuged at 150 × g for 5 min at 4°C and washed with
PBS once. Cell apoptosis was then detected using a flow cytometer.
The data were analyzed using CellQuest 3.0 software (BD
Biosciences, Franklin Lakes, NJ, USA).
Establishment of a xenograft tumor
model of human CRC
In total, 24 male BALB/c nude mice (Silaike
Experimental Animal Center, Shanghai, China; http://www.lascn.net/SupplyDemand/Site/Contact.aspx?id=77)
(weight, 18–22 g) aged 6–7 weeks, were subcutaneously inoculated
with 0.2 ml of the prepared SW480 cell suspension
(1×107/ml). The mice had free access to food and water,
and were maintained in a room at 20–22°C, 40–70% humidity and a 12
h light/dark cycle.
Following this, the general conditions of the
animals, including consciousness, diet and activity, were observed
and recorded. Tumor volume was also observed successively for 14
days and recorded at particular time points to plot a growth curve.
Tumor volume (TV) and relative TV (RTV) were calculated as follows:
TV = ½ × a × b2 (a, b represent long and short diameter
of the tumor tissue, respectively); RTV =
Vt/V0 (Vt represents the tumour
volume at different measurement time points, V0
represents original tumour volume at day 0). All experiments were
conducted in accordance with the National Guidelines for the Care
and Use of Laboratory Animals. This study was approved by the
Ethics Committee of Xinhua Hospital.
Western blotting assay of B-cell
lymphoma-2 (Bcl-2) and Bcl-associated X (Bax) protein expression
levels in transplanted tumor tissue
At the end of the experiment, the animals were
sacrificed and the implanted tumor tissues were isolated and
homogenized. Bax (1:200; cat. no. ab53154; Abcam) and Bcl-2 (1:200;
cat. no. ab59348; Abcam) protein expression was measured by western
blotting at 4°C overnight. The rabbit anti-goat immunoglobulin G
(1:2,000; cat. no. ZDR-5308; Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd.) was used as a secondary antibody at room
temperature for 2 h.
Statistical analysis
All the data are expressed as mean ± standard
deviation. SPSS 17.0 statistics software (SPSS, Inc., Chicago, IL,
USA) was used to analyze differences. The χ2 test was
used to compare the expression of XPC in cancer tissues with
different degrees of differentiation. One-way analysis of variance
assay followed by Dunnett's least significant difference and a
paired Student's t-test was used to compare the difference among
and between groups, respectively. P<0.05 was considered to
indicate a statistically significant difference.
Results
XPC mRNA expression is upregulated in
CRC tissue
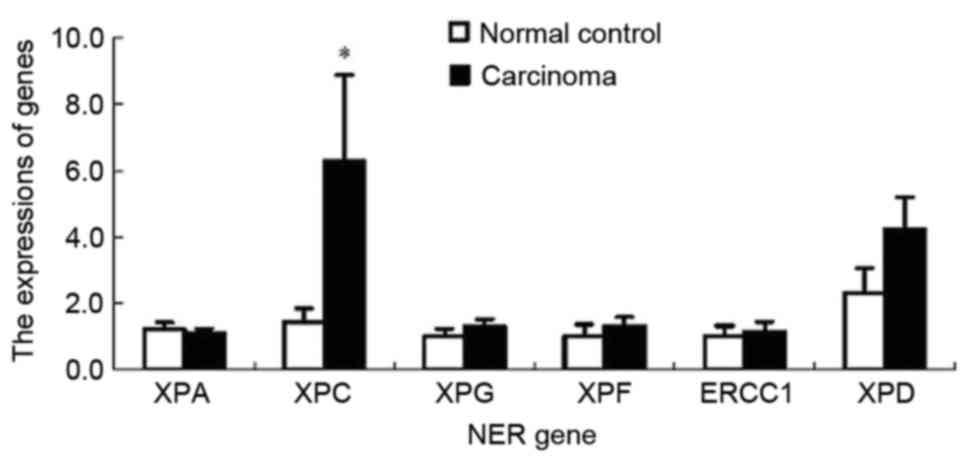
RT-qPCR analysis revealed that only expression of
XPC mRNA in the CRC tissue was significantly increased compared
with that in the normal colorectal tissue (P<0.01). However,
there were no significant differences in the mRNA expression of
other NER genes, including XPA, XPG, XPF, ERCC1 and RAP1 between
the cancerous and normal tissues (P>0.05; Fig. 1).
XPC expression is associated with the
malignancy of CRC
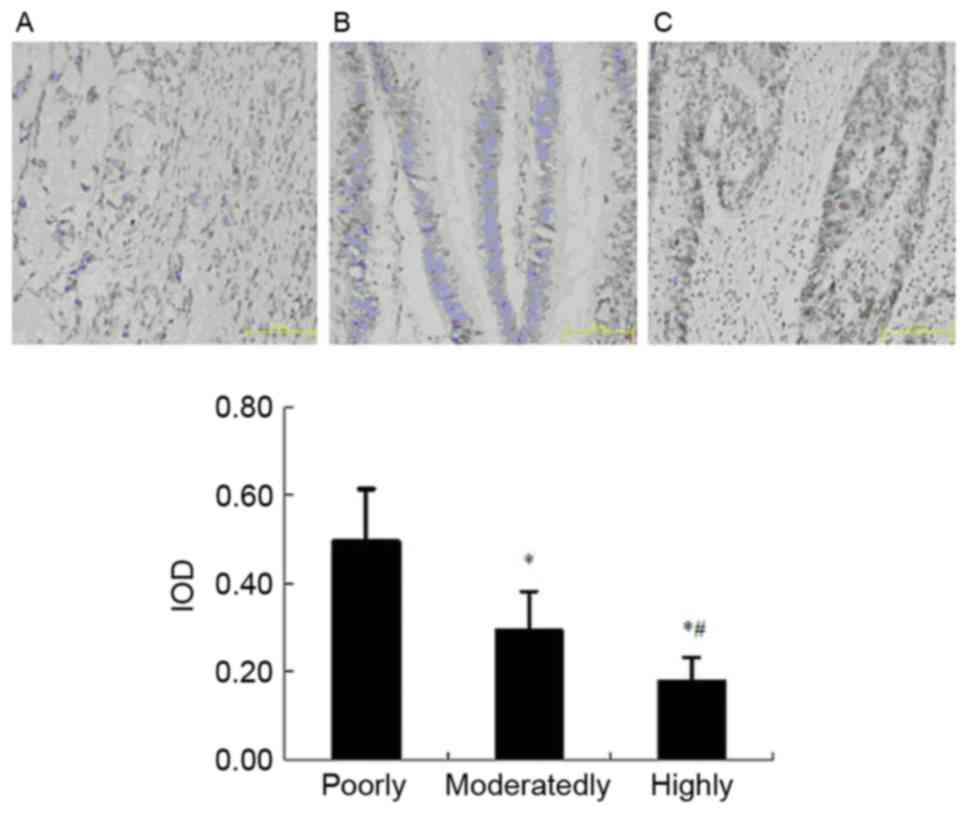
The results of immunohistochemistry indicated that
the XPC-positive expression was present in 72.7% of the poorly
differentiated samples (8/11), 40.0% of the moderately
differentiated samples (8/20) and 20.0% of highly differentiated
samples (3/15) (Table II). The
differences between the two random groups were analyzed using
χ2 test. XPC expression was the highest in the poorly
differentiated samples, then in the moderately differentiated and
lowest in the highly differentiated cancerous tissues (Fig. 2). These results indicated that the XPC
expression was associated with the degree of malignancy in CRC.
 | Table II.Expression of XPC in caner tissue
with different degrees of differentiation. |
Table II.
Expression of XPC in caner tissue
with different degrees of differentiation.
| Differentiation
degree | XPC−,
n | XPC+,
n | Total, n |
|---|
| Poor | 3 | 8 | 11 |
| Moderate | 12 |
8a | 20 |
| High | 12 |
3a,b | 15 |
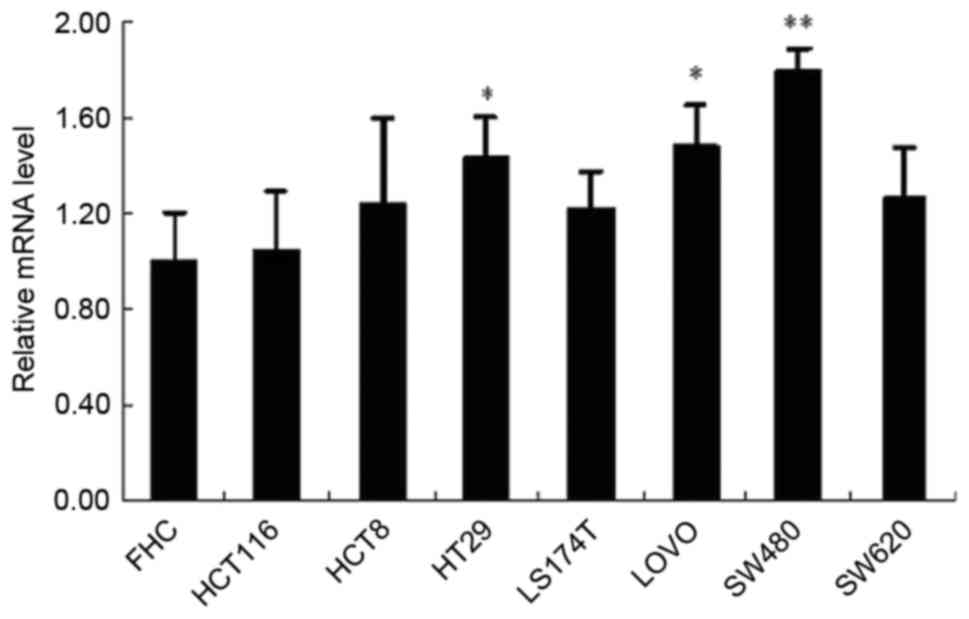
XPC mRNA expression is highest in the
SW480 cell line
The level of XPC mRNA expression was significantly
increased in HT29, LOVO, and SW480 cell lines compared with the FHC
cell line. XPC mRNA was expressed at the highest level in the
SW480. Thus, the SW480 cell line was selected for the following
experiments (Fig. 3).
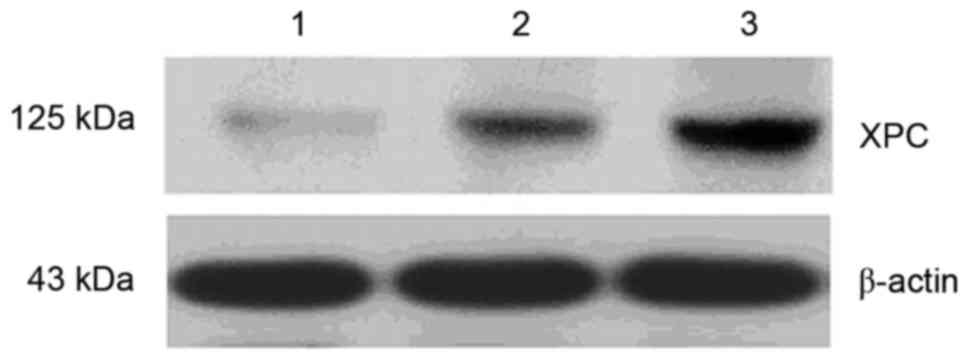
siRNA-XPC increases the
chemotherapeutic sensitivity of SW480 cells to cisplatin
siRNA-XPC transfection reduced the level of XPC
protein expression in the SW480 cells. However, the XPC protein
level was markedly increased in the cells transfected with
pcDNA3-XPC compared with the control (Fig. 4).
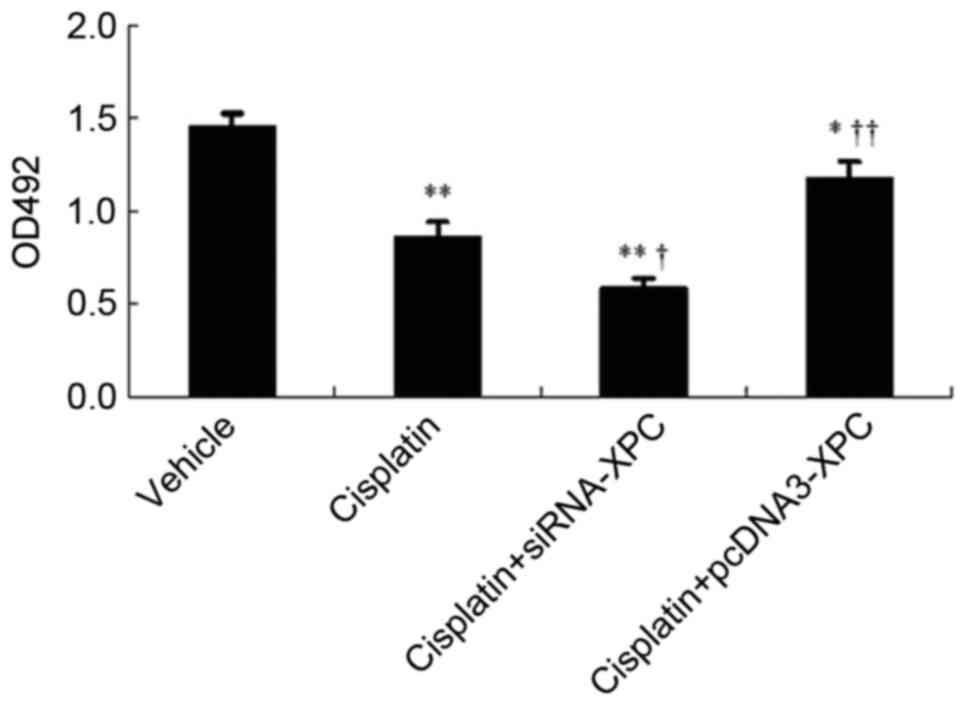
Prior to transfection, cisplatin significantly
inhibited the growth of the tumor cells (P<0.05). Cisplatin in
combination with siRNA-XPC transfection significantly inhibited the
cell growth further, compared with cisplatin treatment alone
(P<0.05; Fig 5). In addition, the
transfected cells overexpressing XPC exhibited reduced sensitivity
to cisplatin compared with cells transfected with the control
vector (P<0.05).
Transfection with siRNA-XPC increases
apoptosis of SW480 cells upon treatment with cisplatin
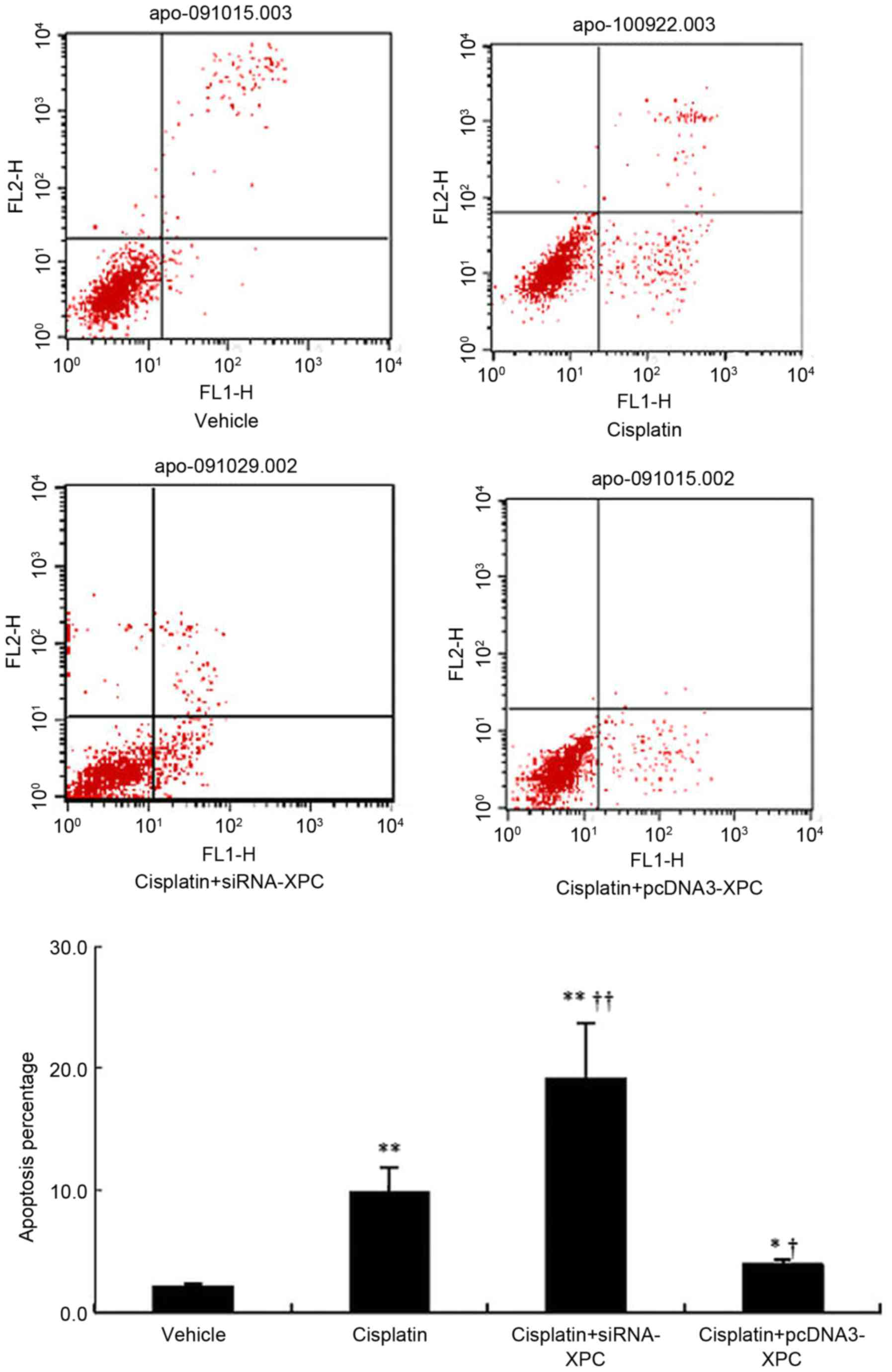
The proportion of cells undergoing apoptosis
significantly increased following cisplatin treatment compared with
the control (P<0.05; Fig. 6).
Notably, siRNA-XPC transfection further increased the proportion of
apoptotic cells in the presence of cisplatin. However, when XPC was
overexpressed, the apoptotic proportion of the transfected cells
was significantly reduced, even in the presence of cisplatin
(Fig. 6).
Transfection with siRNA-XPC inhibits
the growth of implanted tumors in nude mice
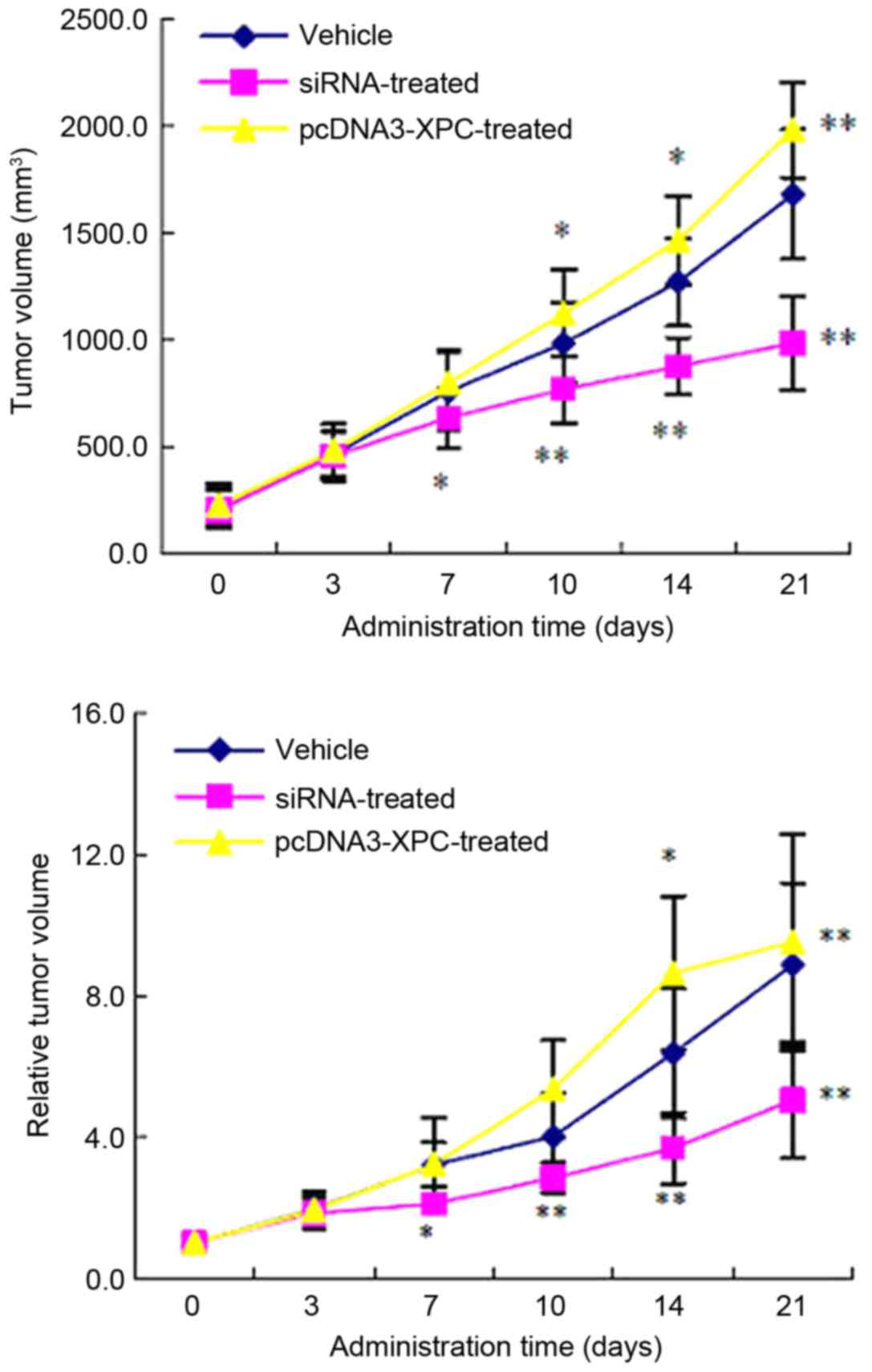
As expected, the growth rate of the implanted tumor
was significantly lower in the group inoculated with siRNA-XPC
SW480 cells compared with that in the control group at 7, 10, 14
and 21 days after inoculation (P<0.05; Fig. 7). However, the growth rate was
significantly faster in the pcDNA3-XPC group compared with that in
the control group 10, 14 and 21 day after the inoculation
(P<0.01; Fig. 7).
Furthermore, the RTV of the nude mice was smaller in
the siRNA-XPC group than that in the control group at 7, 10, 14 and
21 days after inoculation (P<0.05). Compared with the control
group, the RTV of the pcDNA3-XPC group was significantly increased
14 and 21 days after inoculation (P<0.05; Fig. 7).
Transfection with siRNA-XPC
upregulates the level of Bax protein and downregulates that of
Bcl-2 in implanted tumor tissue
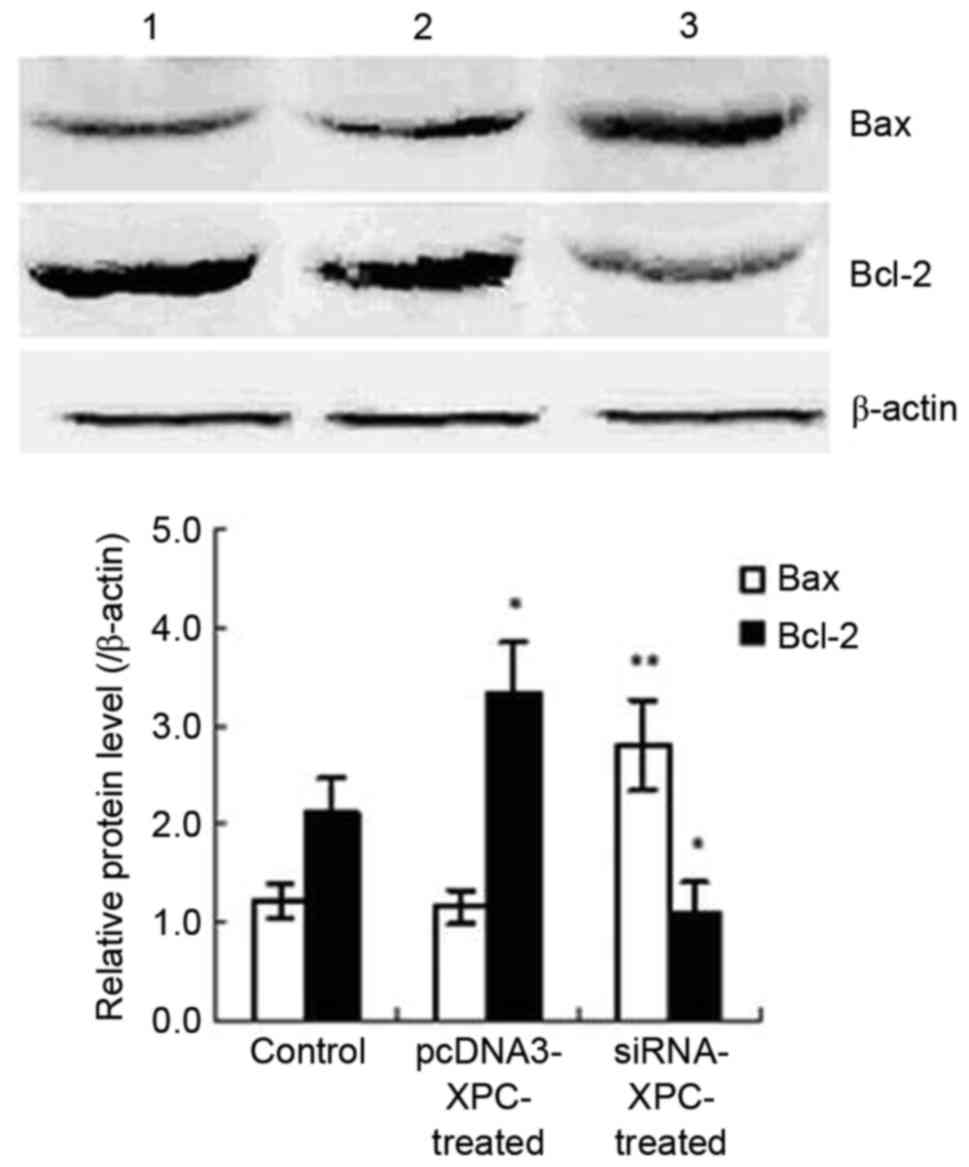
Inoculation with cells transfected with siRNA-XPC
significantly upregulated expression of Bax protein and
downregulated that of Bcl-2 in the implanted tumor tissue (Fig. 8). However, the Bcl-2 protein level was
higher in the group inoculated with cells transfected with
pcDNA3-XPC than that in the control group. These results indicate
that siRNA-XPC significantly altered the expression of
apoptosis-associated genes expressions in vivo, thereby
inhibiting the growth of the implanted tumor (Fig. 8).
Discussion
Generally, abnormal DNA repair is associated with
tumorigenesis and the multi-drug resistance of tumors. NER is a
main mechanism for repairing DNA damage caused by chemotherapeutics
(10,11). Among the large number of
NER-associated proteins, XPC serves an important role in DNA damage
recognition and speed limitation (4,5,21).
Fautrel et al (5) observed that XPC expression in hepatic
carcinoma tissue was significantly higher than that in normal
hepatic tissue. In addition, a high expression of XPC was
associated with decreased chemotherapeutic susceptibility of
hepatic carcinoma (5). Furthermore,
XPC silencing was reported to sensitize glioma cells to arsenic
trioxide via increased oxidative damage (22).
However, it has been confirmed that the incidence of
a variety of tumor types, including CRC, was increased in
XPC-deficient mice (23). Chen et
al (24) found that there was a
direct association between low XPC expression and development of
bladder cancer. Recently, it has been revealed that low XPC
expression and phenotypic variation were involved in the
carcinogenesis of bladder cancer (25). These findings indicated that low XPC
expression was associated with the decreased ability to perform
NER, which serves an important role in carcinogenesis. The
aforementioned studies demonstrate a multiple regulatory role of
XPC in DNA damage in tumors.
H1299, H1355, ovarian cancer cell line 2008, and
MDA-MB-231 provided by Shanghai Institutes for Biological Sciences,
Chinese Academy of Sciences (Shanghai, China) are susceptible to
cisplatin treatment following the knockdown of XPF and ERCC1
(20). Furthermore, the efficacy of
the combined knockdown of XPF and ERCC1 was revealed to be better
than single siRNA (26). When the
XPC-deficient cells were treated with cisplatin, the DNA mutation
frequency was 50 times that observed in normal cells (27). When XPC-deficient cells were treated
with cisplatin, 486 genes in the XPC-deficient cells exhibited
differences in expression at the level of transcription. Notably,
among these genes, 297 were associated with tumorigenesis and DNA
repair (28). This result indicated
that XPC was involved in NER, cell replication and apoptosis. Thus,
the abnormal expression of XPC results in a lack of DNA repair
ability.
Frequently, XPC is a target for the inactivation in
tumors (29). Thus, in the present
study siRNA-XPC was transfected into CRC cells. Assessment of these
transfected cells revealed that their susceptibility to cisplatin
was significantly increased when the XPC gene was silenced. The
proportion of apoptotic XPC-deficient cells was significantly
increased in the presence of cisplatin when compared with the
control. This finding, to a certain degree, agreed with the
hypothesis that XPC overexpression participated in the decreased
susceptibility of CRC to cisplatin.
XPC, located on chromosome 3p25, encoding a
940-amino-acid protein, is involved in DNA damage recognition. XPC
was first documented in the patients with xeroderma pigmentosum
(XP). The incidence of skin cancer in patients with XP was 1,000
times higher than that of normal ones following UV exposure, which
may be due to functional defect of NER (30).
Furthermore, the proportion of apoptotic cells was
significantly decreased when the XPC gene was overexpressed. This
result indicated that the overexpression of XPC attenuated the
sensitivity of the cancer cells to the chemotherapy. However, the
findings of the present study were contradictory to those of Chen
et al (24). In their study,
they found that bladder cancer HT1197 cells expressing low levels
of XPC exhibited a decreased DNA repair capability and were
resistant to cisplatin. However, cisplatin-induced apoptosis
increased when a XPC cDNA-expression vector was stably transfected
into the tumor cells. We hypothesize that this difference may be
due to the difference in the repair ability of the XPC gene in
different cancer cell lines.
The results of the present study indicate that XPC
serves a key role in the chemotherapeutic sensitivity of the CRC
cells to cisplatin. XPC overexpression decreased the sensitivity of
CRC cells to cisplatin. Conversely, transfection with siRNA-XPC
increased the chemotherapeutic sensitivity of these cells, which
was associated with the inhibition of cellular growth and promotion
of apoptosis in these CRC cells.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LX designed all experiments, YZ, JC and YM performed
all experiments, and CQ and FS analyzed the experimental
results.
Ethics approval and consent to
participate
The clinical and experimental studies were approved
by the Ethics Committee of Xinhua Hospital and all patients
provided written informed consent.
Consent for publication
All patients provided written informed consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wood RD, Mitchell M, Sgouros J and Lindahl
T: Human DNA Repair Genes. Science. 291:1284–1289. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fuss JO and Cooper PK: DNA repair: Dynamic
defenders against cancer and aging. PLoS Biol. 4:e203. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Iqbal S, Stoehlmacher J and Lenz HJ:
Tailored chemotherapy for colorectal cancer: A new approach to
therapy. Cancer Invest. 22:762–773. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maltseva EA, Rechkunova NI, Gillet LC,
Petruseva IO, Schärer OD and Lavrik OI: Crosslinking of the NER
damage recognition proteins XPC-HR23B, XPA and RPA to photoreactive
probes that mimic DNA damages. Biochim Biophys Acta. 1770:781–789.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fautrel A, Andrieux L, Musso O, Boudjema
K, Guillouzo A and Langouët S: Overexpression of the two nucleotide
excision repair genes ERCC1 and XPC in human hepatocellular
carcinoma. J Hepatol. 43:288–293. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Okuda M, Nakazawa Y, Guo C, Ogi T and
Nishimura Y: Common TFIIH recruitment mechanism in global genome
and transcription-coupled repair subpathways. Nucleic Acids Res.
45:13043–13055. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spivak G: Nucleotide excision repair in
humans. DNA Repair (Amst). 36:13–18. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hakem R: DNA-damage repair; the good, the
bad, and the ugly. EMBO J. 27:589–605. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee SH: Recognition of DNA damage in
mammals. J Biochem Mol Biol. 34:489–495. 2001.
|
|
10
|
Bartels CL and Lambert MW: Domains in the
XPA protein important in its role as a processivity factor. Biochem
Biophys Res Commun. 356:219–225. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Köberle B, Roginskaya V and Wood RD: XPA
protein as a limiting factor for nucleotide excision repair and UV
sensitivity in human cells. DNA Repair (Amst). 5:641–648. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Worrillow L, Roman E, Adamson PJ, Kane E,
Allan JM and Lightfoot TJ: Polymorphisms in the nucleotide excision
repair gene ERCC2/XPD and risk of non-Hodgkin lymphoma. Cancer
Epidemiol. 33:257–260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zafereo ME, Sturgis EM, Liu Z, Wang LE,
Wei Q and Li G: Nucleotide excision repair core gene polymorphisms
and risk of second primary malignancy in patients with index
squamous cell carcinoma of the head and neck. Carcinogenesis.
30:997–1002. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang WY, Berndt SI, Kang D, Chatterjee N,
Chanock SJ, Yeager M, Welch R, Bresalier RS, Weissfeld JL and Hayes
RB: Nucleotide excision repair gene polymorphisms and risk of
advanced colorectal adenoma: XPC polymorphisms modify
smoking-related risk. Cancer Epidemiol Biomarkers Prev. 15:306–311.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fan W, Zhang HL and Wu XM: Enhancement
effect of nucleotide excision repair gene xeroderma pigmentosun
group an antisense RNA on sensitivity of human lung adenocarcinoma
cell line A549 to cisplatin. Ai Zheng. 24:403–407. 2005.(In
Chinese). PubMed/NCBI
|
|
16
|
Li L, Peterson C and Legerski R: Sequence
of the mouse XPC cDNA and genomic structure of the human XPC gene.
Nucleic Acids Res. 24:1026–1028. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Legerski RJ, Liu P, Li L, Peterson CA,
Zhao Y, Leach RJ, Naylor SL and Siciliano MJ: Assignment of
xeroderma pigmentosum group C (XPC) gene to chromosome 3p25.
Genomics. 21:266–269. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) methods. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sugai T, Habano W, Uesugi N, Jiao YF,
Nakamura S, Sato K, Chiba T and Ishii M: Molecular validation of
the modified Vienna classification of colorectal tumors. J Mol
Diagn. 4:191–200. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maruyama K, Ochiai A, Akimoto S, Nakamura
S, Baba S, Moriya Y and Hirohashi S: Cytoplasmic beta-catenin
accumulation as a predictor of hematogenous metastasis in human
colorectal cancer. Oncology. 59:302–309. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang W: Structure and mechanism for DNA
lesion recognition. Cell Res. 18:184–197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu SY, Wen CY, Lee YJ and Lee TC: XPC
silencing sensitizes glioma cells to arsenic trioxide via increased
oxidative damage. Toxicol Sci. 116:183–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nahari D, McDaniel LD, Task LB, Daniel RL,
Velasco-Miguel S and Friedberg EC: Mutations in Trp53 gene of
UV-irradiated Xpc mutant mice suggest a novel XPC dependent DNA
repair process. DNA Repair. 3:379–386. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Z, Yang J, Wang G, Song B, Li J and
Xu LZ: Attenuated expression of xeroderma pigmentosum group C is
associated with critical events in human bladder cancer
carcinogenesis and progression. Cancer Res. 67:4578–4585. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang J, Xu Z, Li J, Zhang R, Zhang G, Ji
H, Song B and Chen Z: XPC epigenetic silence coupled with p53
alteration has a significant impact on bladder cancer outcome. J
Urol. 184:336–343. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Arora S, Kothandapani A, Tillison K,
Kalman-Maltese V and Patrick SM: Downregulation of XPF-ERCC1
enhances cisplatin efficacy in cancer cells. DNA Repair (Amst).
9:745–753. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Z, Xu XS, Yang J and Wang G: Defining
the function of XPC protein in psoralen and cisplatin-mediated DNA
repair and mutagenesis. Carcinogenesis. 24:1111–1121. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang G, Chuang L, Zhang X, Colton S,
Dombkowski A, Reiners J, Diakiw A and Xu XS: The initiative role of
XPC protein in cisplatin DNA damaging treatment-mediated cell cycle
regulation. Nucleic Acids Res. 32:2231–2240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
De Feraudy S, Ridd K, Richards LM, Kwok
PY, Revet I, Oh D, Feeney L and Cleaver JM: The DNA damage-binding
protein XPC is a frequent target for inactivation in squamous cell
carcinomas. Am J Pathol. 177:555–562. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hanawalt PC, Ford JM and Lloyd DR:
Functional characterization of global genomic DNA repair and its
implications for cancer. Mutat Res. 544:107–114. 2003. View Article : Google Scholar : PubMed/NCBI
|