Introduction
Cervical cancer is the fourth most common cancer in
women worldwide (1–3). Therefore, finding novel potential
targets for diagnosing, prognosing and treating cervical cancer is
important. Potassium (K+) channels are associated with
multiple human diseases, including cardiac arrhythmias, diabetes
and cancer (4,5). Various K+ channels have been
proposed as novel targets for cancer therapy (5). The ATP-sensitive potassium
(KATP) channels are heteromultimers composed of at least
two types of subunit: An inwardly rectifying K+ channel
(Kir6.x, which forms the pore of the channel) and a sulfonylurea
receptor (SUR) (6). Kir6.2 and SUR1
form the characteristic K+ channel in pancreatic
β-cells, thereby coupling the metabolic state of the cell with the
electrical activity (7–9). KATP channels are present in
numerous types of cells and tissues, including pancreatic β-cells
(10), cardiac muscle (11), smooth muscle (12), skeletal muscle and the brain (13). Sulfonylureas inhibit KATP
channels and are used to treat non-insulin dependent diabetes
mellitus (9,14,15). In
addition, KATP channels represent important therapeutic
targets in heart failure (16) and
tissue ischemia (17).
KATP channels are expressed in multiple types of cancer,
including hepatocellular carcinoma (18), human bladder cancer (19), human gastric cancer (20) and glioma (21). Accordingly, KATP channel
openers (minoxidil, cromakalim and pinacidil) increase the
proliferation of HepG2 liver cancer cells, whereas KATP
channel blockers (quinidine and glibenclamide) attenuate cell
proliferation (18). Glibenclamide
inhibits proliferation and induces apoptosis in various types of
cancer, including bladder carcinoma (22), prostate cancer (23) and gastric cancer (20). However, although multiple studies have
described the presence of different K+ channels in
diverse types of human cancer, the association of KATP
channels with cervical cancer remains elusive. Therefore, the
present study assessed whether the Kir6.2, SUR1 and SUR2 subunits
are expressed in cervical cancer cell lines and/or human biopsies,
and the potential associations between subunit expression and tumor
differentiation and invasion were analyzed. The effect of the
KATP channel blocker glibenclamide on the proliferation
and apoptosis of cervical cancer cell lines was also studied.
Materials and methods
Cell culture
Two previously established primary cultures of human
cervical cancer that were characterized as expressing cytokeratins
and the E7 gene from human papilloma virus 16 by
immunocytochemistry and PCR, respectively, were used in the current
study and cultured as previously described (24). The human cervical cancer cell lines
C33A, CaSki, HeLa, INBL and SiHa, and primary epidermal
keratinocytes (normal human neonatal foreskin) were obtained from
the American Type Culture Collection (Manassas, VA, USA) and
cultured according to the supplier's instructions.
Cell viability assays
Cell viability was measured by MTT assay. Cells were
seeded at a density of 3.5×103 cells into 96-well
plates. After 24 h of incubation at 37°C, the cells were cultured
with Dulbecco's modified Eagle's medium for cervical cancer cells
(cat. no. 12430054; Gibco; Thermo Fisher Scientific, Inc., Waltham
MA, USA) and Keratinocyte-SFM (cat. no. 17005042; Gibco; Thermo
Fisher Scientific, Inc.) for epidermal keratinocytes)) and various
concentrations (40–200 µM) of glibenclamide (cat. no. G0639-5G;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for different
incubation times (24–72 h). Subsequently, MTT tetrazolium salt (0.5
mg/ml) was added to each well and cells were incubated for a
further 4 h. Formazan crystals were then dissolved using SDS, and
the absorbance was measured using a Multiskan FC microplate
photometer (Thermo Fisher Scientific, Inc.).
Apoptosis assay
HeLa and primary epidermal keratinocytes cells were
incubated for 48 h at 37°C in culture medium as previously
described, either with or without glibenclamide (100 or 150 µM) or
vehicle (sterile water). Camptothecin (apoptosis inductor, 10
mg/ml) and methanol (necrosis inductor, 39.6 mg/ml) were used as
positive controls. Apoptosis was determined using the Annexin
V-FITC kit (Invitrogen; Thermo Fisher Scientific, Inc.) to measure
binding to phosphatidylserine and DNA staining by propidium iodide
(1 mg/ml), according to manufacturer's protocol. The experiments
were performed using the CyAn ADP flow cytometer (Dako; Agilent
Technologies, Inc., Santa Clara, CA, USA). Quadrant analysis was
performed using the Summit 4.3 software (Beckman Coulter, Inc.,
Fullerton, CA).
Human biopsies
Human cervical cancer biopsies were obtained from
the Instituto Nacional de Cancerología (Mexico City, Mexico). The
protocol was approved by the corresponding Research and Ethics
Committees [protocol no. 013/024/GII (CEI/846)] and written
informed consent was obtained from all the patients. A total of 74
biopsies were studied and the age of the patients ranged from 25 to
82 years old. The present study only included samples from patients
that had not received any anti-cancer therapy and excluded patients
that had vaginal infections. Non-cancerous cervical tissues were
obtained from hysterectomy patients with benign gynecologic
pathology at the Hospital General ‘Dr Manuel Gea Gonzalez’ (Mexico
City, Mexico). The protocol followed the local ethical
considerations (protocol no. 11-84-2013) and was approved by the
corresponding ethics committee. The non-cancerous cervical tissue
group included patients whose cervixes were described as healthy,
but excluded patients taking hormones. The tissues were either
collected in TRIzol and placed in liquid nitrogen to obtain mRNA,
or fixed in formaldehyde (10% w/v) and embedded in paraffin for
subsequent immunohistochemical analysis. The samples were
classified as adenocarcinoma (AC) or squamous cell carcinoma (SCC),
according to the International Federation of Gynecology and
Obstetrics (FIGO) staging system (25). A total of 24 cancer biopsies were
analyzed for Kir6.2 mRNA expression, and 30 tumor tissues were
analyzed for Kir6.2 and SUR protein expression.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was isolated using TRIzol reagent (cat. no.
T9424; Sigma-Aldrich; Merck KGaA). The total RNA (5 µg) was treated
with DNase I (Roche Diagnostics GmbH, Mannheim, Germany;
04716728001) and reverse transcribed using the M-MuLV reverse
transcriptase (New England BioLabs, Inc., Ipswich, MA, USA). The
temperature and time protocol was as follows: 65°C for 5 min, 37°C
for 50 min, 70°C for 15 min and 4°C for 60 min. When small biopsies
were obtained, the total RNA (518 ng) was processed using the
Path-ID Multiplex One-Step RT-PCR kit (cat. no. 4388641; Ambion;
Thermo Fisher Scientific, Inc.). The expression of Kir6.2
(Hs00265026_s1; Applied Biosystems; Thermo Fisher Scientific, Inc.)
was assessed by RT-qPCR using the TaqMan™ Master-mix based
detection system (cat. no. 4304437; Applied Biosystems; Thermo
Fisher Scientific, Inc.). The temperature and cycling protocol were
as follows: 95°C for 10 min, 95°C for 10 sec and 60°C for 1 min.
The first two steps were repeated for 40 cycles. Expression of a
housekeeping gene, hypoxanthine-guanine phosphoribosyltransferase
(cat. no. 4326321E; Applied Biosystems; Thermo Fisher Scientific,
Inc.), was used as an internal control. Relative expression levels
of the gene of interest were determined by the 2−∆∆Cq
method according to the threshold cycle (26), and 2–3 experimental replicates were
performed.
Immunostaining
For immunohistochemistry, serial sections (5 µm)
were mounted on charged glass slides and deparaffinized. Antigen
retrieval and blocking were performed as previously described
(24). The slides were then incubated
for 2 h with the primary antibodies, anti-Kir6.2 (1:100; cat. no.
NBP1-00900), anti-SUR1 (1:50; cat. no. NBP1-59778) or anti-SUR2
(1:300; cat. no. NBP1-84436; all from Novus Biologicals, LLC,
Littleton, CO, USA), at 4°C. The antibodies were diluted in
ImmunoDetector Protein Blocker/Antibody Diluent (cat. no. 0041; Bio
SB, Santa Barbara, CA, USA). Subsequently, the slides were
incubated at room temperature with a biotin-conjugated secondary
antibody for 15 min, followed by 15 min with horseradish peroxidase
(HRP)-conjugated streptavidin polymer (both provided as part of the
Mouse/Rabbit ImmunoDetector DAB HRP Brown kit; cat. no. 0005; Bio
SB). The staining reaction was completed in the presence of
diaminobenzidine in a buffer reaction solution (#0005; Bio SB) and
washed with distilled water. The slides were counterstained with
hematoxylin (Sigma-Aldrich; Merck KGaA) and rinsed with water and
ethanol as previously described (24). The slides were mounted with resin (EMD
Millipore, Billerica, MA, USA) and observed using an Olympus IX51
light microscope and an Olympus DP70 camera (Olympus Corporation,
Tokyo, Japan). Brown immunostaining indicated Kir6.2/SUR1/SUR2
expression.
For immunocytochemistry, cervical cancer cells were
mounted on glass coverslips and fixed in ethanol. Subsequently,
staining was performed as per the protocol previously described for
the tissue samples.
Statistical analysis
Data are presented as the mean ± SD from three
independent experiments with 6–8 replicates for cell proliferation
assays and 2–3 replicates for PCR studies. Data were analyzed by a
Student's t-test or by an analysis of variance followed by a
Tukey-Kramer post hoc test using the GraphPad Prism software
version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
Differential expression of Kir6.2 mRNA
and protein in cervical cancer cell lines
Kir6.2 subunit expression was assessed in two
primary cultures of cervical cancer (PC1 and PC2) and five human
cervical cancer cell lines (C33A, CaSki, HeLa, INBL and SiHa). The
expression of Kir6.2 subunit mRNA was evaluated by RT-qPCR and
normalized relative to KATP channel expression in normal
primary epidermal keratinocytes. KATP channel expression
was present in each of the cell lines and primary cultures
(Fig. 1A). However, the only
statistically significant difference (P<0.05) compared with
normal keratinocytes was KATP channel overexpression in
HeLa cells. Therefore, HeLa cells were used in further analyses of
KATP channel protein expression. Fig. 1B depicts the protein expression of the
Kir6.2 and SUR2 subunits in HeLa cells. Human cerebral cortex and
testis samples were used as positive controls for Kir6.2 and SUR2
protein expression, respectively (data not shown). SUR1 protein
expression was not detected (data not shown), whereas the SUR2
subunit was positively stained. These results suggested that HeLa
cells express KATP channels composed of Kir6.2 and SUR2
subunits, and that these channels may serve a function in cervical
carcinogenesis.
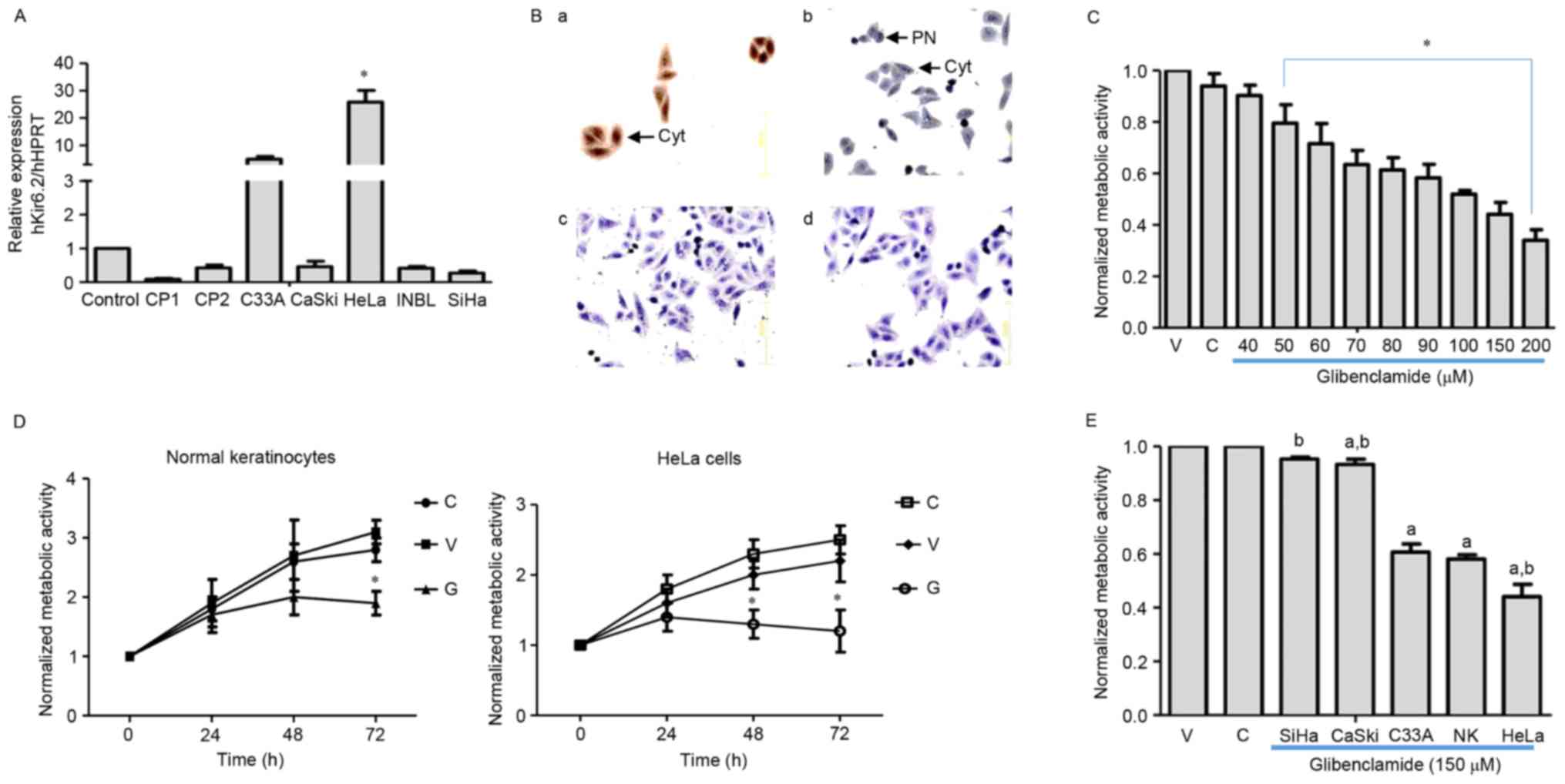 | Figure 1.Expression of KATP in
cultured cervical cancer cells. (A) Relative mRNA expression of the
Kir6.2 subunit in normal keratinocytes (control), primary cultures
(CP1 and CP2) and cervical cancer cell lines (CaSki, HeLa, INBL and
SiHa). Three different cultures were studied for each cell type.
Data are presented as the mean ± standard deviation (*P<0.05 vs.
control). (B) Protein expression of KATP subunits in
HeLa cells. Brown immunostaining reveals the presence of the (a)
Kir6.2 and (b) SUR2 subunits. Kir6.2 was predominantly localized in
the cytoplasm (arrow) while SUR2 was detected in the PN (arrow). No
immunostaining was observed in the absence of the primary antibody
for either Kir6.2 (c) or SUR2 (d) subunits (magnification, ×200).
(C-E) Cells were cultured in medium alone (‘C’), with sterile water
(‘V’) or with glibenclamide (‘G’). Following 48 h of treatment with
glibenclamide, HeLa cell viability decreased (C)
concentration-dependently and (D) time-dependently at 150 µM
glibenclamide. The inhibitory effect was increased in HeLa cells
compared with normal keratinocytes (D). Data are presented as the
mean ± SEM from three independent experiments (*P<0.05 vs.
vehicle). (E) The effect of glibenclamide (150 µM, 48 h) was
increased in cell lines exhibiting increased Kir6.2 mRNA
expression. Data are presented as the mean ± SEM from four
independent experiments (P<0.05 vs. avehicle or
bnormal keratinocytes). HPRT, hypoxanthine-guanine
phosphoribosyltransferase; NK, normal keratinocytes;
KATP, ATP-sensitive K-channels; cyt, cytoplasm; PN,
perinuclear space; SUR, sulfonylurea receptor; SEM, standard error
of the mean; V, vehicle. |
Glibenclamide decreases cervical
cancer and normal keratinocyte cell proliferation
The effect of glibenclamide, a KATP
channel inhibitor, on HeLa cell proliferation was investigated by
assaying the metabolic activity. The HeLa cells were incubated with
various glibenclamide concentrations (40–200 µM) for 48 h, which
resulted in a concentration- and time-dependent decrease in cell
viability of >60% at the highest concentration compared with the
vehicle-treated cells (Fig. 1C and
D). By contrast, glibenclamide (150 µM) decreased the
proliferation of normal keratinocytes by 30% (Fig. 1D). The present study also studied the
effect of glibenclamide on other cervical cancer cell lines that
displayed varying mRNA expression levels of the Kir6.2 subunit. The
results revealed that as the Kir6.2 mRNA expression levels
increased, the inhibitory effect of glibenclamide also increased
(Fig. 1E). Glibenclamide did not
affect apoptosis (data not shown).
KATP channel mRNA
expression in human biopsies and its potential association with
tumor differentiation and invasion
Kir6.2 subunit mRNA expression was assessed in 24
cervical cancer samples (22 SCC and 2 AC biopsies). The samples
were classified as cervical cancer of FIGO stage I (4 samples from
stage IA and 2 samples from stage IB), II (1 sample from stage IIA
and 5 samples from stage IIB), III (7 samples from stage IIIB) or
IV (3 samples from stage IVB), and included 1 AC in situ and
1 invasive AC. The mRNA expression of 10 non-cancerous cervical
biopsies (controls) from other patients was normalized to 1 and
used for comparison. Increased Kir6.2 mRNA expression was detected
in the stage IV SCC biopsies (Fig.
2A), with some of these samples exhibiting mRNA levels ≤60×
those of the controls. Differential Kir6.2 mRNA expression was also
observed in invasive tumors; compared with non-invasive tumors or
non-cancerous biopsies (control group; expression value normalized
to 1), some of the invasive tumors displayed >50× the Kir6.2
mRNA expression (Fig. 2B).
Non-invasive tumors were considered to be those where the invasive
component was delimited to microscopic foci of only a few microns
in length and depth, as observed in in situ carcinoma
biopsies (microinvasive tumors, 4 samples from cervical cancer FIGO
stage IAI and 1 AC in situ). The samples were also divided
according to their differentiation grade into well, moderately or
poorly differentiated groups. Increased Kir6.2 subunit expression
was observed in samples from the moderate and poor differentiation
grades and certain biopsies from these groups displayed ≤50× Kir6.2
mRNA levels compared with the controls (Fig. 2C). The present study also analyzed the
potential association between Kir6.2 and lymphovascular space
invasion. In total, 7 samples exhibited lymphovascular space
invasion and 100% of these samples displayed increased Kir6.2 mRNA
expression. However, 13/23 (56.6%) samples exhibiting no
lymphovascular space invasion also displayed increased Kir6.2 mRNA
expression. No significant association was identified between
lymphovascular invasion and mRNA channel expression.
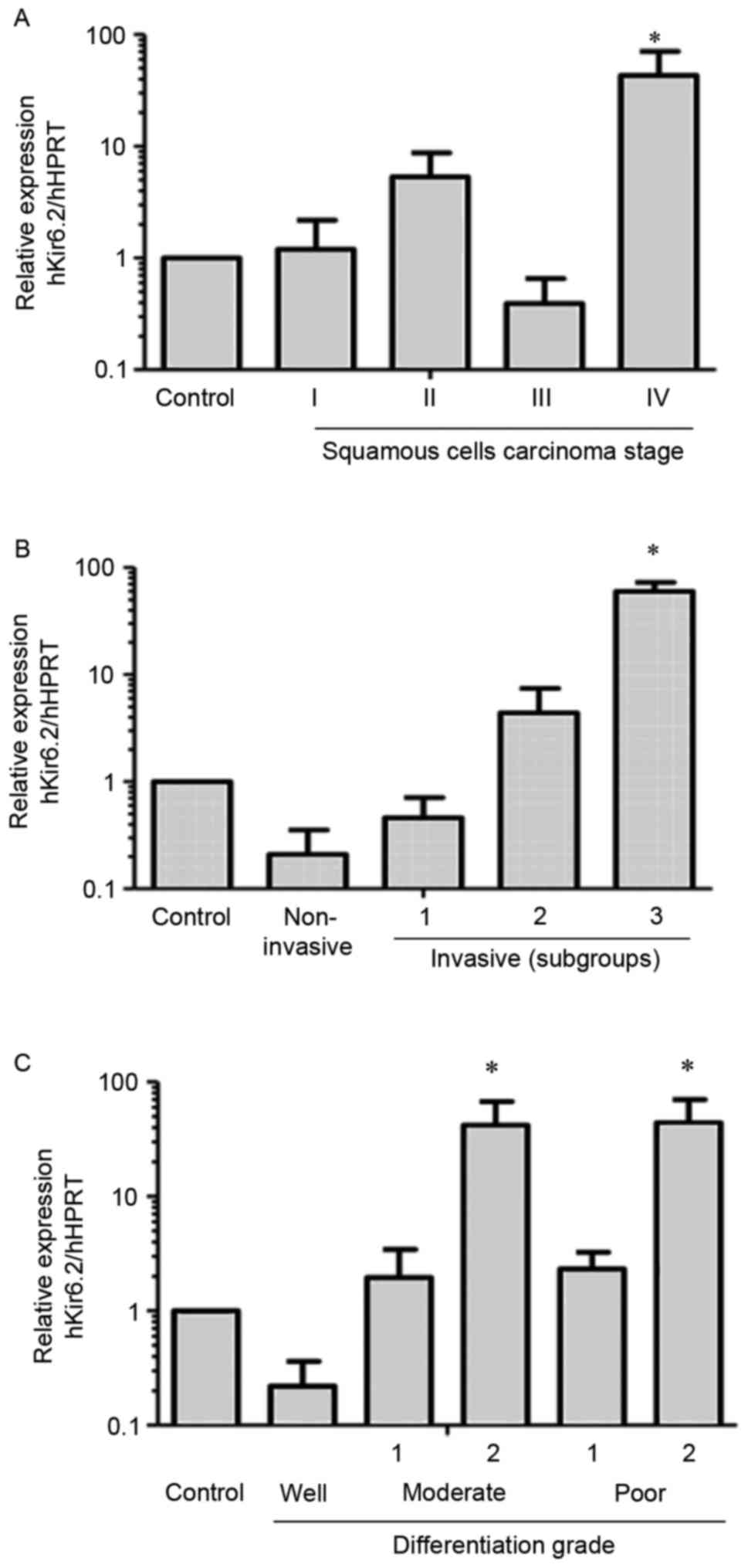 | Figure 2.Kir6.2 mRNA expression in human
cervical biopsies. The values were normalized relative to the
Kir6.2 expression of 10 non-cancerous cervical tissue samples
(control). (A) Kir6.2 subunit mRNA expression in samples from
different clinical stages of squamous cell carcinoma: Stage I
(n=6), stage II (n=6), stage III (n=7) and stage IV (n=3). Samples
from stage IV exhibited increased expression. *P<0.05 vs.
control. (B) Samples were classified as non-invasive (n=5),
invasive 1 (n=7), invasive 2 (n=12) and invasive 3 (n=4). The
relative mRNA expression of the Kir6.2 subunit was increased in
multiple invasive tissues. *P<0.05 vs. control and non-invasive
groups. (C) The biopsies were grouped according to their
differentiation grade into well, moderately or poorly
differentiated, and the moderate and poor groups were each divided
into two subgroups based on their relative expression patterns:
Well (n=4); moderate-1 (n=13); moderate-2 (n=3); poor-1 (n=4);
poor-2 (n=3) *P<0.05 vs. control, well, moderate-1 and poor-1.
HPRT, hypoxanthine-guanine phosphoribosyltransferase; SD, standard
deviation. |
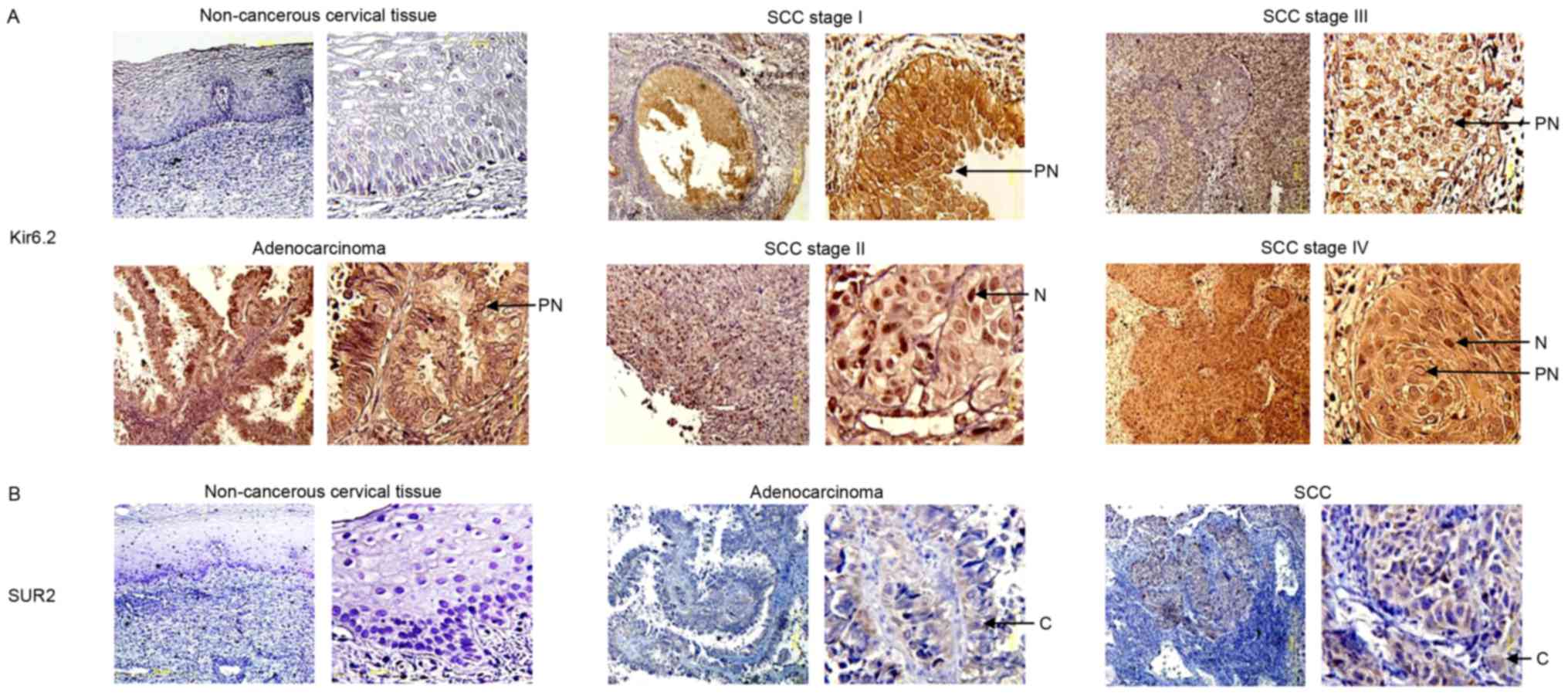
Kir6.2 and SUR protein expression in
human cervical tissues
The present study determined the protein expression
levels of the Kir6.2 and SUR2 subunits in human cervical biopsies
by performing immunohistochemical analysis on 30 cervical cancer
tissues (24 SCCs and 6 ACs) and 10 non-cancerous cervical tissues
(controls). All of these samples were obtained from different
patients from those that participated in the mRNA studies. Kir6.2
subunit immunostaining was not observed in the majority (9/10) of
the non-cancerous cervical tissues (Fig.
3A), while 1 sample exhibited a weak signal. Similarly, SUR2
expression was not observed in any of these non-cancerous cervical
tissues (Fig. 3B). By contrast, 10/30
cervical cancer samples revealed strong Kir6.2 staining (Fig. 3A). The positive biopsies corresponded
to samples classified as cervical cancer FIGO stage I (1 sample
from stage IA and 1 sample from stage IB), stage II (2 samples from
stage IIB), III (2 samples from stage IIIB) and stage IV (1 sample
from stage IVA and 2 samples from stage IVB), and 1 AC from stage
IB. Stromal cells in the tumor tissues also exhibited Kir6.2
staining. SUR2 subunit expression was detected in 4 cancer tissues
(Fig. 3B). Two samples in the
immunochemical analysis were obtained from patients with
lymphovascular space invasion, both of which displayed increased
Kir6.2 protein expression in comparison with controls (data not
shown).
Discussion
The present study demonstrated, for the first time
to the best of our knowledge, the presence of Kir6.2 subunit
expression in cervical cancer cell lines and human biopsies. The
differential Kir6.2 subunit expression observed in cervical cancer
cell lines and human cervical biopsies reflect the heterogeneity
among cervical tumors. The KATP channel blocker
glibenclamide decreased cell proliferation in HeLa cells more than
that in the control cells (normal keratinocytes). Although the
inhibitory function of glibenclamide may be explained by its
ability to block KATP channels (9) in cervical cancer cells, it cannot be
ruled out that other proteins may also be involved. For example,
glibenclamide also inhibits other ATP-binding cassette transporters
(27–36). Peng et al (37) suggested that increased cystic fibrosis
transmembrane conductance regulator expression is associated with
cervical cancer progression and a poor prognosis. Since
glibenclamide did not induce apoptosis, its anti-proliferative
effects may be associated with cell cycle arrest in the G1-S phase,
as reported for human bladder (HTB-9) and breast (MDA-MB-231)
carcinoma cells (22–38). While further studies are required to
reveal the molecular mechanism of the anti-proliferative effects of
glibenclamide, the results of the present study suggested that this
drug could represent a potential therapy for patients with cervical
cancer overexpressing KATP channels. Cervical cancer
incidence in diabetic patients taking glibenclamide requires
further investigation. A limitation of the present study was that
the presence of diabetes mellitus was not an exclusion
criterion.
Human cervical biopsies also displayed differential
expression of Kir6.2 subunit mRNA. Increased mRNA levels were
observed in samples from patients with stage IV SCC in comparison
with control and low stages samples. Increased mRNA expression was
also identified in certain invasive or poorly differentiated
carcinoma tissues in comparison with non-invasive or
well-differentiated samples. Strong Kir6.2 protein expression was
observed in 33% of the cancer samples, while the non-cancerous
cervical tissues displayed weak immunostaining in only 1/10 samples
and no expression of the SUR2 subunit. While Kir6.2 protein
expression was observed in 10/30 cancer tissues, SUR2 subunit
expression was detected in 4 cancer tissues. This could be because
the SUR2 gene encodes different isoforms and splice variants
(7,9),
some of which may not be recognized by the antibody used. Although
the present study did not find an association between Kir6.2
expression and lymphovascular space invasion, all of the samples
exhibiting lymphovascular space invasion displayed increased Kir6.2
mRNA and/or protein expression in comparison with control
samples.
Various K+ channels have been
demonstrated to contribute to cancer cell invasion and metastasis
in other types of cancer, including Kv1.3 in melanoma (39–52).
Further studies are required to elucidate the function of
KATP channels in cervical cancer progression and
malignancy and the potential associated mechanism. In addition,
future clinical studies should aim to increase the number of human
biopsies analyzed. A potential limitation of the present study is
that hysterectomy samples may undergo hypoxia due to the ligation
of the descending uterine vessels during surgery. In addition,
although the control samples were reported as healthy, future
studies should aim to include normal cervical biopsies obtained
without hysterectomy.
Further studies of the association between Kir6.2
expression and overall survival in patients with cervical cancer
are required. In addition, the effect of glibenclamide in in
vivo cervical cancer models and in patient-derived xenografts
should also be investigated. The present study observed increased
KATP channel expression in invasive cervical cancer
samples and in those that were poorly differentiated and of an
advanced stage in comparison with the control and some samples from
the non-invasive or low-stage groups. Furthermore, the present
study revealed a potential function for these channels in cervical
cancer cell proliferation. The results of the present study suggest
that KATP channels may represent potential tools for
cervical cancer diagnosis and therapy. Since glibenclamide
inhibited the proliferation of cervical cancer cells displaying
increased Kir6.2 mRNA expression, KATP channels may be a
novel therapeutic target in patients with cervical cancer
overexpressing this protein.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moore DH: Cervical cancer. Obstet Gynecol.
107:1152–1161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cadron I, Van Gorp T, Amant F, Leunen K,
Neven P and Vergote I: Chemotherapy for recurrent cervical cancer.
Gynecol Oncol. 107 1 Suppl 1:S113–S118. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dubois JM and Rouzaire-Dubois B: Role of
potassium channels in mitogenesis. Prog Biophys Mol Biol. 59:1–21.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pardo LA and Stühmer W: The roles of K(+)
channels in cancer. Nat Rev Cancer. 14:39–48. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clement JP IV, Kunjilwar K, Gonzalez G,
Schwanstecher M, Panten U, Aguilar-Bryan L and Bryan J: Association
and stoichiometry of K(ATP) channel subunits. Neuron. 18:827–838.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Babenko AP, Aguilar-Bryan L and Bryan J: A
view of sur/KIR6.X, KATP channels. Annu Rev Physiol. 60:667–687.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inagaki N, Gonoi T, Clement JP, Wang CZ,
Aguilar-Bryan L, Bryan J and Seino S: A family of sulfonylurea
receptors determines the pharmacological properties of
ATP-sensitive K+ channels. Neuron. 16:1011–1017. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aguilar-Bryan L and Bryan J: Molecular
biology of adenosine triphosphate-sensitive potassium channels.
Endocr Rev. 20:101–135. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aguilar-Bryan L, Nichols CG, Wechsler SW,
Clement JP IV, Boyd AE III, González G, Herrera-Sosa H, Nguy K,
Bryan J and Nelson DA: Cloning of the beta cell high-affinity
sulfonylurea receptor: A regulator of insulin secretion. Science.
268:423–426. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Noma A: ATP-regulated K+
channels in cardiac muscle. Nature. 305:147–148. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Quayle JM, Nelson MT and Standen NB:
ATP-sensitive and inwardly rectifying potassium channels in smooth
muscle. Physiol Rev. 77:1165–1232. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sakura H, Ammälä C, Smith PA, Gribble FM
and Ashcroft FM: Cloning and functional expression of the cDNA
encoding a novel ATP-sensitive potassium channel subunit expressed
in pancreatic beta-cells, brain, heart and skeletal muscle. FEBS
Lett. 377:338–344. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Trube G, Rorsman P and Ohno-Shosaku T:
Opposite effects of tolbutamide and diazoxide on the ATP-dependent
K+ channel in mouse pancreatic beta-cells. Pflugers
Arch. 407:493–499. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sturgess NC, Ashford ML, Cook DL and Hales
CN: The sulphonylurea receptor may be an ATP-sensitive potassium
channel. Lancet. 2:474–475. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kasikcioglu HA and Cam N: A review of
levosimendan in the treatment of heart failure. Vasc Health Risk
Manag. 2:389–400. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Downey JM and Cohen MV: Do mitochondrial
KATP channels serve as triggers rather than end-effectors of
ischemic preconditioning's protection? Basic Res Cardiol.
95:272–274. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Malhi H, Irani AN, Rajvanshi P, Suadicani
SO, Spray DC, McDonald TV and Gupta S: KATP channels regulate
mitogenically induced proliferation in primary rat hepatocytes and
human liver cell lines. Implications for liver growth control and
potential therapeutic targeting. J Biol Chem. 275:26050–26057.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Monen SH, Schmidt PH and Wondergem R:
Membrane potassium channels and human bladder tumor cells. I.
Electrical properties. J Membr Biol. 161:247–256. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qian X, Li J, Ding J, Wang Z, Duan L and
Hu G: Glibenclamide exerts an antitumor activity through reactive
oxygen species-c-jun NH2-terminal kinase pathway in human gastric
cancer cell line MGC-803. Biochem Pharmacol. 76:1705–1715. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang L, Li B, Li W, Guo H and Zou F:
ATP-sensitive potassium channels control glioma cells proliferation
by regulating ERK activity. Carcinogenesis. 30:737–744. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wondergem R, Cregan M, Strickler L, Miller
R and Suttles J: Membrane potassium channels and human bladder
tumor cells: II. Growth properties. J Membr Biol. 161:257–262.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abdul M and Hoosein N: Expression and
activity of potassium ion channels in human prostate cancer. Cancer
Lett. 186:99–105. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Farias LM, Ocaña DB, Díaz L, Larrea F,
Avila-Chávez E, Cadena A, Hinojosa LM, Lara G, Villanueva LA,
Vargas C, et al: Ether a go-go potassium channels as human cervical
cancer markers. Cancer Res. 64:6996–7001. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pecorelli S: Revised FIGO staging for
carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol
Obstet. 105:103–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Payen L, Delugin L, Courtois A, Trinquart
Y, Guillouzo A and Fardel O: The sulphonylurea glibenclamide
inhibits multidrug resistance protein (MRP1) activity in human lung
cancer cells. Br J Pharmacol. 132:778–784. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Borst P, Evers R, Kool M and Wijnholds J:
The multidrug resistance protein family. Biochim Biophys Acta.
1461:347–357. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lautier D, Canitrot Y, Deeley RG and Cole
SP: Multidrug resistance mediated by the multidrug resistance
protein (MRP) gene. Biochem Pharmacol. 52:967–977. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sheppard DN and Robinson KA: Mechanism of
glibenclamide inhibition of cystic fibrosis transmembrane
conductance regulator Cl- channels expressed in a murine cell line.
J Physiol. 503:333–346. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim JA, Kang YS, Lee SH, Lee EH, Yoo BH
and Lee YS: Glibenclamide induces apoptosis through inhibition of
cystic fibrosis transmembrane conductance regulator (CFTR) Cl(-)
channels and intracellular Ca(2+) release in HepG2 human
hepatoblastoma cells. Biochem Biophys Res Commun. 261:682–688.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sheppard DN and Welsh MJ: Effect of
ATP-sensitive K+ channel regulators on cystic fibrosis
transmembrane conductance regulator chloride currents. J Gen
Physiol. 100:573–591. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gerloff T, Stieger B, Hagenbuch B, Madon
J, Landmann L, Roth J, Hofmann AF and Meier PJ: The sister of
P-glycoprotein represents the canalicular bile salt export pump of
mammalian liver. J Biol Chem. 273:10046–10050. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Stieger B, Fattinger K, Madon J,
Kullak-Ublick GA and Meier PJ: Drug- and estrogen-induced
cholestasis through inhibition of the hepatocellular bile salt
export pump (Bsep) of rat liver. Gastroenterology. 118:422–430.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ambudkar SV, Dey S, Hrycyna CA,
Ramachandra M, Pastan I and Gottesman MM: Biochemical, cellular,
and pharmacological aspects of the multidrug transporter. Annu Rev
Pharmacol Toxicol. 39:361–398. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Golstein PE, Boom A, van Geffel J, Jacobs
P, Masereel B and Beauwens R: P-glycoprotein inhibition by
glibenclamide and related compounds. Pflugers Arch. 437:652–660.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Peng X, Wu Z, Yu L, Li J, Xu W, Chan HC,
Zhang Y and Hu L: Overexpression of cystic fibrosis transmembrane
conductance regulator (CFTR) is associated with human cervical
cancer malignancy, progression and prognosis. Gynecol Oncol.
125:470–476. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Núñez M, Medina V, Cricco G, Croci M,
Cocca C, Rivera E, Bergoc R and Martín G: Glibenclamide inhibits
cell growth by inducing G0/G1 arrest in the human breast cancer
cell line MDA-MB-231. BMC Pharmacol Toxicol. 14:62013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Artym VV and Petty HR: Molecular proximity
of Kv1.3 voltage-gated potassium channels and beta(1)-integrins on
the plasma membrane of melanoma cells: Effects of cell adherence
and channel blockers. J Gen Physiol. 120:29–37. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Agarwal JR, Griesinger F, Stühmer W and
Pardo LA: The potassium channel Ether à go-go is a novel prognostic
factor with functional relevance in acute myeloid leukemia. Mol
Cancer. 9:182010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Afrasiabi E, Hietamäki M, Viitanen T,
Sukumaran P, Bergelin N and Törnquist K: Expression and
significance of HERG (KCNH2) potassium channels in the regulation
of MDA-MB-435S melanoma cell proliferation and migration. Cell
Signal. 22:57–64. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pillozzi S, Brizzi MF, Bernabei PA,
Bartolozzi B, Caporale R, Basile V, Boddi V, Pegoraro L, Becchetti
A and Arcangeli A: VEGFR-1 (FLT-1), beta1 integrin, and hERG
K+ channel for a macromolecular signaling complex in
acute myeloid leukemia: Role in cell migration and clinical
outcome. Blood. 110:1238–1250. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cherubini A, Hofmann G, Pillozzi S, Guasti
L, Crociani O, Cilia E, Di Stefano P, Degani S, Balzi M, Olivotto
M, et al: Human ether-a-go-go-related gene 1 channels are
physically linked to beta1 integrins and modulate
adhesion-dependent signaling. Mol Biol Cell. 16:2972–2983. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lastraioli E, Guasti L, Crociani O,
Polvani S, Hofmann G, Witchel H, Bencini L, Calistri M, Messerini
L, Scatizzi M, et al: herg1 gene and HERG1 protein are
overexpressed in colorectal cancers and regulate cell invasion of
tumor cells. Cancer Res. 64:606–611. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Guo Z, Liu J, Zhang L, Su B, Xing Y, He Q,
Ci W, Li X and Zhou L: KCNJ1 inhibits tumor proliferation and
metastasis and is a prognostic factor in clear cell renal cell
carcinoma. Tumour Biol. 36:1251–1259. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Veeravalli KK, Ponnala S, Chetty C, Tsung
AJ, Gujrati M and Rao JS: Integrin α9β1-mediated cell migration in
glioblastoma via SSAT and Kir4.2 potassium channel pathway. Cell
Signal. 24:272–281. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kraft R, Krause P, Jung S, Basrai D,
Liebmann L, Bolz J and Patt S: BK channel openers inhibit migration
of human glioma cells. Pflugers Arch. 446:248–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Weaver AK, Bomben VC and Sontheimer H:
Expression and function of calcium-activated potassium channels in
human glioma cells. Glia. 54:223–233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sciaccaluga M, Fioretti B, Catacuzzeno L,
Pagani F, Bertollini C, Rosito M, Catalano M, D'Alessandro G,
Santoro A, Cantore G, et al: CXCL12-induced glioblastoma cell
migration requires intermediate conductance
Ca2+-activated K+ channel activity. Am J
Physiol Cell Physiol. 299:C175–C184. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cuddapah VA, Turner KL, Seifert S and
Sontheimer H: Bradykinin-induced chemotaxis of human gliomas
requires the activation of KCa3.1 and ClC-3. J Neurosci.
33:1427–1440. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chantôme A, Potier-Cartereau M, Clarysse
L, Fromont G, Marionneau-Lambot S, Guéguinou M, Pagès JC, Collin C,
Oullier T, Girault A, et al: Pivotal role of the lipid Raft
SK3-Orai1 complex in human cancer cell migration and bone
metastases. Cancer Res. 73:4852–4861. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu X and Chang Y, Reinhart PH, Sontheimer
H and Chang Y: Cloning and characterization of glioma BK, a Novel
BK Channel isoform highly expressed in human glioma cells. J
Neurosci. 22:1840–1849. 2002. View Article : Google Scholar : PubMed/NCBI
|