Introduction
Colorectal cancer (CRC) is a major global public
health concern (1). CRC is the second
leading cause of cancer-associated mortality in developed
countries, and the sixth to seventh leading cause of
cancer-associated mortality in developing countries (2). Comprehensive studies investigating CRC
tumorigenesis have made significant progress in previous decades,
elucidating the molecular mechanisms governing CRC initiation,
development and progression (3).
CRC arises through at least two distinct genetic
pathways. One of these pathways involves chromosomal instability
(CIN), and the other involves microsatellite instability (MSI)
(4). Although the majority of
sporadic CRCs exhibit CIN, MSI is only observed in ~15% of all
CRCs, and the majority of CRCs with a high frequency of MSI are
sporadic (5).
MSI indicates a defective DNA mismatch repair (MMR)
system, which is why it is used as a molecular marker. MMR is one
of the best understood molecular pathways involved in the
pathogenesis of inherited and sporadic cancer. The MMR system
serves a function in DNA homeostasis, and is involved in the repair
of specific types of errors that occur during DNA replication in
dividing somatic cells (6). MSI has
been defined as variable microsatellite sequence length caused by
insertions or deletions that occur within a tumor, but not in the
corresponding normal tissue, and MSI tumors are characterized by an
accelerated accumulation of mutations due to a defective MMR system
(7).
When MMR is functioning normally, MuS homolog 2
(MSH2) and MutS homolog 6 (MSH6) bind to form the hMutS-α
heterodimer, which recognizes and binds to mismatched base pairs,
and recruits the MutL homolog 1 (MLH1)/PMS1 homolog 2 (PMS2)
hMutL-α heterodimer to repair them. MSH2 and MLH1 are required to
stabilize MSH6 and PMS2, respectively, and the loss of either MSH2
or MLH1 results in the degradation of its binding partner (8).
MMR-deficient tumors are typically identified via
immunohistochemistry (IHC), which detects the loss of expression of
one or more MMR proteins (including MLH1, MSH2, MSH6 and PMS2) and
MSI testing to identify tumors with elevated MSI (MSI-H) (9). Reduced DNA repair protein expression is
associated with MSI; therefore, numerous laboratories routinely use
IHC with a panel of antibodies (specific to MLH1, PMS2, MSH2 and
MSH6) as surrogate MSI markers (10).
IHC has the advantage of being flexible, with the ability to target
any tissue type (including frozen and fixed samples, irrespective
of the fixative used) and is performed by clinical pathology
laboratories as a routine diagnostic test for MSI in patient
tissues (11). Therefore, IHC
detection of MMR proteins may be a faster, easier and cheaper
method of CRC detection when compared with genetic analysis for
MSI.
In the present study, IHC analysis of MLH1, MSH2,
MSH6 and PMS2 protein expression was performed to evaluate the
prognostic significance of MMR status in patients with a series of
stage II and III sporadic CRC.
Materials and methods
Patients and tissues
Tissue samples from 153 patients with primary
sporadic colorectal adenocarcinoma who underwent curative resection
at the Civil Aviation General Hospital (Beijing, China) were
obtained from a prospectively collected database between January
2004 and December 2013. The protocol for the present study was
approved by the Institutional Review Board of the Civil Aviation
General Hospital, and written informed consent was obtained from
all patients in accordance with institutional regulations. All
patients included in the present study were diagnosed with stage II
or stage III sporadic colorectal adenocarcinoma. None of the
patients received prior therapy, including radiotherapy or
chemotherapy. All hematoxylin and eosin-stained sections were
reviewed for tissue quality, and the highest quality section from
each specimen was selected. Following an independent review by two
pathologists, all tissues were histologically confirmed to be CRC.
Clinical data, including sex and age, were obtained by chart
review. Patients with incomplete data were not included in the
present study.
Clinicopathological characteristics
and histopathological review of tumor samples
The cancer-specific data evaluated for each patient
included the tumor stage at presentation, tumor grade, specific
histology, tumor location, angiolymphatic invasion status and the
number of positive lymph nodes. The stage of each tumor was coded
according to the AJCC 6th edition TNM staging system (12) (Table I),
as follows: T1, tumor invades submucosa; T2, tumor invades
muscularis propria; T3, tumor invades through the muscularis
propria into the subserosa, or into nonperitonealized pericolic
tissues; T4, tumor directly invades other organs or structures, or
perforates visceral peritoneum; N0, no regional lymph node
metastasis; N1, metastasis to one to three regional lymph nodes;
N2, metastasis to four or more regional lymph nodes; M0, no distant
metastasis; and M1, distant metastasis.
 | Table I.Tumor classification. |
Table I.
Tumor classification.
| Stage | T Stage | N Stage | M Stage |
|---|
| I | T1 or T2 | N0 | M0 |
| IIa | T3 | N0 | M0 |
| IIb | T4 | N0 | M0 |
| IIIa | T1 or T2 | N1 | M0 |
| IIIb | T3 or T4 | N1 | M0 |
| IIIc | Any T | N2 | M0 |
| IIId | Any T | N3 | M0 |
| IIIc | Any T | N4 | M0 |
| IV | Any T | Any N | M1 |
IHC analysis of MLH1, MSH2, MSH6 and
PMS2 protein expression
IHC analysis of MLH1, MSH2, MSH6 and PMS2 protein
expression was performed using a streptavidin-peroxidase IHC kit
according to the manufacturer's protocol (Fuzhou Maixin
Biotechnology Co., Ltd., Fuzhou, China). IHC staining was performed
using 4-µm thick, paraffin-embedded CRC tissue sections mounted on
positively charged slides. Tissue sections were deparaffinized with
xylene and rehydrated through a gradient alcohol series. Sections
were subsequently immersed in 10 mmol/l citrate buffer, pH 6.0, and
heated in an autoclave at 121°C for 5 min for non-enzymatic antigen
retrieval. Next, to block the endogenous peroxidase activity, dual
endogenous enzyme by incubating the sections in 3% hydrogen
peroxide (H2O2; Fuzhou Maixin Biotech. Co.,
Ltd. Fuzhou, China) blocking solution was applied to tissue
sections for 10 min at 37°C. Primary mouse monoclonal antibodies
against human MLH1 (MAB-0642), MSH2 (MAB-0291), MSH6 (MAB-0643) and
PMS2 (MAB-0656; Fuzhou Maixin Biotechnology Co., Ltd.) were
incubated with the tissue sections for 20 min at room temperature.
Next, the slides were washed three times using 0.1 M
phosphate-buffered saline (PBS; pH 7.4) and incubated with a
biotinylated horseradish peroxidase conjugated Rabbit anti-mouse
IgG secondary antibodies (working liquid; cat no., KIT9710) (Fuzhou
Maixin Biotechnology Co., Ltd.) for 20 min at 37°C. Slides were
washed three times following incubation with the secondary antibody
using 0.1 M PBS (pH 7.4). The IHC reaction was visualized by 5–10
min of diaminobenzidine chromogen staining and 5 min hematoxylin
counterstaining at room temperature. Negative controls were
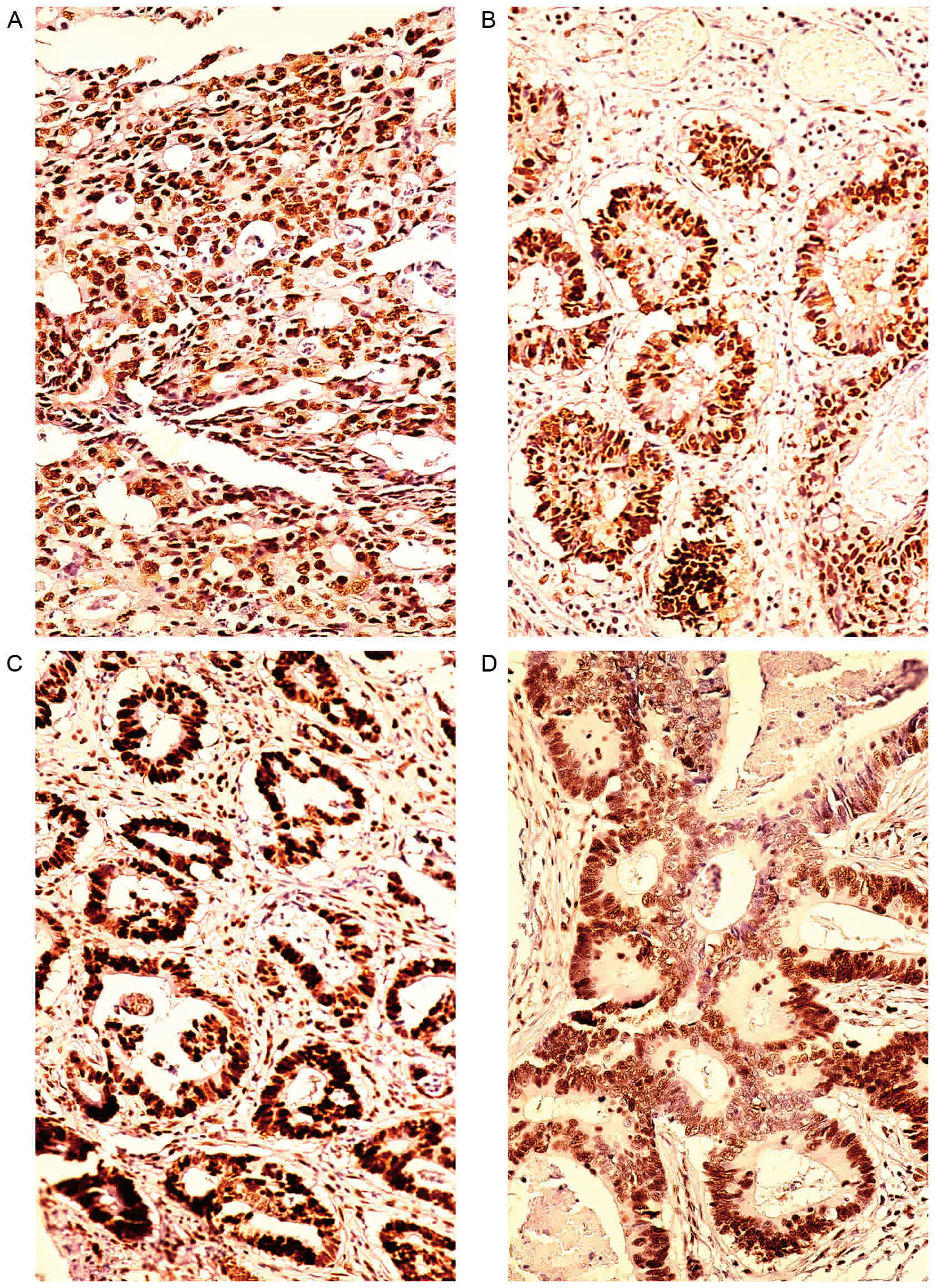
performed by omission of the primary antibody. Fig. 1 revealed that MLH1, MSH2, MSH6 and
PMS2 staining was localized to the cell nuclei. Tissues with
positive nuclear staining in the tumor cells were scored as
positive. Tissue specimens with a complete absence of nuclear
staining were scored as negative. The sections were counterstained
with hematoxylin for nuclear counterstaining and examined for the
extent and intensity of nuclear staining in tumor cells under a
200× magnification light microscope by two independent observers in
a blinded manner. Discordant scores were resolved by review and
consensus agreement or use of a third observer. Non-neoplastic
colonic tissues, stroma and infiltrating lymphocytes normally
demonstrated positive nuclear staining; therefore, these tissues
were used as internal positive controls.
Statistical analysis
The Kruskal-Wallis test was applied
(differentiation, tumor size, invasive depth and lymph node
metastasis which are 3 categories). Cross-table analysis employing
χ2 test, as appropriate, was used to analyze
associations between MMR defects (identified by IHC) and sex, ages,
type, angiolymphatic invasion characteristics. Potential variables
were verified by multivariate analysis using binary logistic
regression. P<0.05 was considered to indicate a statistically
significant difference for all analyses, and all calculations were
performed using SPSS software (version 13.0; SPSS Inc., Chicago,
IL, USA).
Results
Patient demographics
A total of 153 patients with CRC were included in
the present study. Out of these, 51 patients were ≤50 years old. A
further 102 were >50 years old. Patient age at diagnosis ranged
between 23–84 years, with a mean age of 58 years. Out of the 153
patients included in the present study, 97 (63.0%) were males and
56 (37.0%) were females.
Tumor histology
Of the 153 patients with CRC included in the present
study, 92 had stage II and 61 had stage III cancer. The
localization of all tumors was noted (right colon, defined as cecum
through transverse colon; left colon, defined as descending colon
through rectum; and rectum). Information concerning morphological
features (tumor size, tumor grade, angiolymphatic invasion, tumor
differentiation and depth of invasion) was also collected. Tumor
localization was as follows: 47/153 (31.0%) were located in the
left colon, 38/153 (25.0%) were located in the right colon and
68/153 (44.0%) were located in the rectum. Of these, 21/153 (14.0%)
patients had tumors that were smaller than 3 cm, 92/153 (60.0%) had
tumors between 3 and 5 cm and 40/153 (26.0%) had tumors larger than
5 cm.
IHC analysis of MLH1, MSH2, MSH6 and
PMS2 protein expression
MLH1, MSH2, MSH6 and PMS2 protein expression in
stage II and III CRC was examined (Table
II). Of the 153 tumors included in the present study, 120
(78.4%) were positive for MLH1, 115 (75.2%) were positive for MSH2,
68 (44.4%) were positive for MSH6 and 122 (79.7%) were positive for
PMS2. Of the 120 MLH1 positive tumors, 65 (54.0%) were stage II and
55 (46.0%) were stage III. Of the 115 MSH2 positive tumors, 66
(57.0%) were stage II and 49 (43.0%) were stage III. Of the 68 MSH6
tumors, 31 (46.0%) were stage II and 37 (54.0%) were stage III. Of
the 122 PMS2 positive tumors, 70 (57.0%) were stage II and 52
(43.0%) were stage III.
 | Table II.MLH1, MSH2, MSH6 and PMS2 protein
expression in stage II and III colorectal cancers. |
Table II.
MLH1, MSH2, MSH6 and PMS2 protein
expression in stage II and III colorectal cancers.
|
| Stage II (92) | Stage III (61) |
|
|
|---|
|
|
|
|
|
|
|---|
| Protein | − (%) | + (%) | − (%) | + (%) | χ2 | P-value |
|---|
| MLH1 | 27 (29.3) | 65 (70.7) | 6 (9.88) | 55 (90.2) | 8.225 | 0.004 |
| MSH2 | 26 (28.2) | 66 (71.8) | 12 (19.7) | 49 (80.3) | 1.449 | 0.229 |
| MSH6 | 61 (66.3) | 31 (33.7) | 24 (39.3) | 37 (60.7) | 10.798 | 0.001 |
| PMS2 | 22 (23.9) | 70 (76.1) | 9 (14.8) | 52 (85.2) | 1.904 | 0.168 |
Clinicopathological characteristics
associated with MLH1, MSH2, MSH6 and PMS2 protein expression
Associations between the expression of MLH1, MSH2,
MSH6 and PMS2 in stage II and stage III sporadic CRC and several
standard clinicopathological patient and tumor characteristics are
detailed in Tables III–VI. No significant associations were
observed between MLH1, MSH2, MSH6 or PMS2 protein expression and
sex, age, tumor location, tumor type or angiolymphatic invasion.
There were significant associations between MLH1 and MSH6 protein
expression and larger tumor size, poor differentiation, infrequent
lymph node metastasis and invasive depth (P<0.05). MSH2 and PMS2
protein expression was positively associated with larger tumor
size, tumor invasion depth and infrequent lymph node metastasis
(P<0.05).
 | Table III.MLH1 protein expression and the
clinicopathological characteristics of patients and their
tumors. |
Table III.
MLH1 protein expression and the
clinicopathological characteristics of patients and their
tumors.
|
|
| MLH1 |
|
|
|
|
|
|
|
|
| Variable | n (153) | − (%) | + (%) | χ2 | P-value |
|---|
| Sex |
|
|
|
|
|
| Male | 97 | 24 (24.7) | 73 (75.3) | 1.578 | 0.209 |
|
Female | 56 | 9 (16.1) | 47 (83.9) |
|
|
| Age, years |
|
|
|
|
|
| ≤50 | 51 | 13 (25.5) | 38 (74.5) | 0.695 | 0.404 |
|
>50 | 102 | 20 (19.6) | 82 (80.4) |
|
|
| Type |
|
|
|
|
|
|
Ulcerated | 139 | 3 (22.3) | 108 (77.7) | 0.843 | 0.487 |
|
Protruded | 14 | 2 (14.2) | 12 (85.8) |
|
|
| Tumor size, cm |
|
|
|
|
|
|
<3 | 21 | 2 (9.5) | 19 (90.5) | 11.267 | 0.004 |
|
3–5 | 92 | 15 (16.3) | 77 (83.7) |
|
|
|
>5 | 40 | 16 (40) | 24 (60.0) |
|
|
| Localization |
|
|
|
|
|
| Right
colon | 38 | 9 (23.7) | 29 (76.3) | 2.300 | 0.317 |
| Left
colon | 47 | 13 (27.7) | 34 (72.3) |
|
|
|
Rectum | 68 | 11 (16.2) | 57 (83.8) |
|
|
|
Differentiation |
|
|
|
|
|
|
Well | 36 | 4 (11.1) | 32 (88.9) | 7.814 | 0.020 |
|
Moderate | 64 | 11 (17.2) | 53 (82.8) |
|
|
|
Poor | 53 | 18 (33.9) | 35 (66.1) |
|
|
| Invasive depth |
|
|
|
|
|
| Intra
muscularia | 11 | 2 (18.2) | 9 (81.8) | 10.408 | 0.005 |
| Intra
subserosa | 112 | 18 (16.1) | 94 (83.9) |
|
|
| Extra
subserosa | 30 | 13 (43.3) | 17 (56.7) |
|
|
| Angiolymphatic
invasion |
|
|
|
|
|
|
Without | 104 | 19 (18.3) | 85 (81.7) | 2.090 | 0.148 |
|
With | 49 | 14 (28.6) | 35 (71.4) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Without | 44 | 17 (38.7) | 27 (61.4) | 10.586 | 0.005 |
|
1–3 | 73 | 11 (15.1) | 62 (84.9) |
|
|
| ≥4 | 36 | 5 (13.9) | 31 (86.1) |
|
|
 | Table VI.PMS2 protein expression and the
clinicopathological characteristics of patients and their
tumors. |
Table VI.
PMS2 protein expression and the
clinicopathological characteristics of patients and their
tumors.
|
|
| PMS2 |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | n | − (%) | + (%) | χ2 | P-value |
|---|
| Sex |
|
|
|
|
|
|
Male | 97 | 20 (20.6) | 77 (79.4) | 0.021 | 0.885 |
|
Female | 56 | 11 (19.6) | 45 (80.4) |
|
|
| Age, years |
|
|
|
|
|
|
≤50 | 51 | 14 (27.4) | 37 (72.5) | 2.448 | 0.118 |
|
>50 | 102 | 17 (16.7) | 85 (83.3) |
|
|
| Type |
|
|
|
|
|
|
Ulcerated | 139 | 29 (20.9) | 110 (79.1) | 0.341 | 0.559 |
|
Protruded | 14 | 2 (14.3) | 12 (85.7) |
|
|
| Tumor size, cm |
|
|
|
|
|
|
<3 | 21 | 2 (9.5) | 19 (90.5) | 7.718 | 0.021 |
|
3–5 | 92 | 15 (16.3) | 77 (83.7) |
|
|
|
>5 | 40 | 14 (35.0) | 26 (65.0) |
|
|
| Localization |
|
|
|
|
|
| Right
colon | 38 | 8 (21.1) | 30 (78.9) | 1.525 | 0.466 |
| Left
colon | 47 | 12 (25.5) | 35 (74.5) |
|
|
|
Rectum | 68 | 11 (16.2) | 57 (83.8) |
|
|
|
Differentiation |
|
|
|
|
|
|
Well | 36 | 5 (13.9) | 31 (86.1) | 3.378 | 0.185 |
|
Moderate | 64 | 11 (17.2) | 53 (82.8) |
|
|
|
Poor | 53 | 15 (28.3) | 38 (71.7) |
|
|
| Invasive depth |
|
|
|
|
|
| Intra
muscularia | 11 | 2 (18.2) | 9 (81.8) | 12.308 | 0.002 |
| Intra
subserosa | 112 | 16 (14.3) | 96 (85.7) |
|
|
| Extra
subserosa | 30 | 13 (43.3) | 17 (56.7) |
|
|
| Angiolymphatic
invasion |
|
|
|
|
|
|
Without | 104 | 18 (17.3) | 86 (82.7) | 1.754 | 0.185 |
|
With | 49 | 13 (26.5) | 36 (73.5) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Without | 44 | 15 (34.1) | 29 (65.9) | 9.256 | 0.010 |
|
1–3 | 73 | 12 (16.4) | 61 (83.6) |
|
|
| ≥4 | 36 | 4 (11.1) | 32 (88.9) |
|
|
Poorly differentiated tumors demonstrated lower MLH1
expression when compared with moderately and well differentiated
tumors, with a statistically significant difference between them
(poor vs. moderate, P=0.036; poor vs. well-differentiated,
P=0.014). MLH1 positive expression was detected in 32/36 (88.9%)
well differentiated tumors, 53/64 (82.8%) moderately differentiated
tumors and 35/53 (66.0%) poorly differentiated tumors.
A statistically significant association was observed
between MLH1 expression and tumor size (χ2=11.267;
P=0.004). MLH1 expression was frequently observed in tumors <3
cm, and 19/21 (90.5%) tumors were positive for MLH1. In tumors
between 3–5 cm, 77/92 (83.7%) tumors were MLH1 positive. In tumors
>5 cm, 24/40 (60.0%) were identified to be MLH1 positive.
Negative expression of MLH1 was associated with the
invasive depth of tumors, and the differences in expression were
statistically significant (χ2=10.408, P=0.005). MLH1
expression was observed in 9/11 (81.8%) intra muscularia, 94/112
(83.9%) intra subserosa and 17/30 (56.7%) extra subserosa
tumors.
MLH1 expression was higher in tumors with multiple
lymph node metastases than in tumors without lymph node metastasis,
with a statistically significant difference between them
(χ2=10.586; P=0.005). MLH1 expression was detected in
27/44 (61.4%) of tumors without lymph node metastasis, 62/73
(84.9%) of tumors with one to three lymph node metastases and 31/36
(86.1%) of tumors with four or more lymph node metastases.
MSH2 protein expression was observed in 18/21
(85.7%) of tumors <3 cm, 74/92 (80.4%) of tumors between 3 and 5
cm and 23/40 (57.2%) of tumors >5 cm. These differences were
statistically significant (χ2=9.246; P=0.01).
MSH2 negative expression was also associated with
the invasive depth of tumors, and the differences in expression
were statistically significant (χ2=6.824; P=0.033). MSH2
protein expression was observed in 9/11 (81.5%) intra muscularia,
89/112 (79.5%) intra subserosa and 17/30 (56.7%) extra subserosa
tumors.
MSH2 expression was detected in 27/44 (61.4%) tumors
without lymph node metastasis, 58/73 (79.5%) with one to three
metastases and 30/36 (83.8%) with four or more lymph node
metastases. These differences in expression were statistically
significant (χ2=7.305; P=0.026).
MSH6 expression was more common in
well-differentiated tumors than in moderately or poorly
differentiated tumors, and this difference was statistically
significant (poor vs. moderately differentiated, P=0.145; poor vs.
well differentiated, P=0.007). MSH6 expression was detected in
22/36 (61.1%) well differentiated, 29/64 (45.3%) moderately
differentiated and 17/54 (32.1%) poorly differentiated tumors.
MSH6 expression was observed in 15/21 (71.4%) tumors
<3 cm, 39/92 (42.4%) tumors between 3–5 cm and 14/40 (35.0%)
tumors >5 cm. These differences were statistically significant
(χ2=5.818; P=0.05).
The frequency of MSH6 positive tumors significantly
decreased with increasing invasive depth, and this difference in
expression was revealed to be statistically significant
(χ2=6.051, P=0.049). MSH6 expression was observed in
7/11 (63.6%) intra muscularia, 53/112 (47.3%) intra subserosa and
8/30 (26.7%) extra subserosa tumors.
MSH6 expression was detected in 13/44 (29.5%) tumors
without lymph node metastasis, 34/73 (46.6%) of tumors with one to
three metastases and 21/36 (58.3%) of tumors with four or more
lymph node metastases. These differences were statistically
significant (χ2=8.905; P=0.012).
Larger tumors were less frequently positive for PMS2
protein expression, and this difference was statistically
significant (χ2=7.718; P=0.021). PMS2 expression was
observed in 19/21 (90.5%) tumors <3 cm, 77/92 (83.7%) tumors
between 3 and 5 cm and 26/40 (65.0%) tumors >5 cm.
The frequency of PMS2 positive tumors was
significantly lower in tumors, which had invaded out of the
subserosa, compared with those that had not, and this difference
was statistically significant (χ2=12.308; P=0.002). PMS2
expression was observed in 9/11 (81.8%) intra muscularia, 96/112
(85.7%) intra subserosa and 17/30 (56.7%) extra subserosa
tumors.
PMS2 expression was observed in 29/44 (65.9%) tumors
without lymph node metastasis, 61/73 (83.6%) tumors with one to
three metastases and 32/36 (88.9%) tumors with four or more lymph
node metastases. These differences were statistically significant
(χ2=9.256; P=0.010).
Discussion
MSI was first reported in 1993, as the presence of
thousands of somatic alterations in the length of DNA
microsatellite repeats in sporadic and familial colorectal tumors
(13). MSI is the result of defects
in the MMR system and the MSH2, MLH1, PMS2 and MSH6 genes (14).
The MMR system recognizes and corrects base-pair
mismatches and small nucleotide (1–4 base pair) insertion or
deletion mutations within the duplex DNA that arise from nucleotide
misincorporation during DNA replication (15,16). A
properly functioning MMR system is essential for the maintenance of
genomic stability, and the mutation rates in tumor cells with MMR
deficiencies are 100- to 1,000-fold higher than in normal cells
(17). Consequently, a malfunctioning
MMR system leads to genome-wide instability.
Previous studies have identified that colorectal
tumors with MSI accumulate mutations at microsatellite sequences in
the coding regions of tumor progression genes (15). MSI is detected in tumor tissue by
examining a panel of five markers: BAT25, BAT26, D2S123, D5S346 and
D17S250, known as the Bethesda markers. This method requires
specific laboratory expertise and equipment, and is routinely used
in clinical pathology laboratories (18). It has been demonstrated that loss of
MLH1, MSH2, PMS2 and MSH6 protein expression is associated with a
defective MMR system. Therefore, IHC analysis of the expression of
these markers may potentially be used to detect MSI tumors
(19).
In the present study, MLH1, MSH2, MSH6 and PMS2
expression was observed at 70.7, 71.7, 33.7 and 76.1% of stage II
colon cancer tissues, respectively. MLH1, MSH2, MSH6 and PMS2
expression was higher in stage III tumors, at 90.2, 80.3, 60.7 and
85.2% respectively. The differences in expression were
statistically significant for MLH1 and MSH6, and were significantly
higher in stage III colon cancer than in stage II tumors
(χ2=8.225 and 10.798, respectively; P<0.05). MMR
deficiency in sporadic cancers is primarily due to MLH1 expression
loss due to the somatic hypermethylation of its promoter; however,
in the present study, MMR deficiencies in examined sporadic CRCs
were primarily due to the loss of MSH6. In the present study, MLH1
protein was detected in 70.7% of stage II and 90.2% of stage III
colon cancer samples, and MSH6 protein was detected in 33.7% of
stage II and 60.7% of stage III colon cancer samples.
In the present study, in tumors >5 cm, MLH1,
MSH2, MSH6 and PMS2 expression was lower than the expression of
these proteins observed in the tumors <5 cm. This result is in
accordance with the results of several previous studies (20). For example, Lanza et al
(11) demonstrated that patients with
MMR defective colorectal tumors were younger and had tumors that
were localized in the right-colon. MMR defective colorectal tumors
exhibited infrequent lymph node metastasis, were larger and were
poorly differentiated or of mucinous histology. Additionally,
patients with MMR colorectal tumors had distinct
clinicopathological characteristics, including a lower risk of
recurrence. In another study by Sinicrope et al (21), the prevalence of a defective MMR
system in stage II and III colon cancers was 15%, and the MMR
phenotype was significantly associated with higher tumor stage,
proximal site, poor or undifferentiated histology, female sex and
older age.
In the present study, it was observed that the loss
of MLH1, MSH2, MSH6 and PMS2 expression was associated with
advanced tumors. The loss of MLH1 and MSH6 protein expression was
significantly associated with large, poorly differentiated tumors
characterized by extraserosal invasion and infrequent lymph node
metastasis. Similarly, the loss of MLH2 and PMS2 protein expression
was significantly associated with large tumors characterized by
extraserosal invasion and infrequent lymph node metastasis. No
significant associations between the loss of MLH1, MSH2, MSH6 or
PMS2 expression and the age, sex, tumor location or angiolymphatic
invasion of patients with CRC were observed.
Unlike patients with intact MMR, patients with MMR
deficient colon cancers do not benefit from 5-fluorouracil-based
adjuvant therapy (22). Therefore,
the identification of patients with MMR deficient tumors is
critical for the selection of an appropriate and effective
treatment strategy. The results of the present study may help
improve patient outcomes by assisting the identification of
patients who possess CRCs exhibiting defective MMR. The use of this
information in clinical decision-making would represent an
important step toward individualized cancer therapy.
Acknowledgements
The present study was supported by the Civil
Aviation Hospital Foundation of Hospital-level Topics (grant no.,
2013013).
References
|
1
|
Weitz J, Koch M, Debus J, Höhler T, Galle
PR and Büchler MW: Colorectal cancer. Lancet. 365:153–165. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Black RJ, Bray F, Ferlay J and Parkin DM:
Cancer incidence and mortality in the European Union: Cancer
registry data and estimates of national incidence for 1990. Eur J
Cancer. 33:1075–1107. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mundade R, Imperiale TF, Prabhu L, Loehrer
PJ and Lu T: Genetic pathways, prevention, and treatment of
sporadic colorectal cancer. Oncoscience. 1:400–406. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee JH, Cragun D, Thompson Z, Coppola D,
Nicosia SV, Akbari M, Zhang S, McLaughlin J, Narod S, Schildkraut
J, et al: Association between IHC and MSI testing to identify
mismatch repair-deficient patients with ovarian cancer. Genet Test
Mol Biomarkers. 18:229–235. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Haydon AM and Jass JR: Emerging pathways
in colorectal-cancer development. Lancet Oncol. 3:83–88. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jascur T and Boland CR: Structure and
function of the components of the human DNA mismatch repair system.
Int J Cancer. 119:2030–2035. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pawlik TM, Raut CP and Rodriguez-Bigas MA:
Colorectal carcinogenesis: MSI-H versus MSI-L. Dis Markers.
20:199–206. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wheeler JM, Bodmer WF and Mortensen NJ:
DNA mismatch repair genes and colorectal cancer. Gut. 47:148–153.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yoon YS, Yu CS, Kim TW, Kim JH, Jang SJ,
Cho DH, Roh SA and Kim JC: Mismatch repair status in sporadic
colorectal cancer: Immunohistochemistry and microsatellite
instability analyses. J Gastroenterol Hepatol. 26:1733–1739. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lindor NM, Burgart LJ, Leontovich O,
Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen
GM, Walsh MD, Leggett BA, et al: Immunohistochemistry versus
microsatellite instability testing in phenotyping colorectal
tumors. J Clin Oncol. 20:1043–1048. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lanza G, Gafà R, Maestri I, Santini A,
Matteuzzi M and Cavazzini L: Immunohistochemical pattern of
MLH1/MSH2 expression is related to clinical and pathological
features in colorectal adenocarcinomas with microsatellite
instability. Mod Pathol. 15:741–749. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smith NJ, Bees N, Barbachano Y, Norman AR,
Swift RI and Brown G: Preoperative computed tomography staging of
nonmetastatic colon cancer predicts outcome: Implications for
clinical trials. Br J Cancer. 96:1030–1036. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boland CR and Goel A: Microsatellite
instability in colorectal cancer. Gastroenterology. 138:2073–2087.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iacopetta B, Grieu F and Amanuel B:
Microsatellite instability in colorectal cancer. Asia Pac J Clin
Oncol. 6:260–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sameer AS, Nissar S and Fatima K: Mismatch
repair pathway: Molecules, functions, and role in colorectal
carcinogenesis. Eur J Cancer Prev. 23:246–257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Michailidi C, Papavassiliou AG and
Troungos C: DNA repair mechanisms in colorectal carcinogenesis.
Curr Mol Med. 12:237–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Raut CP, Pawlik TM and Rodriguez-Bigas MA:
Clinicopathologic features in colorectal cancer patients with
microsatellite instability. Mutat Res. 568:275–282. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Boland CR, Thibodeau SN, Hamilton SR,
Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA,
Fodde R, Ranzani GN and Srivastava S: A National cancer institute
workshop on microsatellite instability for cancer detection and
familial predisposition: Development of international criteria for
the determination of microsatellite instability in colorectal
cancer. Cancer Res. 58:5248–5257. 1998.PubMed/NCBI
|
|
19
|
Bao F, Panarelli NC, Rennert H, Sherr DL
and Yantiss RK: Neoadjuvant therapy induces loss of MSH6 expression
in colorectal carcinoma. Am J Surg Pathol. 34:1798–1804. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim JC, Cho YK, Roh SA, Yu CS, Gong G,
Jang SJ, Kim SY and Kim YS: Individual tumorigenesis pathways of
sporadic colorectal adenocarcinomas are associated with the
biological behavior of tumors. Cancer Sci. 99:1348–1354. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sinicrope F, Foster NR, Sargent DJ,
Thibodeau SN, Smyrk TC and O'Connell MJ: North Central Cancer
Treatment Group: Model-based prediction of defective DNA mismatch
repair using clinicopathological variables in sporadic colon cancer
patients. Cancer. 116:1691–1698. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jover R, Zapater P, Castells A, Llor X,
Andreu M, Cubiella J, Balaguer F, Sempere L, Xicola RM, Bujanda L,
et al: The efficacy of adjuvant chemotherapy with 5-fluorouracil in
colorectal cancer depends on the mismatch repair status. Eur J
Cancer. 45:365–373. 2009. View Article : Google Scholar : PubMed/NCBI
|