Introduction
Primary liver cancers (PLC), including
cholangiocarcinoma (CC) and hepatocellular carcinoma (HCC), are the
third and sixth most common causes of cancer-associated mortalities
for men and women, respectively. Worldwide, >560,000 people are
diagnosed with PLC annually (1,2). Of these,
55% of the cases are in China, where PLC was the second most common
cause of cancer mortality in 2002 (1–3). HCC
accounts for 85–90% of all PLC worldwide (4); HCC and PLC often are used
interchangeably. The median survival length in unresectable cases
is >4 months, and >1 year for untreated patients with less
advanced disease, which demonstrates the poor prognosis of this
type of cancer (1,4,5).
Therefore, it is crucial to understand the mechanisms that regulate
the progression of HCC for the development of novel and effective
therapeutic approaches.
Integrins mediate the adhesion between the cellular
cytoskeleton and extracellular matrix. They serve a key role in
outside-in and inside-out signaling, and also control a variety of
vital cell functions including: Adhesion; differentiation;
migration; cell division; and apoptosis (6–8). αvβ6
integrin (αvβ6) is an important member of the integrin family,
which is unique as it is expressed exclusively in epithelial cells
(9). During embryogenesis, αvβ6 is
expressed at high levels in the developing lung, skin and kidney
epithelia cells, while its expression is downregulated in healthy
adult epithelia cells (10–12). Previously, αvβ6 integrin was
demonstrated to be highly upregulated in carcinomas of the breast,
cholangiocarcinoma, pancreatic ductal, colon, stomach, ovary and
endometrium, and also in liver metastases derived from colorectal
and pancreatic carcinomas (13–19).
However, the expression and role of αvβ6 has not been thoroughly
investigated in HCC.
We previously indicated that Lysophosphatidic acid
(LPA) induces αvβ6-mediated transforming growth factor-β1 (TGF-β1)
activation (20), and Geng et
al (21) demonstrated similar
results. TGF-β1 is involved in the development of tumor-initiating
cells, contributes to angiogenesis and promotes liver cancer
development (22–24). There is significant evidence that also
indicates its important role for Autotaxin-LPA signaling in human
HCC (25–27). Furthermore, β6 only partners with αv,
forming a single heterodimer, and its synthesis is rate-limiting
for αvβ6 expression (9). Based on
results from these previous studies, we hypothesized that the
integrin β6 subunit (Itgβ6) is upregulated in HCC, and that there
is an association between TGF-β1 and LPA in the expression of
Itgβ6. In the present study, the expression levels for Itgβ6 in HCC
tissue samples and tumor-derived cell lines were evaluated, and the
role of LPA and TGF-β1 in the regulation of Itgβ6 expression and
transcriptional activation of Itgβ6 gene promoter was investigated
in Hep-3B.
Materials and methods
Cell culture
Cell lines were purchased from the Shanghai
Institute of Biochemistry and Cell Biology, Shanghai Institutes of
Biological Sciences, Chinese Academy of Sciences (Shanghai, China).
Human hepatic cancer Hep-3B cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.). Human hepatocyte
HL-7702 cells were maintained in RPMI-1640 (RPMI-1640; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 20% FBS
containing penicillin (100 U/ml) and streptomycin sulfate (100
µg/ml). Cell lines were maintained at 37°C with 5% CO2
in a humid incubator.
Human samples
Paraffin-embedded human liver and colorectal
carcinoma tissues were obtained from the Second Affiliated Hospital
of Shantou Medical College (Shantou, China). Samples were collected
from consecutive patients undergoing surgical resection for HCC
(n=23), hepatic metastatic adenoma [n=2; 1 male, 1 female, age
range 50–52 years (mean, 51.00±1.41)] and colorectal carcinoma
[n=5; 4 males, 1 female, age range 54–78 years (mean,
64.80±11.30)]. Histologically normal liver tissues were collected
from patients undergoing resection for other carcinomas [n=4; 3
males, 1 female, age range 54–78 years (mean, 62.00±10.86)], and
served as controls. The population of patients with HCC included 21
men and 2 women ranging in age between 37–78 years (mean,
50.04±2.45). HCC was histologically confirmed, and tumor staging
was performed using the Cancer of the Liver Italian Program (CLIP;
which incorporates Child stage, tumor morphology, AFP levels, and
the presence of portal vein thrombosis) system (28). Patient characteristics are summarized
in Table I; no patients had received
preoperative chemotherapy. The Ethical Committees of the Second
Affiliated Hospital of Shantou Medical College and the Medical
College of Shantou University approved the present study, and all
patients provided written informed consent.
 | Table I.Characteristics of patients with
HCC. |
Table I.
Characteristics of patients with
HCC.
| Variables | HCC (n=23) |
|---|
| Age, years |
|
| Mean ±
standard error of the mean | 50.04±2.445 |
| Sex |
|
|
Male | 21 |
|
Female | 2 |
| CLIP stages
(0–6) |
|
| 0 | 5 |
| 1 | 3 |
| 2 | 11 |
| 3 | 4 |
Immunohistochemistry
The histological sections were de-paraffinized in
100% xylene for 30 min at room temperature, and then incubated in
graded ethanol (100% ethanol for 15 min, 95% ethanol for 5 min, 90%
ethanol for 5 min, then 70% ethanol for 5 min, successively) for 30
min at room temperature. Subsequent to blocking endogenous
peroxidase activity in 3% H2O2 for 30 min at
room temperature, the sections were incubated at 4°C overnight with
the primary anti-Itgβ6 mouse monoclonal antibody (Merck KGaA,
Darmstadt, Germany) applied at a dilution rate of 1:50 in antibody
diluents (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA),
followed by PV9000 reagent (ZSGB-BIO; OriGene Technologies, Inc.,
Beijing, China) according to the manufacturer's protocol. A
3-amino-9-ethylcarbazole kit (ZSGB-BIO; OriGene Technologies, Inc.)
was used to visualize positive staining according to the
manufacturer's protocol. Nuclei were stained with 0.5% hematoxylin
for 10 sec at room temperature and covered with glycerogelatin. The
primary antibodies were replaced by antibody diluent to serve as
controls (50:1; DAKO; Agilent Technologies, Inc., Santa Clara, CA,
USA). The anti-Itgβ6 mouse monoclonal antibody (mAb)
immunoreactivity colorectal carcinoma specimen that is known to be
positive for β6 was used as a positive control. Staining was scored
using the following scale: Negative (−, 0% of cytoplasmic stain),
weak (+, <20%), moderate (++, 20–50%), strong (+++, >50%). A
total of 10 random views were taken of each section and the
proportions of cytoplasmic stain were calculated in these views.
Representative images were captured at ×200 original magnification
using a light microscope (BX-51TRF; Olympus Corporation, Tokyo,
Japan).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Hep-3B cells were stimulated with either: i) TGF-β1
(5 ng/ml) for 8, 12 and 24 h; ii) LPA (10 µM) for 12, 24, 48 and 72
h; iii) DMEM/0.1% bovine serum albumin (BSA) alone (serving as a
negative control) in 6 well plates. Hep-3B cells were stimulated
with either TGF-β (5 ng/ml) for 8 h, LPA (10 µM) for 24 h, TGF-β (5
ng/ml) for 8 h + LPA (10 µM) for 24 h, or DMEM/0.1% BSA alone
(serving as a negative control) in 6 well plates. Total RNA was
extracted using universal RNA Pure reagent (Guangzhou Dongsheng
Biotech, Guangzhou, China) according to the manufacturer's
protocol. PCR conditions were: 94°C for 2 min, followed by 35–40
cycles of 94°C for 30 sec, 58°C for 30 sec and 72°C for 1 min,
finished at 72°C for 5–10 min. Template cDNA was obtained using
reverse transcription of 1 µg of the total RNA with EasyScript
First-Strand cDNA Synthesis SuperMix (Beijing TransGen Biotech Co.,
Ltd., Beijing, China), and relative transcript levels were
quantified using qPCR on an ABI PRISM® 7300 Fast
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. The expression of
Itgβ6 and the housekeeping gene human 18s rRNA (18s) gene were
determined using the primer sequences: Itgβ6 forward primer
5′-GCAAGCTGCTGTGTGTAAGGAA-3′ and reverse primer
5′-CTTGGGTTACAGCGAAGATCAA-3′; 18s forward primer
5′-TTGGTGGAGCGATTTGTCTG-3′ and reverse primer
5′-AATGGGGTTCAACGGGTTAC-3′. Itgβ6 and 18s transcripts were
amplified using the SYBR® Premix Ex Taq™ kit
(Takara Biotechnology Co., Ltd., Dalian, China), according to
manufacturer's protocol. The thermocycler conditions were as
follows: Initial denaturation at 94°C for 30 sec, followed by 40
cycles of 95°C for 5 sec and 60°C for 30 sec. The relative
expression levels of Itgβ6 mRNAs were determined using the
comparative cycle threshold (Cq) method (29) that utilized 18s as the endogenous
control. Quantity One 4.4 (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) was used for analysis.
Western blot assays
Hep-3B cells were seeded in 25 cm2 flasks
(Corning Incorporated, NY, USA) containing DMEM supplemented with
10% FBS. Cells were grown to 60–70% confluence and the medium was
changed to DMEM with 0.1% BSA for 30 h in the absence (control) or
presence of TGF-β1 (5 ng/ml) for 14 h, LPA (10 µM) for 30 h, or
TGF-β1 (5 ng/ml) for 14 h + LPA (10 µM) for 30 h. Cells were then
washed with ice-cold PBS 3 times and lysed in
radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 1%
Nonidet P-40, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl
fluoride, 1 µg/ml aprotinin and leupeptin, 1 mM sodium
orthovanadate and 1 mM NaF). Following centrifugation (13,400 × g
at 4°C for 15 min), the supernatant was collected for protein
analysis. Protein concentrations were determined using a BCA
Protein Assay kit (Merck KGaA) according to the manufacturer's
protocol. Protein samples (80 µg) were separated by 10% SDS-PAGE
and transferred onto nitrocellulose membranes (Bio TraceTMNT; Pall
Corporation, Port Washington, NY, USA), and incubated with
anti-integrin β6 mouse mAb (cat. no., 442.5C4; Merck KGaA;
dilution, 1:200) in TBST at 4°C overnight prior to washing and
incubation at room temperature for 1 h with horseradish
peroxidase-conjugated goat anti-mouse secondary antibody (sc-2031;
Santa Cruz Biotechnology, Inc., Hercules, CA, USA) diluted to
1:1,000 in TBST with 1% BSA. Immunoreactive bands were visualized
using an EasySee™ Western Blot kit (TransGen Biotech,
Beijing, China). GAPDH (1:1,000) was used as a loading control. The
optical densities for each band and the density ratio for β6 to
GAPDH were analyzed using the Quantity One software 4.62 (Bio-Rad
Laboratories, Inc.).
Transfection and luciferase
assays
The sequence containing the full-length β6 promoter
region (−946/+189 bp, 1,135 bp) was generated by PCR as
aforementioned. The amplified fragment was digested with Kpn I/Xho
I and inserted into the Kpn I/Xho I sites of pGL2-Basic luciferase
reporter gene, and the resulting plasmid was identified as pGLB-FL.
Promoter deletions were generated from pGLB-FL by PCR as
aforementioned, and inserted into the pGL2-Basic plasmid as
previously described (30). Plasmids
were identified as pGLB-112 (+60/+172; 112 bp), pGLB-207 (−35/+172;
207 bp), pGLB-359 (−187/+172; 359 bp), pGLB-630 (−458/+172; 630
bp), pGLB-A (−470/-157; 313 bp), pGLB-B (−388/-157; 231 bp) and
pGLB-C (−326/−157; 169 bp), respectively.
Hep-3B cells cultured to 70–80% confluency were
transfected using METAFECTENE®PRO reagent (Biontex
Laboratories GmbH, München, Germany) with 0.2 µg for each of the
Itgβ6 constructs, and mixed at ratio of 200:1 with a control
Renilla construct (Biontex Laboratories GmbH) according to the
manufacturer's protocol. Cell extracts were evaluated for promoter
activity at 48 h following transfection. Prior to collecting cells
for extracts, the cells were treated with either TGF-β1 (5 ng/ml)
for 24 h, LPA (10 µM) for 24 h, or DMEM/0.1% BSA alone as a
negative control. Luciferase and Renilla activities were determined
using a Dual-Luciferase® Reporter Assay System (Promega
Corporation, Madison, USA) according to manufacturer's protocol.
Luminescence was measured using an EG&G Berthold Lumat LB9507
luminometer (Berthold Technologies GmbH & Co. KG, Bad Wildbad,
Germany). For each sample, luciferase relative light units were
adjusted based on Renilla activity to normalize for transfection
efficiency.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). Analyses
included one-tailed Pearson tests, two-tailed Student t-tests for
single comparisons and one-way analysis of variance for comparing
multiple treatment groups with a control group. Bonferroni post hoc
test was used to determine which results differed significantly.
The data are expressed as the mean ± standard error of the mean,
unless stated otherwise. P<0.05 was considered to indicate a
statistically significant difference.
Results
Itgβ6 immunohistochemistry in human
liver tumors
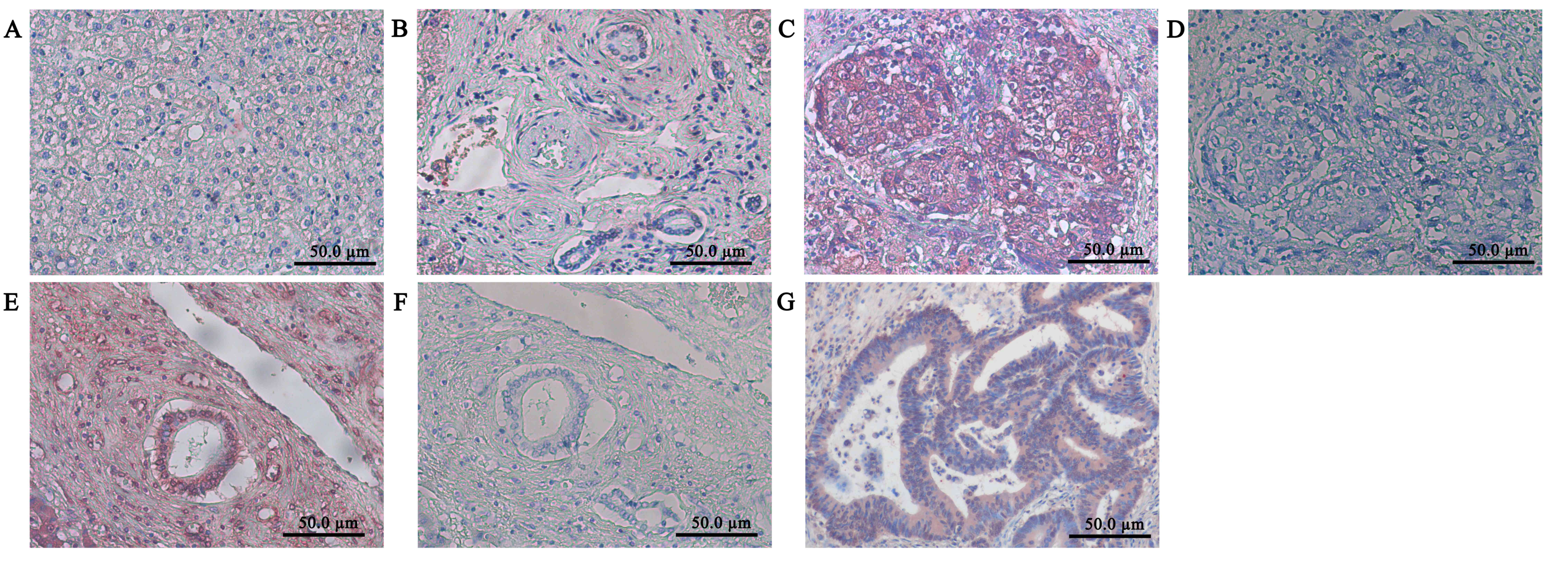
Immunoreactivity for Itgβ6 was absent in normal
liver tissue, liver cells, and intrahepatic bile ducts, which was
similar to the negative controls (Fig. 1A
and B). In contrast, strong staining was present in the nests
and intrahepatic bile ducts of the HCC samples (Fig. 1C and D); blank controls are presented
in Fig. 1E and F. Itgβ6
immunoreactivity was present in the positive control colorectal
carcinoma sections (Fig. 1G) and
detected in hepatic metastases adenoma samples (data not shown).
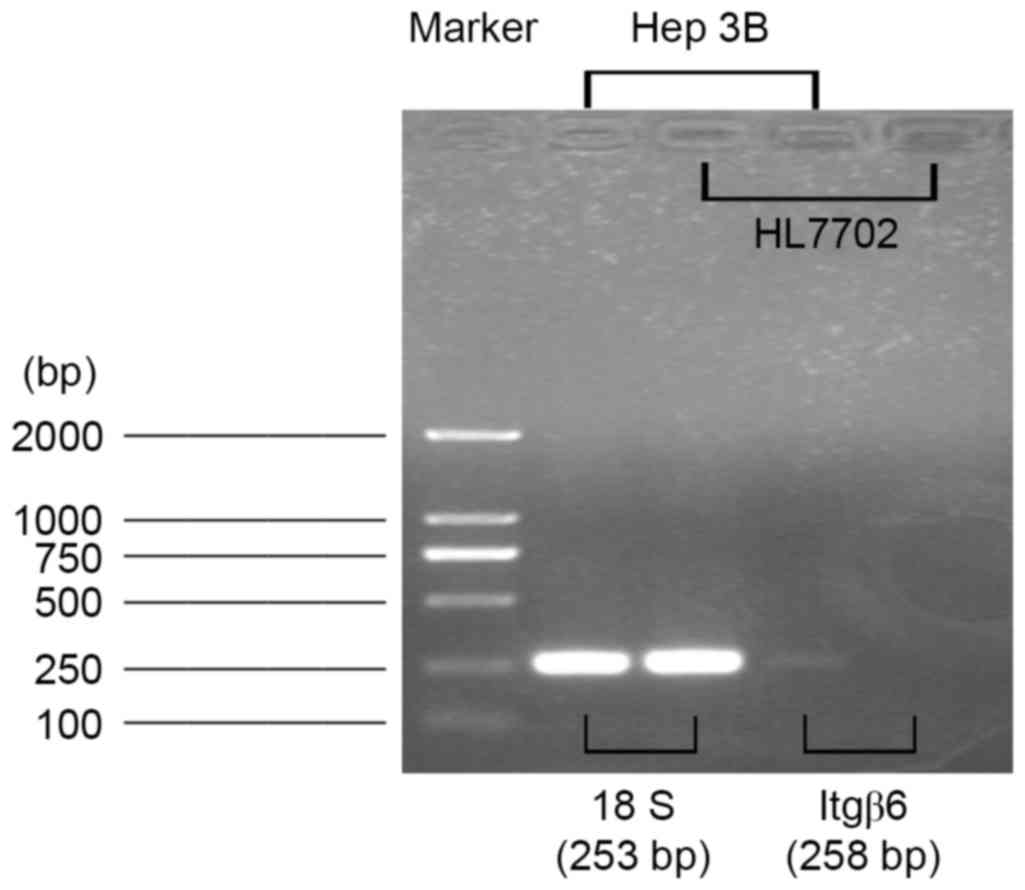
The expression of Itgβ6 was measured at mRNA levels in the human
normal hepatocyte HL-7702 and human hepatic cancer Hep-3B cell
lines. Itgβ6 mRNA was detected in Hep-3B cells, but Itgβ6
expression was not detected in HL-7702 cells (Fig. 2).
No clear association between Itgβ6
immunoreactivity and the grade of HCC
No clear association between Itgβ6 immunoreactivity
and the CLIP grade was observed. There was also no significant
correlation between Itgβ6 expression and tumor grade (r=−0.284)
observed, probably due to the small group size (n=23; Table II).
 | Table II.Integrin β6 expression in HCC. |
Table II.
Integrin β6 expression in HCC.
|
| Integrin β6
expression, n (%) |
|---|
|
|
|
|---|
| Variables | – | + | ++ | +++ |
|---|
| HCC (n=23) | 0 | 3 (13) | 9 (39) | 11 (48) |
TGF-β1 and LPA induce Itgβ6 expression
in Hep-3B cells
The association between the Itgβ6 mRNA levels and
TGF-β1/LPA was evaluated in Hep-3B cells treated with or without
TGF-β1 or LPA for the time periods indicated (Fig. 3A and B). TGF-β1 and LPA induced an
increase in Itgβ6 expression at mRNA and protein levels in Hep-3B
cells (Fig. 3C and D). When compared
with the non-treated cells, TGF-β1, LPA and TGF-β1+LPA treatments
increased β6 mRNA levels by 1.87-, 2.00- and 4.53-fold,
respectively (Fig. 3E), and protein
levels by 2.68-, 2.76- and 4.29-fold, respectively (Fig. 3F).
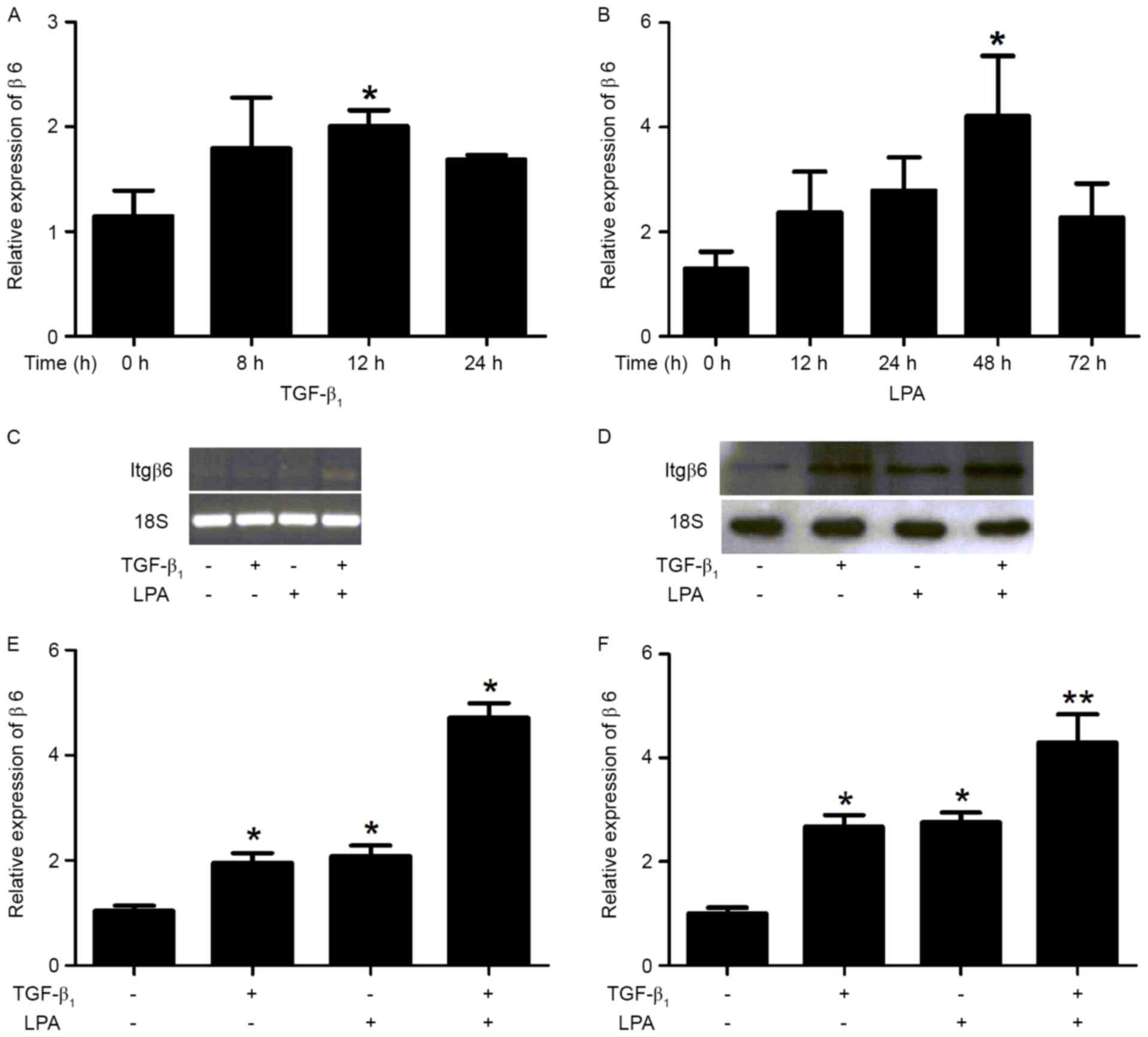 | Figure 3.TGF-β1 and LPA induces the Itgβ6
expression in Hep-3B cells. Itgβ6 expression was examined using
reverse transcription PCR and western blot analysis in Hep-3B
cells. Hep-3B cells were serum-starved for 24 h, then treated with
either TGF-β1 or LPA for various time periods. Hep-3B cells were
serum-starved for 24 h, then treated with either TGF-β (5 ng/ml)
for 8 h, LPA (10 µM) for 24 h, TGF-β (5 ng/ml) for 8 h + LPA (10
µM) for 24 h, or DMEM/0.1% BSA alone (serving as a negative
control). (A) TGF-β1- and (B) LPA-induced Itgβ6 expression for
various time periods in Hep-3B cells. (C) Itgβ6 and 18s mRNA
expression by PCR that included 28 cycles. Ethidium bromide
staining of β6 and 18s electrophoresis gels was included to
indicate equal loading among the same set of RNA samples. (D)
Hep-3B cells were serum-starved for 24 h then treated with either
TGF-β (5 ng/ml) for 14 h, LPA (10 µM) for 30 h or TGF-β (5 ng/ml)
for 14 h+ LPA (10 µM) for 30 h, or DMEM/0.1% BSA alone (serving as
a negative control), followed by western blot analysis with the
indicated antibodies. (E) When compared with non-treated cells, the
increased β6 mRNA expression induced by TGF-β1, LPA or TGF-β1+LPA
was 1.87-, 2.00- and 4.53-fold, respectively. (F) When compared to
non-treated cells, the increased β6 protein expression induced by
TGF-β1, LPA or TGF-β1+LPA was 2.68-, 2.76- and 4.29-fold,
respectively. Data are presented as the mean ± standard error of
the mean for three independent experiments performed in triplicate
with similar results. *P<0.05 and **P<0.01 vs. control.
TGF-β1, transforming growth factor-β1; LPA, lysophosphatidic acid;
Itgβ6/β6, integrin β6; PCR, polymerase chain reaction. |
TGF-β1 and LPA activate the Itgβ6 gene
promoter
To confirm that TGF-β1 and LPA induce
transcriptional activation of the human Itgβ6 gene, the Itgβ6
promoter was cloned and luciferase reporter gene assays measured
the activity by the generation of a series of Itgβ6 promoter
deletion mutants. Hep-3B cells were transiently transfected with
pGLB-112, pGLB-207, pGLB-359, pGLB-630, pGLB-FL, and
pGL2-basic (pGLB, control). The pGLB-630 (−458/+172 bp
region) was identified to stimulate the maximal luciferase activity
(Fig. 4A). Sequence deletion from
−458 to −187 bp resulted in an ~50% reduction in luciferase
activity. To investigate the role of the specific regions in
stimulation of Itgβ6 promoter activity, the region from −470 to
−157 bp (containing −458 to −187 bp) was amplified, and additional
deletion constructs were generated for the region and named pGLB-A
(−470/-157 bp), pGLB-B (−388/-157 bp), and pGLB-C (−326/-157 bp).
Hep-3B cells were transiently transfected with the plasmids
containing these deletion constructs. The transfected cells treated
with TGF-β1 or LPA markedly increased luciferase activity (Fig. 4B), thus confirming that TGF-β1 and LPA
trigger transcriptional activation of the Itgβ6 gene promoter.
Notably, the −326 to −157 bp proximal fragment was identified to be
the minimum sequence that retained the full response to TGF-β1 or
LPA. Transcriptional Element Search System (TESS; http://www.cbil.upenn.edu/tess) analysis
indicated that the cloned fragment contained several well-defined
transcription factor-binding sites, including Activator protein
(AP-1), transcriptional activator (c-Myb), hepatocyte nuclear
factor (HNF)-1/HNF-1B, serum response factor (SRF), and Yin Yang 1
(YY1) that have been implicated in hepatocarcinogenesis (Fig. 4C).
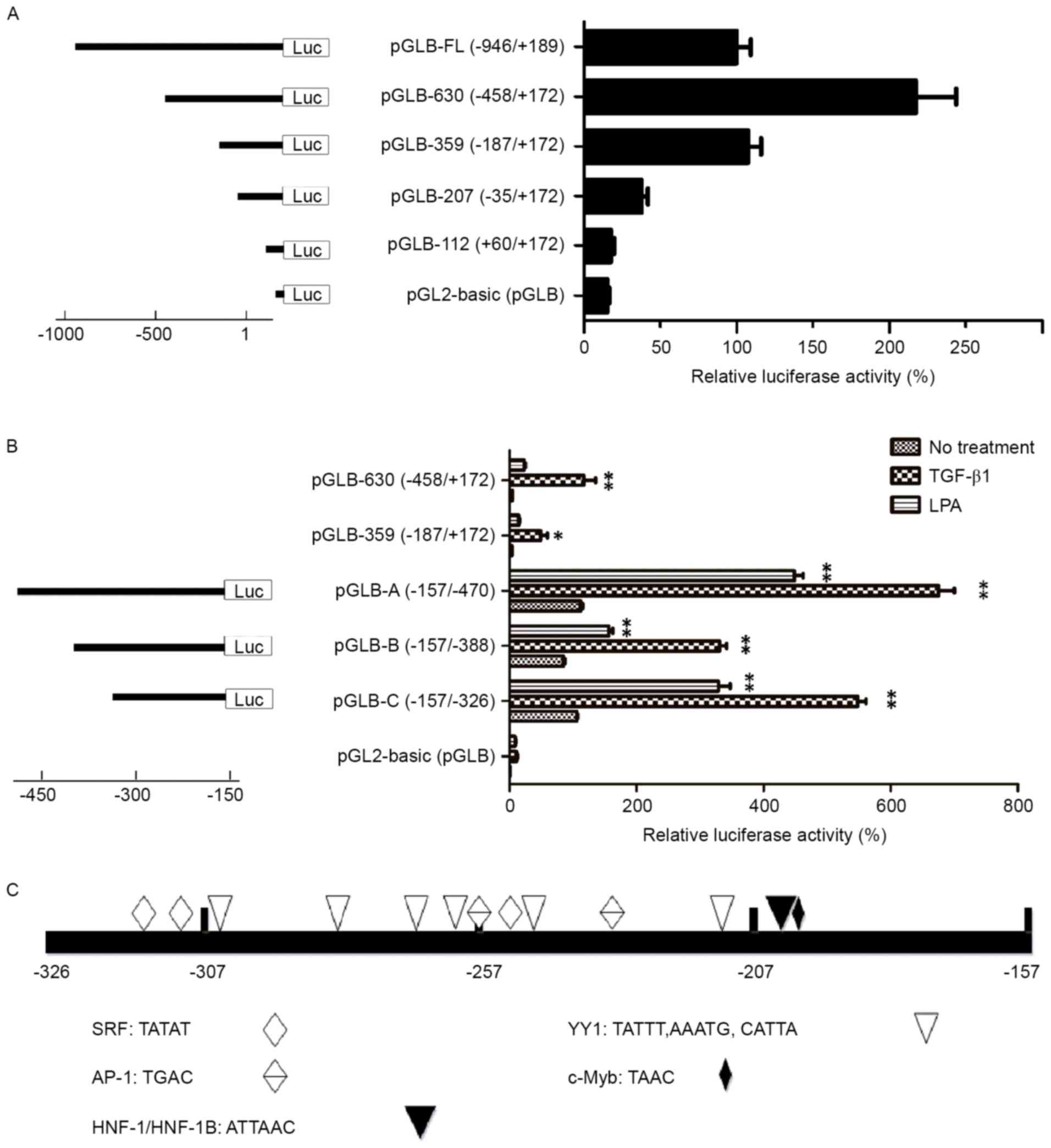 | Figure 4.TGF-β1 and LPA activates the Itgβ6
gene promoter. (A) Deletion analysis of the Itgβ6 gene promoter:
Hep-3B cells were transfected with luciferase reporter constructs
containing different lengths of the β6 promoter as indicated. In
the right column, luciferase activity was converted to percent
activity normalized to cells transfected with the construct
containing the full-length Itgβ6 promoter (−946 to +189 bp),
defined as 100%. The pGLB-630 (−458/+172 bp region) was identified
to exhibit maximum luciferase activity. Data are the mean ± SEM of
triplicate assays, representative of three independent experiments.
(B) Hep-3B cells were transfected with luciferase reporter
constructs containing the −470 to −157 bp fragment (pGLB-A) or its
deletion forms, pGLB-B (−388/-157 bp) and pGLB-C (−326/-157 bp).
Fold increases in luciferase activity induced by TGF-β1 and LPA in
transfected cells were determined using one-way analysis of
variance with *P<0.05 and **P<0.01 vs. control (no
treatment). Data are the mean ± SEM of triplicate assays,
representative of two independent experiments. (C) TESS analysis
indicated that the cloned fragment harbored certain well-defined
transcription factor-binding sites: AP-1, c-Myb, HNF-1/HNF-1B, SRF
and YY1; these were implicated in hepatocarcinogenesis. AP-1,
Activator protein; c-Myb, transcriptional activator; HNF-1/-1B,
hepatocyte nuclear factor −1/-1B; SRF serum response factor; YY1,
Yin Yang 1; TGF-β1, transforming growth factor-β1; LPA,
lysophosphatidic acid; Itgβ6/β6, integrin β6; SEM, standard error
of the mean. |
Discussion
In the present study, the expression levels of Itgβ6
were evaluated using hepatocellular carcinoma. Itgβ6 expression was
increased in HCC cell lines and tissue samples compared with normal
liver cells and liver tissue; TGF-β1 and LPA induced Itgβ6
expression and activated the Itgβ6 gene promoter. The positive
regulatory region of the promoter at −326 to −157 bp was identified
by deletion analysis.
Itgβ6 was expressed in human HCC tissues and the
Hep-3B hepatocellular carcinoma cell line, while Itgβ6 expression
was absent in human normal liver tissue and the normal HL-7702
hepatocyte cell line. These results contrast the results
demonstrated by Patsenker et al (14), which demonstrated the complete absence
of Itgβ6 in HCC in samples obtained from the Department of Visceral
Surgery and Medicine, University of Bern (Bern, Switzerland). The
difference in the expression of Itgβ6 may be due to race and
hepatitis infection status. For example, hepatitis C virus is
involved in the development of HCC in the Swiss population, while
hepatitis B virus-associated HCC occurs at a higher rate in China
(31,32).
A previous study indicated that the expression of
Itgβ6 was associated with poor prognosis and survival in colorectal
carcinoma (33). However, in
non-small lung carcinoma, expression of Itgβ6 was predominantly
observed in well-differentiated tumors, and considered to be an
indicator of good prognosis (34). In
the present study, there was no significant correlation between
Itgβ6 immunoreactivity and CLIP stage in patients with HCC, which
may be due to the small group size (n=23).
TGF-β1 and LPA treatment in Hep-3B cells increased
Itgβ6 mRNA and protein expression. Notably, the additive
stimulatory effect of the combination of TGF-β1 and LPA suggests a
potential for multiple mechanisms regulating Itgβ6 expression.
These results are consistent with previous studies that
demonstrated TGF-β1 and LPA may facilitate the development of human
HCC (22–26).
Although it has been well-established that Itgβ6 is
implicated in numerous aspects of cancer cell biology (13–19,35–37),
the transcription factors and signal transduction pathways involved
remain poorly understood. The present study describes experiments
investigating the Itgβ6 promoter. Truncation experiments indicated
that the cloned human Itgβ6 promoter region (−946/+189, 1,135 bp)
contained a positive regulatory region from −326 to −157 bp. TESS
analysis demonstrated that this region harbored certain
well-defined transcription factor-binding sites that are associated
with hepatocarcinogenesis, including AP-1, c-Myb, HNF-1/HNF-1B,
SRF, and YY1.
Additionally, activation of Itgβ6 by TGF-β1 and LPA
were revealed to significantly increase Itgβ6 promoter activity,
therefore confirming that TGF-β1 and LPA triggered transcriptional
activation of the Itgβ6 gene promoter. This observation was in
accordance with induced increases in Itgβ6 mRNA and protein levels
by TGF-β1 and LPA in Hep-3B cells, and highlights the possibility
that TGF-β1 and LPA may contribute to the transcriptional
activation of Itgβ6 through the regulation of the aforementioned
transcription factor-binding sites. In conclusion, to the best of
our knowledge, the present study is the first to identify Itgβ6
expression in HCC. TGF-β1 and LPA induced the Itgβ6 expression in
Hep-3B cells and activated the Itgβ6 gene promoter. Future studies
will include examination of therapeutic strategies for
hepatocellular carcinoma by Itgβ6 genetic intervention.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 30900661).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RX, MX, GC and YF made substantial contributions to
conception and design, and designed the outline and revised the
manuscript. RX, MX, XD, HH, XC and WH contributed to the
acquisition and analysis of data for the study. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The Ethical Committees of the Second Affiliated
Hospital of Shantou Medical College and the Medical College of
Shantou University approved the present study, and all patients
provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wilson JF: Liver cancer on the rise. Ann
Intern Med. 142:1029–1032. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al: Sorafenib in advanced hepatocellular carcinoma. N Engl J
Med. 359:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sherman M: Hepatocellular Carcinoma:
Epidemiology, surveillance, and diagnosis. Semin Liver Dis.
30:3–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nordenstedt H, White DL and El-Serag HB:
The changing pattern of epidemiology in hepatocellular carcinoma.
Dig Liver Dis. 42 Suppl 3:S206–S214. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hynes RO: Integrins: A family of cell
surface receptors. Cell. 48:549–554. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Giancotti FG and Ruoslahti E: Integrin
signaling. Science. 285:1028–1032. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hynes RO: Integrins: Bidirectional,
allosteric signaling machines. Cell. 110:673–687. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Patsenker E, Popov Y, Stickel F, Jonczyk
A, Goodman SL and Schuppan D: Inhibition of integrin alphavbeta6 on
cholangiocytes blocks transforming growth factor-beta activation
and retards biliary fibrosis progression. Gastroenterology.
135:660–670. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Breuss JM, Gallo J, DeLisser HM,
Klimanskaya IV, Folkesson HG, Pittet JF, Nishimura SL, Aldape K,
Landers DV, Carpenter W, et al: Expression of the beta 6 integrin
subunit in development, neoplasia and tissue repair suggests a role
in epithelial remodeling. J Cell Sci. 108:2241–2251.
1995.PubMed/NCBI
|
|
11
|
Breuss JM, Gillett N, Lu L, Sheppard D and
Pytela R: Restricted distribution of integrin beta 6 mRNA in
primate epithelial tissues. J Histochem Cytochem. 41:1521–1527.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Busk M, Pytela R and Sheppard D:
Characterization of the integrin alpha v beta 6 as a
fibronectin-binding protein. J Biol Chem. 267:5790–5796.
1992.PubMed/NCBI
|
|
13
|
Yang GY, Xu KS, Pan ZQ, Zhang ZY, Mi YT,
Wang JS, Chen R and Niu J: Integrin alpha v beta 6 mediates the
potential for colon cancer cells to colonize in and metastasize to
the liver. Cancer Sci. 99:879–887. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Patsenker E, Wilkens L, Banz V,
Osterreicher CH, Weimann R, Eisele S, Keogh A, Stroka D, Zimmermann
A and Stickel F: The alphavbeta6 integrin is a highly specific
immunohistochemical marker for cholangiocarcinoma. J Hepatol.
52:362–369. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ahmed N, Pansino F, Clyde R, Murthi P,
Quinn MA, Rice GE, Agrez MV, Mok S and Baker MS: Overexpression of
alpha(v)beta6 integrin in serous epithelial ovarian cancer
regulates extracellular matrix degradation via the plasminogen
activation cascade. Carcinogenesis. 23:237–244. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hecht JL, Dolinski BM, Gardner HA,
Violette SM and Weinreb PH: Overexpression of the alphavbeta6
integrin in endometrial cancer. Appl Immunohistochem Mol Morphol.
16:543–547. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang ZY, Xu KS, Wang JS, Yang GY, Wang W,
Wang JY, Niu WB, Liu EY, Mi YT and Niu J: Integrin alphanvbeta6
acts as a prognostic indicator in gastric carcinoma. Clin Oncol (R
Coll Radiol). 20:61–66. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Azare J, Leslie K, Al-Ahmadie H, Gerald W,
Weinreb PH, Violette SM and Bromberg J: Constitutively activated
Stat3 induces tumorigenesis and enhances cell motility of prostate
epithelial cells through integrin beta 6. Mol Cell Biol.
27:4444–4453. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marsh D, Dickinson S, Neill GW, Marshall
JF, Hart IR and Thomas GJ: alpha vbeta 6 Integrin promotes the
invasion of morphoeic basal cell carcinoma through stromal
modulation. Cancer Res. 68:3295–3303. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu MY, Porte J, Knox AJ, Weinreb PH, Maher
TM, Violette SM, McAnulty RJ, Sheppard D and Jenkins G:
Lysophosphatidic acid induces alphavbeta6 integrin-mediated
TGF-beta activation via the LPA2 receptor and the small G protein G
alpha(q). Am J Pathol. 174:1264–1279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Geng H, Lan R, Singha PK, Gilchrist A,
Weinreb PH, Violette SM, Weinberg JM, Saikumar P and Venkatachalam
MA: Lysophosphatidic acid increases proximal tubule cell secretion
of profibrotic cytokines PDGF-B and CTGF through LPA2- and
Gαq-mediated Rho and αvβ6 integrin-dependent activation of TGF-β.
Am J Pathol. 181:1236–1249. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu K, Ding J, Chen C, Sun W, Ning BF, Wen
W, Huang L, Han T, Yang W, Wang C, et al: Hepatic transforming
growth factor beta gives rise to tumor-initiating cells and
promotes liver cancer development. Hepatology. 56:2255–2267. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Balzarini P, Benetti A, Invernici G,
Cristini S, Zicari S, Caruso A, Gatta LB, Berenzi A, Imberti L,
Zanotti C, et al: Transforming growth factor-beta1 induces
microvascular abnormalities through a down-modulation of neural
cell adhesion molecule in human hepatocellular carcinoma. Lab
Invest. 92:1297–1309. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kitisin K, Ganesan N, Tang Y, Jogunoori W,
Volpe EA, Kim SS, Katuri V, Kallakury B, Pishvaian M, Albanese C,
et al: Disruption of transforming growth factor-beta signaling
through beta-spectrin ELF leads to hepatocellular cancer through
cyclin D1 activation. Oncogene. 26:7103–7110. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang R, Zhang Z, Pan X, Huang X, Huang Z
and Zhang G: ATX-LPA axis induces expression of OPN in hepatic
cancer cell SMMC7721. Anat Rec (Hoboken). 294:406–411. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Park SY, Jeong KJ, Panupinthu N, Yu S, Lee
J, Han JW, Kim JM, Lee JS, Kang J, Park CG, et al: Lysophosphatidic
acid augments human hepatocellular carcinoma cell invasion through
LPA1 receptor and MMP-9 expression. Oncogene. 30:1351–1359. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mills GB and Moolenaar WH: The emerging
role of lysophosphatidic acid in cancer. Nat Rev Cancer. 3:582–591.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
A new prognostic system for hepatocellular
carcinoma: A retrospective study of 435 patients: The cancer of the
liver Italian program (CLIP) investigators. Hepatology. 28:751–755.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Knoche K and Kephart D: Cloning blunt-end
PfuDNA Polymerase-generated PCR fragments into pGEM®-T
vector systems. Promega Notes. 71:101999.
|
|
31
|
Kuske L, Mensen A, Müllhaupt B, Negro F,
Semela D, Moradpour D, Male PJ, Heim MH, Malinverni R, Cerny A and
Dufour JF: Swiss Hepatitis C Cohort Study: Characteristics of
patients with chronic hepatitis C who develop hepatocellular
carcinoma. Swiss Med Wkly. 142:w136512012.PubMed/NCBI
|
|
32
|
Tan YJ: Hepatitis B virus infection and
the risk of hepatocellular carcinoma. World J Gastroentero.
17:4853–4857. 2011. View Article : Google Scholar
|
|
33
|
Bates RC, Bellovin DI, Brown C, Maynard E,
Wu B, Kawakatsu H, Sheppard D, Oettgen P and Mercurio AM:
Transcriptional activation of integrin beta6 during the
epithelial-mesenchymal transition defines a novel prognostic
indicator of aggressive colon carcinoma. J Clin Invest.
115:339–347. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Smythe WR, LeBel E, Bavaria JE, Kaiser LR
and Albelda SM: Integrin expression in non-small cell carcinoma of
the lung. Cancer Metastasis Rev. 14:229–239. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Yang Y, Hu Y, Dang D, Regezi J,
Schmidt BL, Atakilit A, Chen B, Ellis D and Ramos DM:
Alphavbeta6-Fyn signaling promotes oral cancer progression. J Biol
Chem. 278:41646–41653. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thomas GJ, Nyström ML and Marshall JF:
Alphavbeta6 integrin in wound healing and cancer of the oral
cavity. J Oral Pathol Med. 35:1–10. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ramos DM, Dang D and Sadler S: The role of
the integrin alpha v beta6 in regulating the epithelial to
mesenchymal transition in oral cancer. Anticancer Res. 29:125–130.
2009.PubMed/NCBI
|