Introduction
Colorectal carcinoma (CRC) is the most common of the
gastrointestinal malignancies, and the second leading cause of
cancer death in most countries. Colon cancer tissues commonly
produce secreted and cell surface-bound mucins (1), which are glycoproteins carrying large
numbers of O-linked oligosaccharides, accounting for up to 80% of
the molecular mass. Alteration in O-glycan profiles is a
hallmark of cancer development, which determines the expression of
truncated O-glycosylated tumor-associated antigens (2,3). These
glycoproteins are involved in fundamental biological processes,
such as invasion and metastasis (4)
as well as in the epithelial-mesenchymal transition (EMT) process
(5). Mucins of human colon cancer
cells commonly express several types of short O-glycan
antigens, that can be identified using monoclonal antibodies, but
not detected in normal colorectal cells (6). The most extensively characterized are
Tn, STn and T antigens (7,8), but the expression by colon cancer
tissues of others unusual truncated O-glycans, such the Tk
antigen (9) and the core6 structure
(10) was also reported.
Cancer-associated mucin antigens can be exploited in diagnosis and
prognosis (3), and for the
development of cancer vaccines (11,12). The
synthesis of O-linked glycosylation is started in the Golgi
apparatus by the covalent linkage of an α-N-acetylgalactosamine
residue (GalNAc) to the hydroxyl group of Ser/Thr residues. This
reaction is catalyzed by the family of
UDP-N-acetyl-D-galactosamine:polypeptide
N-acetylgalactosaminyltransferase (GalNAc-Ts, EC 2.4.2.41) composed
at least by 20 members in humans (13). Following the synthesis of
GalNAcα-O-Ser/Thr (Tn antigen) other sugar residues are
incorporated. In total, 8 mucin-type core structures can be
distinguished, depending on the second sugar and its sugar linkage,
in a process controlled by specific glycosyltransferases (14).
The colon mucus layer is formed by polymerized
mucins, primarily MUC2, that are produced by goblet cells (15). Unlike core 1-derived O-glycans
that are present in most tissues (16), core 3-derived O-glycans are
expressed predominantly in normal colonic epithelial cells
(17). Several evidences support the
concept that normal mucins and O-glycans are key components
in colon tissues and that defects in their expression may be
associated with an increase in the susceptibility for inflammatory
diseases and cancer development (18,19). For
example: i) The loss of core 1-derived O-glycans led to a
rapid induction of severe spontaneous colitis in mice by 2 weeks
after birth (20); ii) the absence of
membrane-bound mucin Muc1 leads to the exacerbation of chronic
inflammations in both Th1-mediated and Th2-mediated colitis models
(21); iii) Muc2-deficient mice
(Muc2-/-) developed adenomas at 6 months of age, which progressed
to invasive adenocarcinoma in the small intestine as well as rectal
tumors at an older age (22),
suggesting that Muc2 play a role in the suppression of intestinal
cancer; iv) mice lacking core 3
β1,3-N-acetylglucosaminyltransferase (C3GnT), an important enzyme
for the synthesis of core 3-derived O-glycans, exhibited an
increased susceptibility to experimental colitis and colorectal
adenocarcinoma (23); v) in addition,
it was found that inactivating somatic and germline mutations in
the gene encoding for GalNAc-T12 (GALNT12) (a gene highly
expressed in the normal colon), are associated with colon cancer
development (24).
One of the underlying causes of glycosylation
changes in cancer is deregulated expression or localization of
glycosyltransferases and associated proteins within the tumor cell.
In CRC mutations in several genes of glycosylation pathway has been
demonstrated (25). Among
glycosyltransferases, GalNAc-Ts have been found to be
differentially expressed in malignant tissue compared to normal
tissue (26–28). Numerous evidences support the role of
some GalNAc-Ts in cancer biology (29). It was found that overexpression of
GALNT3 promotes pancreatic cancer cell growth (30). Increasing evidences suggest that these
enzymes might be useful tumor markers. For example, GalNAc-T3
expression has been shown to correlate with prognosis in patients
with lung (31,32) and gallbladder cancer (33). GalNAc-T5 expression was markedly
reduced in gastric cancer tissues compared with non-malignant
gastric mucosa and was an independent prognostic marker for the
overall survival of gastric cancer patients (34). GALNT14 expression was highly
associated with lower recurrence-free survival in non-small cell
lung cancer (NSCLC) patients (35).
GalNAc-T14 also promotes invasive properties of lung adenocarcinoma
cells through Wnt dependent HOXB9 expression (36). We demonstrated that GALNT6
expression in bone marrow samples correlated with poor clinical
outcome in lymph node-negative breast cancer patients (37). We have also shown, in a human
neuroblastoma model, that GALNT13 was the most strongly
up-regulated gene in metastatic neuroblasts compared with primary
tumor xenograft (38). In the same
study, we demonstrated that GALNT13 expression in bone
marrow of neuroblastoma patients at diagnosis was a strong
predictor of poor clinical outcome. In contrast we found that the
brain specific gene GALNT9 is present in neuroblasts derived
from primary tumor but absent in bone marrow metastatic ones. In a
cohort of 122 neuroblatoma patients, GALNT9 expression in
primary tumor was associated with higher overall survival,
independently of the standard risk-stratification covariates
(39). In this context GalNAc-Ts are
emerging as novel prognostic markers and potential new targets for
tumor treatments.
Expression of GalNAc-T1, -T2, -T3, -T4 and -T12 was
reported in normal colon tissues (13,40–42).
Several observations support an important role of some of these
isoenzymes in colon carcinogenesis and tumor behavior. For example,
some evidences confer to GalNAc-T12 a protective role against colon
cancer development (24). Loss of
GalNAc-T3 expression was correlated with a higher metastatic
potential in a mouse colon cancer model (43) and, in the same way, GalNAc-T3
expression in human CRC significantly enhanced the likelihood of
patient survival (44). GalNAc-T6
expression was found in LS174T and T84 human colon cancer cell
lines (45,46) but not in normal human colon cells
(47). Our aim in the present work
was to evaluate the potential role of this enzyme as an
immunohistochemical colon cancer marker. We found that GalNAc-T6
expression in colon cancer is an independent prognostic factor
indicating better overall survival.
Materials and methods
Cell culture
Human colon cancer cell lines HT29 (ATCC
HTB38™), SW480 [SW-480] (ATCC CCL228™) and
SW620 [SW620] (ATCC CCL227™) were cultured in Dulbecco's Modified
Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum
(FBS), 2 mM L-glutamine and 1 mM pyruvate. Subculture of cells
grown in monolayer was carry out after washing once with PBS,
incubating them with 0.53 mM EDTA and 0.05% trypsin in PBS (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) for 5 min at
37°C. Cell pellets were washed in PBS, resuspended in 1 ml of
Tri-Reagent (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and
stocked at −80°C until use.
Patient information and tumor
specimens
Eighty-one formalin fixed paraffin-embedded colon
cancer tissues were studied. Twenty-eight primary tumors with
histopathological diagnosis of colon cancer (all stages), as well
as 10 normal colon tissues from distal or proximal resection margin
and 8 adenomas with different degrees of dysplastic lesions were
obtained from the Department of Pathology, Maciel Hospital,
Montevideo. Approval from the Institutional Ethical Committee at
the University de la Republica (Comité de Etica para Proyectos de
Investigación, Facultad de Medicina, Universidad de la República)
was obtained prior to beginning. All participants provided signed
written informed consent previous to enrollment in the study.
Furthermore, 53 came from a commercial human colorectal cancer
tissue-array (IMH-306; Imgenex Corporation, San Diego, CA,
USA).
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was extracted from colon cancer cell lines
with Tri-Reagent (Sigma-Aldrich; Merck KGaA) according to the
manufacturer's instructions. Two µg of total RNA were included for
first strand cDNA synthesis by using 200 units of M-MLV reverse
transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) in the
presence of 2 µl 10 mM of each deoxynucleotide triphosphate (dNTPs)
and 200 ng of random hexamers (Thermo Fisher Scientific, Inc.) in a
20 µl total reaction volume. After incubation at 37°C for 1 h, the
mixture was heated to 70°C, snap-cooled and stored at −20°C.
Amplification of a 499 bp fragment of GALNT6 transcripts was
performed using the follow specific primers:
5′-TCCAAATCAGGGCTCCAGAAG-3′ and 5′-CACCTGCAGCTGCTTCACGTAC-3′
(accession no.: Y08565). The PCR mixture (total reaction volume of
25 µl) includes 1× enzyme buffer, 2.5 mM MgCl2, 200 µM
dNTPs, 400 nM of each primer and 1 unit of Taq DNA polymerase
(Thermo Fisher Scientific, Inc.). Amplification was performed for
35 cycles under the following conditions: 30 sec at 94°C, 30 sec at
60°C and 1 min at 72°C. Fifteen µl of PCR products were analyzed by
electrophoresis on 2% agarose gels by direct visualization after
ethidium bromide staining. In order to verify cDNA quality, a 596
bp fragment of β2-microglobulin transcripts was amplified under
same conditions using the primers 5′-ATGTCTCGCTCCGTGGCCTTAG-3′ and
5′-AAGTTGCCAGCCCTCCTAGAGC-3′ (accession no.: AB021288).
Immunofluorescence microscopy
Cells plated on glass coverslips were washed with
PBS, fixed in methanol-acetone 50% for 10 min and stored a −20°C
until use. Coverslips were then defrosted, rehydrated in PBS, and
blocked in 30% goat serum for 20 min. Anti-GalNAc-T6 MAb T6.3
(27) (10 µg/ml) was incubated for 1
hr at room temperature, followed by three washes for 5 min each in
PBS. Secondary antibody Alexa FluorR 488 goat-anti mouse IgG
(A11029; Invitrogen; Thermo Fisher Scientific, Inc.) was incubated
for 1 h at room temperature and after three washes, monolayers were
counterstained with DAPI 1 µg/ml, mounted in PBS-glycerol 50% and
analyzed by epifluorescence microscopy.
Analysis of GalNAc-T6 expression on
cancer cell lines by flow cytometry
Cells were fixed-permeabilized (4% PFA, 1% FBS, 0.1%
Tween 20) and incubated with anti-GalNAc-T6 (T6.3) monoclonal
antibody. The specific binding of primary MAb T6.3 to the cell
lines was developed with an anti-mouse polyclonal antibody
FITC-conjugated (Sigma-Aldrich; Merck KGaA) and further analyzed
using a CyAnTM ADP Flow Cytometer (Beckman Coulter, Inc., Brea, CA,
USA) and Summit v4.3 software. For each analysis 10,000 counts,
gated on a FSC vs SSC dot plot excluding doublets populations, were
recorded. Results were expressed as percentage of FITC positive
cells and FITC mean fluorescence intensity (MFI). Data were
expressed as the mean +/-standard deviation. Statistical analysis
was determined using one-way analysis of variance (ANOVA) and
consequently the Tukey's Multiple Camparison test using GraphPad
Prism Sofware v5.00 Demo (GraphPad Software, Inc., La Jolla, CA,
USA).
Immunohistochemistry
Parafin-embedded sections were prepared following
standard protocols. Sections were deparaffinized and rehydrated,
followed by endogenous peroxidase blocking with 3%
H2O2 in PBS for 30 min at room temperature.
After three washes, tissues were blocked with 10% goat serum, 1%
BSA in PBS for 30 min at room temperature, followed by first
antibody incubation at 4°C overnight (MAb T6.3 diluted at 10 µg/ml
in PBS, 0.1% Tween 20, 1% BSA). For every assay, a negative control
was performed omitting MAb T6.3. A well characterized strong
positive tumor was added in each experiment in order to ensure
reproducibility of technical conditions. After washing with PBS,
sections were incubated for 1 h at room temperature with a
polyclonal rabbit anti-mouse IgG peroxidase conjugated (Dako,
catalogue no.: P 0161) diluted at 1:100 in PBS. Staining reaction
was performed with 3,3-diaminobenzidine (DAB) 0.5 mg/ml in TBS,
0.3% hydrogen peroxide for 30 min at room temperature. Sections
were then counterstained in Mayer's hematoxylin, dehydrated in
ethanol and xylene, and mounted. The immunostaining frequency for
each tumor was scored, as previously described (27), based on a 0–3 scale for staining
extension [(0) for negative samples or <10% stained tumor
tissue; (1) for samples stained
between 10 and 39% of tumor tissue; (2) for tumor tissues stained between 40 and
79%; and (3) for tumors with 80% or
more of stained tumor cells]. Signal intensity was scored as strong
(3), moderate (2), weak (1),
and null (0). Total immunostaining score was obtained adding up
both parameters in a 0–6 scale. Scores were established jointly by
four observers (LU, DM, EO, NB) in a penta-head microscope.
Statistical analysis
Statistical analysis for FACS experiments was
determined using One-way analysis of variance (ANOVA) followed by
Tukey's Multiple Camparison test using GraphPad Prism Sofware v5.00
(GraphPad Software, Inc.). Results are from three independent
experiments and P<0.05 was considered to indicate a
statistically significant difference. Univariate survival analysis
was performed using the Kaplan-Meier method and compared with the
log-rank test. The cut-off for GalNAc-T6 expression was established
as negative (0–1) and positive (2–6). Hazard
ratios and 95% confidence intervals (95% CIs) were estimated using
univariate or multivariate Cox proportional-hazard models. All
statistical tests were two-tailed. Proportional hazard assumption
was tested by Schoenfeld's method and plotting (-log (-log S(t))).
All statistical calculations were performed using STATA v14
(StataCorp LP, College Station, TX, USA).
Results
GalNac-T6 expression in colon cancer
cell lines
Considering that GalNAc-T6 expression was previously
found in the T84 and LS174T human colon cancer cell lines (45,46), we
evaluated here other colon cancer cells such as HT29, SW480 and
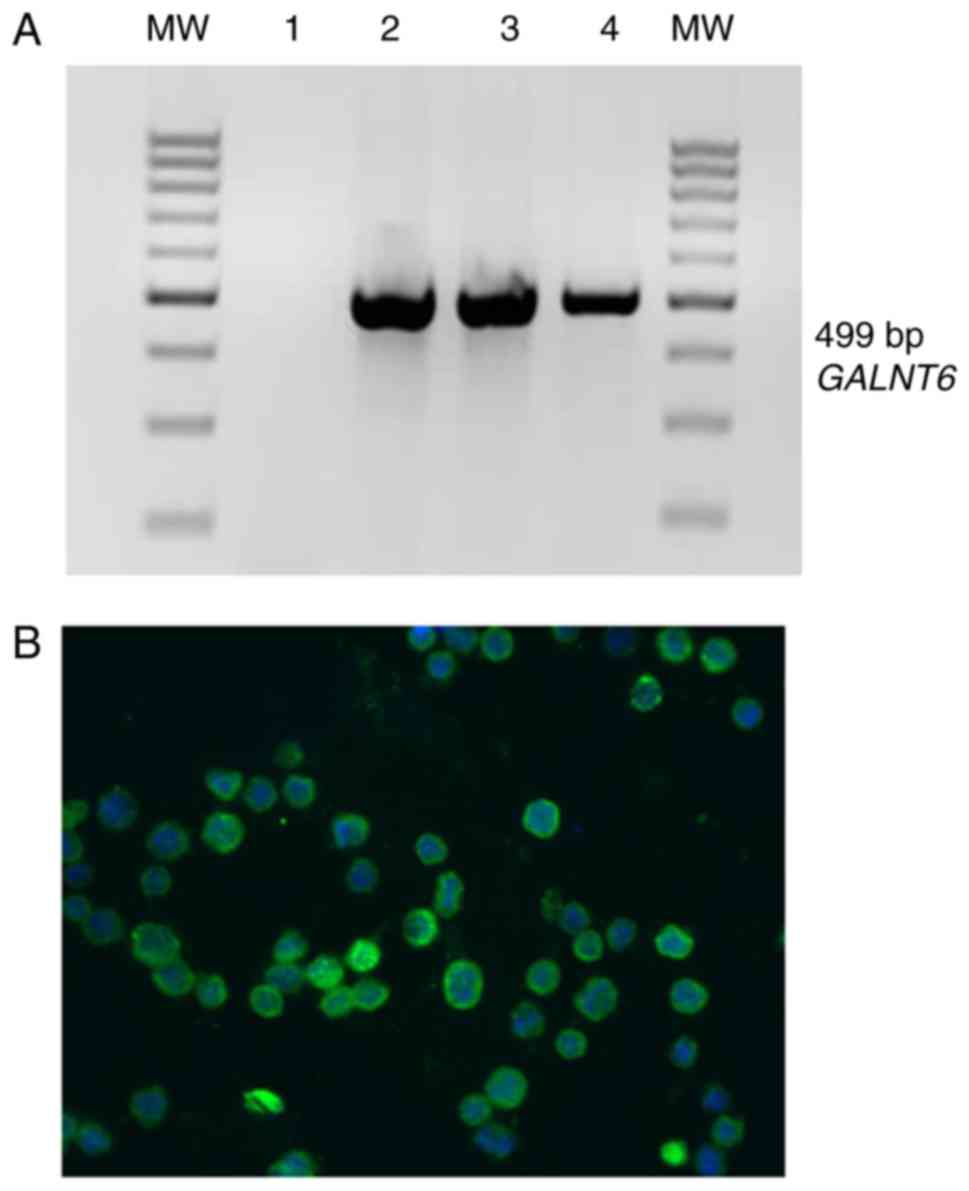
SW620. Using RT-PCR we found the mRNA encoding GalNAc-T6 in all
three cell lines (Fig. 1A), and these
results were confirmed at protein level by immunofluorescence
staining, observing a marked expression of GalNAc-T6 (Fig. 1B). The percentage of GalNAc-T6
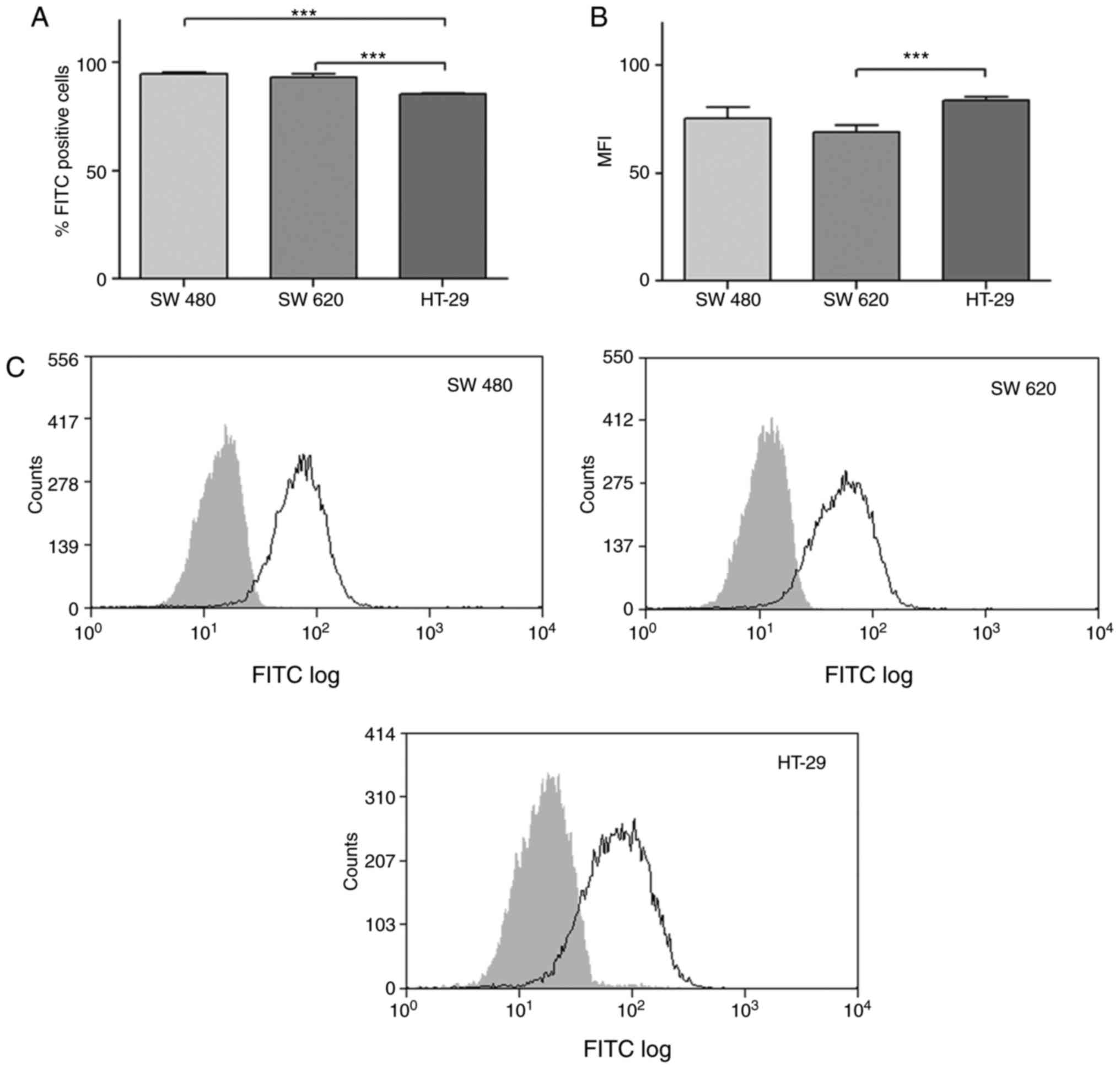
positive cells was evaluated by flow cytometry (Fig. 2). We found that most colon cancer
cells expressed this enzyme: SW480 (96.1%), SW620 (96.1%) and HT-29
(85.4%). The MFI (mean fluorescence intensity) value of
GalNAc-T6/FITC positive cells was comparable among these cell
lines: SW480 (75.3), SW620 (69.3) and HT-29 (83.8).
GalNAc-T6 is expressed in colon cancer
but not in normal colon
The study included 81 patients (53 men and 28 women)
with colorectal cancer diagnosis (Table
I). The average age at surgery was 62 years (range 35–86). Most
patients (88.8%) had operable disease at diagnosis and most tumors
were graded as well to moderately differentiated type (17.3 and
72.8% respectively). Based on the American Joint Committee on
Cancer (AJCC) criteria, the majority of patients had stage II
disease (45.6%) followed by stage III disease (39.5%). MAb T6.3
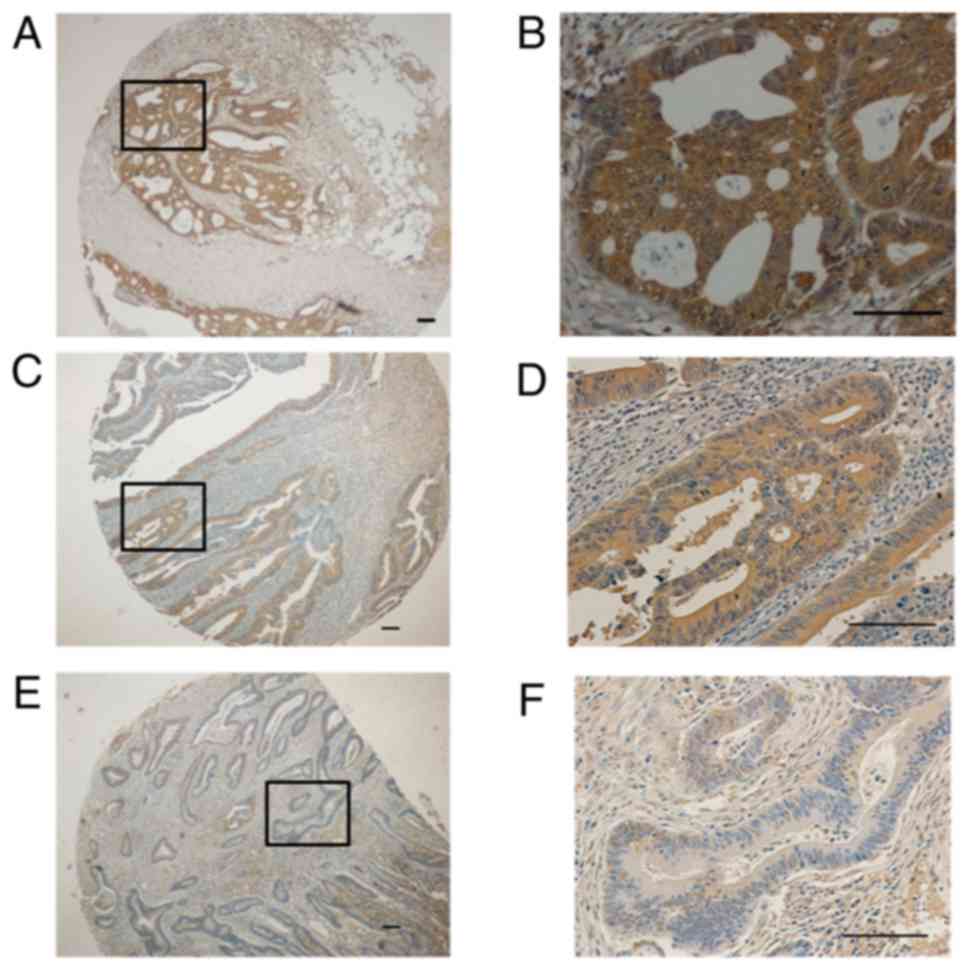
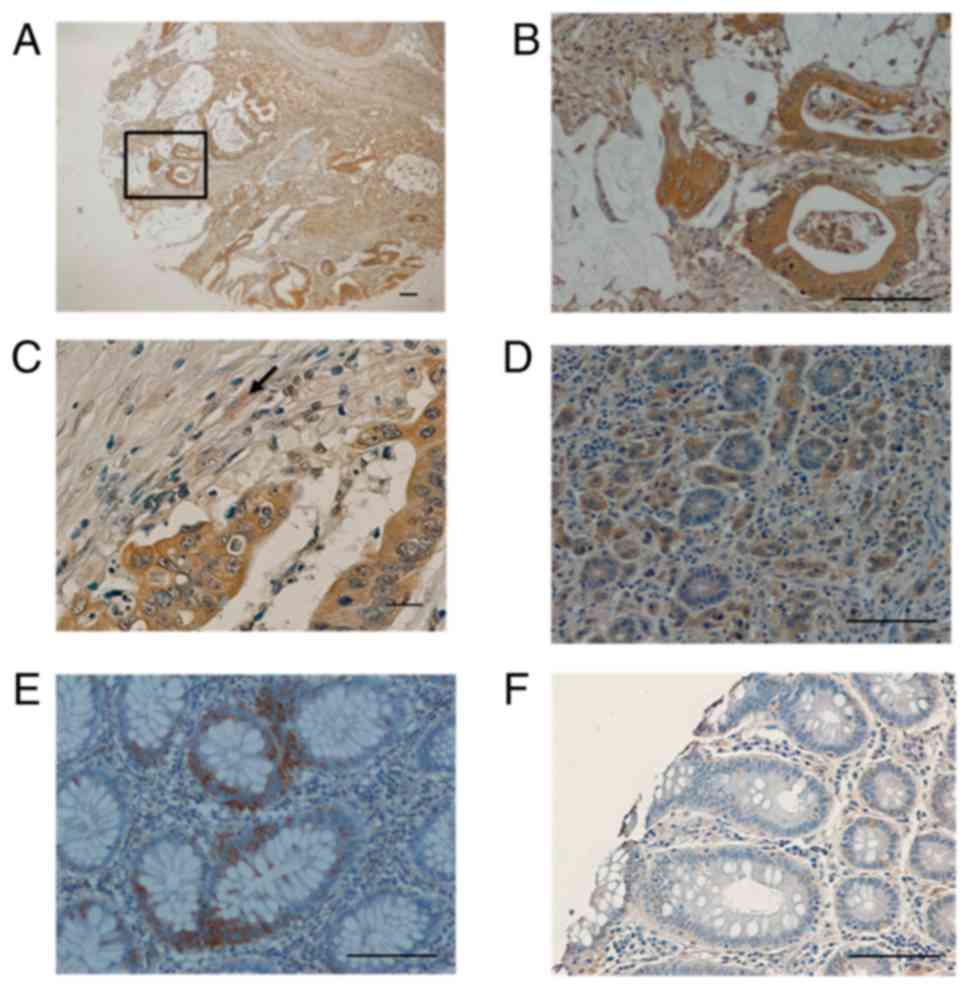
immunostaining was detected in 35 of 81 tumors (43.2%), in 8/8
specimens from adenomas, and 0/10 normal colon tissues from
proximal and distal margin of colonic surgery specimens. This
antibody always showed a diffuse cytoplasmic staining pattern with
different signal intensity (Figs. 3
and 4). Microwave treatment strongly
reduced immunostaining as seen previously for breast cancer
(27). No relationship between
GalNAc-T6 expression and clinic-pathological factors as age, sex,
histology or stage was found (Table
I). Regarding disease stage, GalNAc-T6 was expressed in 1/3
(33.3%) tumors from patient stage I, in 18/37 (48.6%) stage II, in
14/32 (43.7%) stage III and in 2/9 (22.2%) of stage IV (differences
were not statistically significant). No correlation was found
between GalNAc-T6 expression and tumor grading, either between
tumor staining intensity and the pathological parameter evaluated.
Adjacent normal tissue was observed in 16 invasive cancers, and 5
(16%) showed a focal minimal GalNAc-T6 expression, being all other
completely negative (Fig. 4D and F).
All ten normal colon tissue obtained from distal or proximal
resection margins were negative 0/10 (data not shown).
 | Table I.Characteristic of patients and
tissues. |
Table I.
Characteristic of patients and
tissues.
|
Characteristics | n (%) | GalNAc-T6 positive
n (%) | P-value |
|---|
| Age, years |
|
|
|
| Median
(range) | 62 (35–86) |
|
|
|
≤60 | 37 (45.5) | 18 (48.6) |
0.36 |
|
>60 | 44 (54.5) | 17 (38.6) |
|
| Sex |
|
|
|
|
Male | 53 (65.5) | 24 (45) | 0.6 |
|
Female | 28 (34.5) | 11 (39) |
|
| Tumor stage |
|
|
|
| I | 3 (3.8) | 1 (33.3) | 0.53 |
| II | 37 (45.6) | 18 (48.6) |
|
|
III | 32 (39.5) | 14 (43.7) |
|
| IV | 9 (11.1) | 2 (22.2) |
|
| Histological
differentiation |
|
|
|
|
Well | 14 (17.3) | 8 (57) | 0.12 |
|
Moderate | 59 (72.8) | 26 (44) |
|
|
Poor | 8 (9.9) | 1 (12.5) |
|
GalNAc-T6 is an independent prognostic
indicator in colorectal cancer
Patients with GalNAc-T6 expression had significantly
longer overall postoperative survival (median not reached) compared
with those who had GalNAc-T6 negative tumors (median, 58 months,
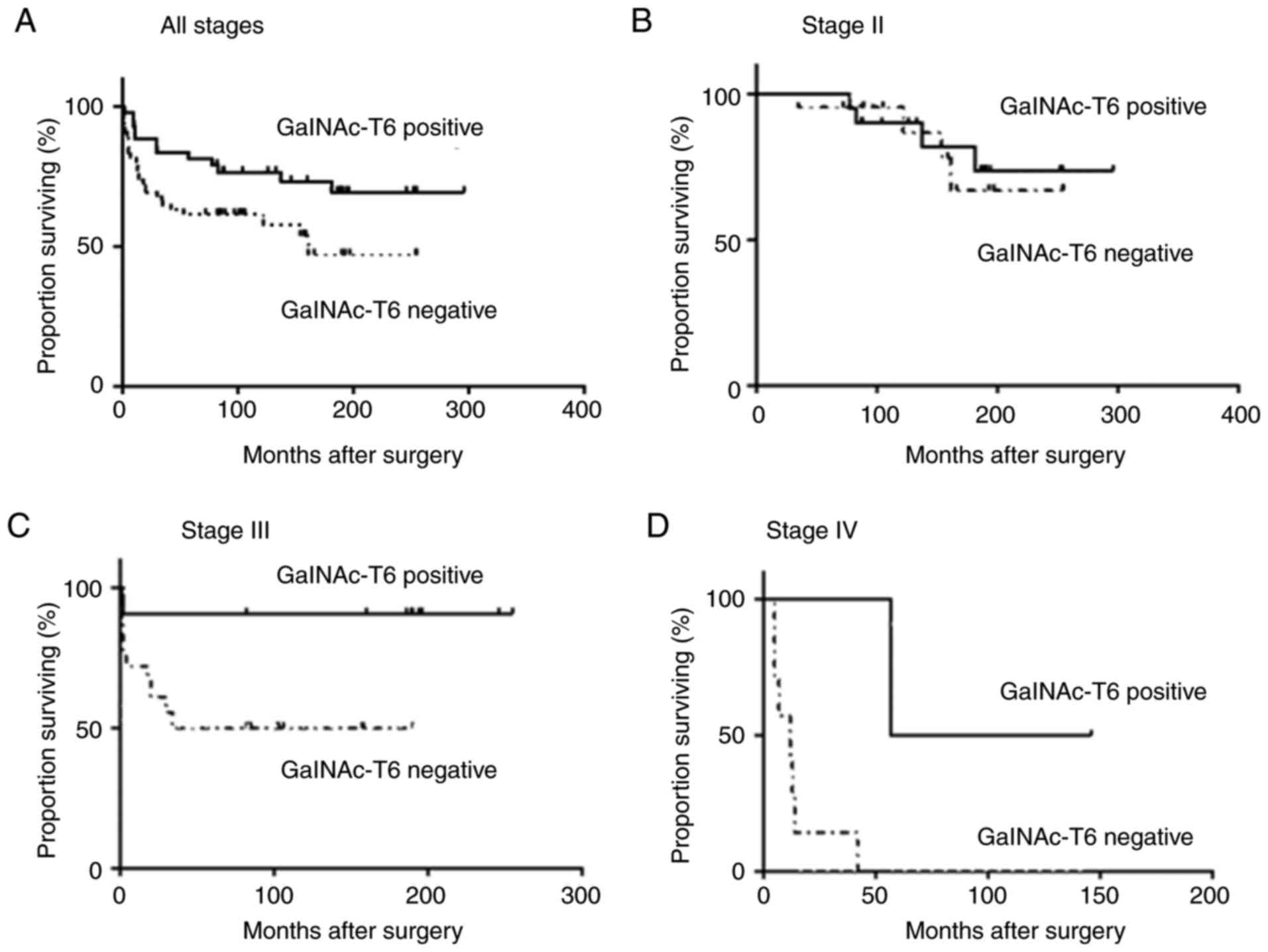
P<0.001; Fig. 5A). In the same
way, this benefit was also observed for stage III patients (median
survival not reached yet for GalNAc-T6-positive and 36 months for
GalNAc-T6-negative, P<0.03) and for stage IV, despite the low
number of cases, with a median survival of 77 months for
GalNAc-T6-positive and 12 months for GalNAc-T6-negative (P<0.02;
Fig. 5C and D respectively). No
relationship was observed between GalNAc-T6 expression and clinical
outcome in stages I and II (P=0.63). To assess whether GalNAc-T6
expression was an independent predictor of overall postoperative
survival, a Cox proportional hazards model was created. Univariate
analysis demonstrated that poor tumor differentiation, advanced
stage and GalNAc-T6-negative status were significant predictors of
poorer survival (P=0.028, 0.007, and <0.0001, respectively). Cox
model showed that patients expressing GalNAc-T6 have a hazard ratio
of 0.22 (P=0.003) adjusted by stage, as previously defined.
Patients who do not express GalNAc-T6 in their tumors seem to have
more than four times higher risk of death compared with those who
express it, even considering staging (Table II).
 | Table II.Univariate and multivariate
analysis. |
Table II.
Univariate and multivariate
analysis.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Risk factor | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Sex | 1.18 | 0.43–3.16 | 0.831 |
|
|
|
| Stage | 2.52 | 0.99–6.37 | 0.007 | 2.74 | 1.11–6.75 | 0.028 |
| Side | 1.23 | 0.51–2.97 | 0.470 |
|
|
|
| Histology | 0.31 | 0.02–3.50 | 0.029 |
|
|
|
| GalNAc-T6 | 0.14 | 0.05–0.38 | <0.0001 | 0.22 | 0.08–0.59 | 0.003 |
Discussion
The GALNT6 gene (encoding a type II
trans-membrane protein GalNAc-T6), located on chromosome 12q13, is
expressed in a restricted pattern, mainly in normal placenta,
trachea, brain, pancreas and fibroblast cells (47). Regarding its expression in cancer,
GalNAc-T6 level was found significantly higher in breast cancer
cells comparing with normal or benign mammary cells (27,48–49). Using
a RT-PCR assay, we identified GalNAc-T6 expression in bone marrow
samples related to poor clinical outcome in lymph node-negative
breast cancer patients (37). It has
been demonstrated that high expression of GalNAc-T6 in breast
cancer cells correlates with increased glycosylation of the mucin
MUC1 and knockdown of GalNAc-T6 suppressed the growth of breast
cancer cells (50). On the other
hand, O-glycosylation of fibronectin (a major constituent of the
extracelullar matrix) by overexpressed GalNAc-T6 in breast cancer
cell lines causes higher invasiveness related to an EMΤ-like
process (51). In addition, high
GalNAc-T6 expression in lung adenocarcinoma was closely related
with advanced tumor stage, and independently predicts reduced
overall survival of patients (52).
In contrast, we observed an opposite significance in colon cancer,
for which high GalNAc-T6 expression was correlated with better
outcome similar to results reported by Li et al (53), for pancreatic cancer. These apparent
contradictory observations could be explained by diverse
repertoires of protein acceptor substrates in each tumor type,
displaying different biological functions after the incorporation
of O-GalNAc residues by GalNAc-T6. Similar behavior was reported
for GalNAc-T3. High expression of this enzyme correlated with tumor
aggressiveness and poor clinical outcome in patients with
gallbladder cancer (33), renal cell
carcinomas (54) and ovarian cancer
(55), while for patients with colon
cancer (44), gastric carcinoma
(56), and lung adenocarcinoma
(32), GalNAc-T3 was a marker of good
prognosis. Sometimes this prognosis significance was related to
subcellular localization of the enzyme. Miyahara et al
(33), reported more intense staining
of GalNAc-T3 in gallbladder cancer compared with normal tissue and
benign lesions, and also distinguished two well defined staining
patterns. While non cancerous tissues always showed a granular
perinuclear staining, in gallbladder carcinomas localization was
heterogeneous, with granular or diffuse type of subcellular
distribution. Importantly, the authors found that postsurgical
survival rate of patients with diffuse-type of staining was
significantly lower than for patients with granular type. It has
been proven that following growth factors stimulation, some
GalNAc-Ts (including GalNAc-T6) might be relocalized from the Golgi
apparatus to the endoplasmic reticulum (ER), leading to modify
O-glycosylation of proteins (57).
Precision oncology is becoming increasingly
important in CRC management since the great molecular heterogeneity
determines large variations in prognosis and response to
chemotherapy of individual patients (58,59).
Certain biomarkers let to predict clinical outcome beyond staging
as well as help in treatment selection (60). The mutational status of RAS and
BRAFV600 combined with analysis of the DNA mismatch repair
system with/without CpG island methylator phenotype have shown
utility to identify colon cancer subtypes with distinct clinical
features and prognosis (61). In this
work we found that GalNAc-T6 is an independent prognosis biomarker
in colon cancer patients. This enzyme was found in the three colon
cancer cell lines evaluated (HT-29, SW480 and SW620), both at mRNA
and protein level. Immunohistochemical staining of formalin-fixed
paraffin embedded tissues using the MAb T6.3 revealed GalNAc-T6
expression in 35/81 (43%) of cases, without clinical or
histological parameters association. We did not observed GalNAc-T6
expression in normal human colon, which is in agreement with
results reported by Bennett et al (13) as well as Lavrsen et al
(62), who found GalNAc-T6 expression
in colon adenocarcinoma but not in normal colon, using another MAb
UH7 (2F3). GalNAc-T6 expression predicts improved overall survival
in both, univariate and multivariate analysis. This benefit
observed for overall population is maintained in more advance
stages (stages III and IV of AJCC), but it was not observed in
early stages (I and II of AJCC). A possible explanation for
different clinical impact between pathological stages could be the
good prognosis of low stages colorectal cancer itself (I and II)
and the higher systemic risk of advanced stages (III and IV).
Furthermore, stages III and IV have a confirmed systemic spread
(nodal or visceral), and also have a demonstrated benefit of
systemic therapeutic approaches like adjuvant (stage III) or
palliative (stage IV) chemotherapy (63), this is also suggested by the null or
marginal benefit of adjuvant chemotherapy in stages I and II
(64).
The molecular mechanism underlying the less
aggressive behavior of colon cancer cells expressing GalNAc-T6
remains to be elucidated. Several studies had revealed the role of
GalNAc-Ts in cancer biology, modulating several biological
functions as cell adhesion, invasiveness and metastasis (29). GalNAc-T3 and GalNAc-T6 are close
paralogs that exhibit very high sequence similarity throughout the
coding region, identical genomic structure and encode enzymes with
similar substrate specificities (13,47). As we
previously exposed, GalNAc-T3 is constitutively expressed in normal
colon cells. It is reasonable to think that its glycosylated
products could impact in cell-cell and cell-matrix interaction, and
its lower expression in colon cancer could affect the way that
cancer cell relates with its environment, allowing the cellular
escape and metastases. We could hypothesize that if GalNAc-T6 is
functional, it could complement the insufficient glycosylation of
the lacking GalNAc-T3. This hypothesis is reinforced by the
observation that high GalNAc-T3 expression in colon cancer is
associated with good clinical outcome (44).
It is too soon to suggest that GalNAc-T6 expression
could determine a different treatment in colorectal cancer
patients. The retrospective condition of our study, the restricted
number of cases, as well as the lack of information about
correlation with treatment´s response, is main limitation of our
work. A prospective trial in a large cohort is needed to confirm
its role as prognostic marker. Furthermore the predictive value of
response to chemotherapy or biological agents of GalNAc-T6 should
be demonstrated prospectively compared with other known markers as
RAS and BRAF status. Recently, a molecular
classification of colorectal cancer has been proposed: CMS1
(immune), CMS2 (canonical), CMS3 (metabolic) and CMS4
(mesenchymal), which could be useful in therapeutic decisions
(65). Defining molecular subgroups
may identify patients who could benefit from aggressive and
targeted therapies, and might be used to select specific treatment
approaches for patients with colon cancer. It could be interesting
to investigate how GalNAc-T6, other GalNAc-transferases and its
products might behave in this molecular classification.
In summary, to the best of our knowledge, this study
is the first to prove that GalNAc-T6 expression in colon cancer is
an independent predictor for better overall survival, especially in
patients with advanced disease (AJCC stages III and IV). It will be
necessary to confirm these findings in prospective studies, with
larger cohorts, comparing GalNAc-T6 and GalNAc-T3 status and other
currently used molecular markers as KRAS, CEA and CA19.9.
Additional research is warranted to elucidate the molecular and
cellular mechanisms involved in the role of GalNAc-T6 in colon
cancer biology.
Acknowledgements
Not applicable.
Funding
This work was partially supported by ‘Programa
Grupos de Investigación’ (CSIC, Universidad de la República,
Montevideo, Uruguay; grant no. 908) and FOCEM (Fondo para la
Convergencia Estructural del MERCOSUR; grant no. COF 03-11).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LU performed the experimental work. EBe provided the
clinical samples and acquired the data. LU, DM, EO and NB
interpreted the data. SV performed the flow cytometry experiments
and interpreted the data. EBa performed statistical analysis. EO
and NB drafted the manuscript and provided overall supervision of
the project. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethical Committee at the University de la Republica and all
participants provided written informed consent prior to their
enrollment in the present study.
Consent for publication
All participants provided written informed consent
for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Byrd JC and Bresalier RS: Mucins and mucin
binding proteins in colorectal cancer. Cancer Metastasis Rev.
23:77–99. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dube DH and Bertozzi CR: Glycans in cancer
and inflammation-potential for therapeutics and diagnostics. Nat
Rev Drug Discov. 4:477–488. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pinho SS and Reis CA: Glycosylation in
cancer: Mechanisms and clinical implications. Nat Rev Cancer.
15:540–555. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kufe DW: Mucins in cancer: Function,
prognosis and therapy. Nat Rev Cancer. 9:874–885. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Freire-de-Lima L, Gelfenbeyn K, Ding Y,
Mandel U, Clausen H, Handa K and Hakomori SI: Involvement of
O-glycosylation defining oncofetal fibronectin in
epithelial-mesenchymal transition process. Proc Natl Acad Sci USA.
108:17690–17695. 2001. View Article : Google Scholar
|
|
6
|
Brockhausen I: Mucin-type O-glycans in
human colon and breast cancer: Glycodynamics and functions. EMBO
Rep. 7:599–604. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Itzkowitz SH, Yuan M, Montgomery CK,
Kjeldsen T, Takahashi HK, Bigbee WL and Kim YS: Expression of Tn,
sialosyl-Tn, and T antigens in human colon cancer. Cancer Res.
49:197–204. 1989.PubMed/NCBI
|
|
8
|
Fu C, Zhao H, Wang Y, Cai H, Xiao Y, Zeng
Y and Chen H: Tumor-associated antigens: Tn antigen, sTn antigen,
and T antigen. HLA. 88:275–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meichenin M, Rocher J, Galanina O, Bovin
N, Nifantev N, Sherman A, Cassagnau E, Heymann MF, Bara J, Fraser
RH and Le Pendu J: Tk, a new colon tumor-associated antigen
resulting from altered O-glycosylation. Cancer Res. 60:5499–5507.
2000.PubMed/NCBI
|
|
10
|
Medina M, Vélez D, Asenjo JA, Egea G, Real
FX, Gil J and Subiza JL: Human colon adenocarcinomas express a
MUC1-associated novel carbohydrate epitope on core mucin glycans
defined by a monoclonal antibody (A10) raised against murine
Ehrlich tumor cells. Cancer Res. 59:1061–1070. 1999.PubMed/NCBI
|
|
11
|
Tarp MA and Clausen H: Mucin-type
O-glycosylation and its potential use in drug and vaccine
development. Biochim Biophys Acta. 1780:546–563. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kimura T, McKolanis JR, Dzubinski LA,
Islam K, Potter DM, Salazar AM, Schoen RE and Finn OJ: MUC1 vaccine
for individuals with advanced adenoma of the colon: A cancer
immunoprevention feasibility study. Cancer Prev Res (Phila).
6:18–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bennett EP, Mandel U, Clausen H, Gerken
TA, Fritz TA and Tabak LA: Control of mucin-type O-glycosylation: A
classification of the polypeptide GalNAc-transferase gene family.
Glycobiology. 22:736–756. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Van den Steen P, Rudd PM, Dwek RA and
Opdenakker G: Concepts and principles of O-linked glycosylation.
Crit Rev Biochem Mol Biol. 33:151–208. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Johansson ME, Larsson JM and Hansson GC:
The two mucus layers of colon are organized by the MUC2 mucin,
whereas the outer layer is a legislator of host-microbial
interactions. Proc Natl Acad Sci USA. 108 Suppl 1:S4659–S4665.
2011. View Article : Google Scholar
|
|
16
|
Brockhausen I, Schachter H and Stanley P:
Chapter 9 O-GalNAc glycansEssentials of Glycobiology. 2nd edition.
Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR,
Hart GW and Etxler ME: Cold Spring Harbor, New York: Laboratory
Press; 2009
|
|
17
|
Xia L: Core 3-derived O-glycans are
essential for intestinal mucus barrier function. Methods Enzymol.
479:123–141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Holst S, Wuhrer M and Rombouts Y:
Glycosylation characteristics of colorectal cancer. Adv Cancer Res.
126:203–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bergstrom K, Liu X, Zhao Y, Gao N, Wu Q,
Song K, Cui Y, Li Y, McDaniel JM, McGee S, et al: Defective
intestinal mucin-type O-glycosylation causes spontaneous
colitis-associated cancer in mice. Gastroenterology.
151:152–164.e11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fu J, Wei B, Wen T, Johansson ME, Liu X,
Bradford E, Thomsson KA, McGee S, Mansour L, Tong M, et al: Loss of
intestinal core 1-derived O-glycans causes spontaneous colitis in
mice. J Clin Invest. 121:1657–1666. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nishida A, Lau CW, Zhang M, Andoh A, Shi
HN, Mizoguchi E and Mizoguchi A: The membrane-bound mucin Muc1
regulates T helper 17-cell responses and colitis in mice.
Gastroenterology. 142:865–874.e2. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Velcich A, Yang W, Heyer J, Fragale A,
Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K and
Augenlicht L: Colorectal cancer in mice genetically deficient in
the mucin Muc2. Science. 295:1726–1729. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
An G, Wei B, Xia B, McDaniel JM, Ju T,
Cummings RD, Braun J and Xia L: Increased susceptibility to colitis
and colorectal tumors in mice lacking core 3-derived O-glycans. J
Exp Med. 204:1417–1429. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guda K, Moinova H, He J, Jamison O, Ravi
L, Natale L, Lutterbaugh J, Lawrence E, Lewis S, Willson JK, et al:
Inactivating germ-line and somatic mutations in polypeptide
N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc
Natl Acad Sci USA. 106:12921–12925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Venkitachalam S and Guda K: Altered
glycosyltransferases in colorectal cancer. Expert Rev Gastroenterol
Hepatol. 11:5–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mandel U, Hassan H, Therkildsen MH,
Rygaard J, Jakobsen MH, Juhl BR, Dabelsteen E and Clausen H:
Expression of polypeptide GalNAc-transferases in stratified
epithelia and squamous cell carcinomas: Immunohistological
evaluation using monoclonal antibodies to three members of the
GalNAc-transferase family. Glycobiology. 9:43–52. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Berois N, Mazal D, Ubillos L, Trajtenberg
F, Nicolas A, Sastre-Garau X, Magdelenat H and Osinaga E:
UDP-N-acetyl-D-galactosamine:polypeptide
N-acetylgalactosaminyltransferase-6 as a new immunohistochemical
breast cancer marker. J Histochem Cytochem. 54:317–328. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu C, Guo X, Wang W, Wang Y, Shan Y, Zhang
B, Song W, Ma S, Ge J, Deng H and Zhu M:
N-Acetylgalactosaminyltransferase-14 as a potential biomarker for
breast cancer by immunohistochemistry. BMC Cancer. 10:1232010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hussain MR, Hoessli DC and Fang M:
N-acetylgalacto-saminyltransferases in cancer. Oncotarget.
7:54067–54081. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Taniuchi K, Cerny RL, Tanouchi A, Kohno K,
Kotani N, Honke K, Saibara T and Hollingsworth MA: Overexpression
of GalNAc-transferase GalNAc-T3 promotes pancreatic cancer cell
growth. Oncogene. 30:4843–5484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gu C, Oyama T, Osaki T, Li J, Takenoyama
M, Izumi H, Sugio K, Kohno K and Yasumoto K: Low expression of
polypeptide GalNAc N-acetylgalactosaminyl transferase-3 in lung
adenocarcinoma: Impact on poor prognosis and early recurrence. Br J
Cancer. 90:436–442. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao S, Guo T, Li J, Uramoto H, Guan H,
Deng W and Gu C: Expression and prognostic value of GalNAc-T3 in
patients with completely resected small (≤2 cm) peripheral lung
adenocarcinoma after IASLC/ATS/ERS classification. Onco Targets
Ther. 8:3143–3152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miyahara N, Shoda J, Kawamoto T, Furukawa
M, Ueda T, Todoroki T, Tanaka N, Matsuo K, Yamada Y, Kohno K and
Irimura T: Expression of
UDP-N-acetyl-alpha-D-galactosamine-polypeptide
N-acetylgalactosaminyltransferase isozyme 3 in the subserosal layer
correlates with postsurgical survival of pathological tumor stage 2
carcinoma of the gallbladder. Clin Cancer Res. 10:2090–2099. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
He H, Shen Z, Zhang H, Wang X, Tang Z, Xu
J and Sun Y: Clinical significance of polypeptide
N-acetylgalactosaminyl transferase-5 (GalNAc-T5) expression in
patients with gastric cancer. Br J Cancer. 110:2021–2029. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee ES, Son DS, Kim SH, Lee J, Jo J, Han
J, Kim H, Lee HJ, Choi HY, Jung Y, et al: Prediction of
recurrence-free survival in postoperative non-small cell lung
cancer patients by using an integrated model of clinical
information and gene expression. Clin Cancer Res. 14:7397–7404.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kwon OS, Oh E, Park JR, Lee JS, Bae GY,
Koo JH, Kim H, Choi YL, Choi YS, Kim J and Cha HJ: GalNAc-T14
promotes metastasis through Wnt dependent HOXB9 expression in lung
adenocarcinoma. Oncotarget. 6:41916–41928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Freire T, Berois N, Sóñora C, Varangot M,
Barrios E and Osinaga E: UDP-N-acetyl-D-galactosamine:polypeptide
N-acetylgalactosaminyltransferase 6 (ppGalNAc-T6) mRNA as a
potential new marker for detection of bone marrow-disseminated
breast cancer cells. Int J Cancer. 119:1383–1388. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Berois N, Blanc E, Ripoche H, Mergui X,
Trajtenberg F, Cantais S, Barrois M, Dessen P, Kågedal B, Bénard J,
et al: ppGalNAc-T13: A new molecular marker of bone marrow
involvement in neuroblastoma. Clin Chem. 52:1701–1172. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Berois N, Gattolliat CH, Barrios E,
Capandeguy L, Douc-Rasy S, Valteau-Couanet D, Bénard J and Osinaga
E: GALNT9 gene expression is a prognostic marker in neuroblastoma
patients. Clin Chem. 59:225–233. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kohsaki T, Nishimori I, Nakayama H,
Miyazaki E, Enzan H, Nomoto M, Hollingsworth MA and Onishi S:
Expression of UDP-GalNAc:polypeptide
N-acetylgalactosaminyltransferase isozymes T1 and T2 in human
colorectal cancer. J Gastroenterol. 35:840–848. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bennett EP, Hassan H and Clausen H: cDNA
cloning and expression of a novel human
UDP-N-acetyl-alpha-D-galactosamine. Polypeptide
N-acetylgalactosaminyltransferase, GalNAc-t3. J Biol Chem.
271:17006–17012. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo JM, Chen HL, Wang GM, Zhang YK and
Narimatsu H: Expression of UDP-GalNAc:polypeptide
N-acetyl-galactosaminyltransferase-12 in gastric and colonic cancer
cell lines and in human colorectal cancer. Oncology. 67:271–276.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kato K, Takeuchi H, Kanoh A, Miyahara N,
Nemoto-Sasaki Y, Morimoto-Tomita M, Matsubara A, Ohashi Y, Waki M,
Usami K, et al: Loss of UDP-GalNAc:polypeptide
N-acetyl-galactosaminyltransferase 3 and reduced O-glycosylation in
colon carcinoma cells selected for hepatic metastasis. Glycoconj J.
27:267–276. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shibao K, Izumi H, Nakayama Y, Ohta R,
Nagata N, Nomoto M, Matsuo K, Yamada Y, Kitazato K, Itoh H and
Kohno K: Expression of
UDP-N-acetyl-alpha-D-galactosamine-polypeptide galNAc
N-acetylgalactosaminyl transferase-3 in relation to differentiation
and prognosis in patients with colorectal carcinoma. Cancer.
94:1939–1946. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kato K, Takeuchi H, Kanoh A, Mandel U,
Hassan H, Clausen H and Irimura T: N-acetylgalactosamine
incorporation into a peptide containing consecutive threonine
residues by UDP-N-acetyl-D-galactosaminide:polypeptide
N-acetylgalactosaminyltransferases. Glycobiology. 11:821–829. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kanoh A, Takeuchi H, Kato K, Waki M, Usami
K and Irimura T: Interleukin-4 induces specific pp-GalNAc-T
expression and alterations in mucin O-glycosylation in colonic
epithelial cells. Biochim Biophys Acta. 1780:577–584. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bennett EP, Hassan H, Mandel U,
Hollingsworth MA, Akisawa N, Ikematsu Y, Merkx G, van Kessel AG,
Olofsson S and Clausen H: Cloning and characterization of a close
homologue of human UDP-N-acetyl-alpha-D-galactosamine:polypeptide
N-acetylgalactosaminyltransferase-T3, designated GalNAc-T6.
Evidence for genetic but not functional redundancy. J Biol Chem.
274:25362–15370. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Brooks SA, Carter TM, Bennett EP, Clausen
H and Mandel U: Immunolocalisation of members of the polypeptide
N-acetyl-galactosaminyl transferase (ppGalNAc-T) family is
consistent with biologically relevant altered cell surface
glycosylation in breast cancer. Acta Histochem. 109:273–284. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Patani N, Jiang W and Mokbel K: Prognostic
utility of glycosyltransferase expression in breast cancer. Cancer
Genomics Proteomics. 5:333–340. 2008.PubMed/NCBI
|
|
50
|
Park JH, Nishidate T, Kijima K, Ohashi T,
Takegawa K, Fujikane T, Hirata K, Nakamura Y and Katagiri T:
Critical roles of mucin 1 glycosylation by transactivated
polypeptide N-acetylgalactosaminyltransferase 6 in mammary
carcinogenesis. Cancer Res. 70:2759–2769. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Park JH, Katagiri T, Chung S, Kijima K and
Nakamura Y: Polypeptide N-acetylgalactosaminyltransferase 6
disrupts mammary acinar morphogenesis through O-glycosylation of
fibronectin. Neoplasia. 13:320–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li Z, Yamada S, Wu Y, Wang KY, Liu YP,
Uramoto H, Kohno K and Sasaguri Y: Polypeptide
N-acetylgalactosaminyltransferase-6 expression independently
predicts poor overall survival in patients with lung adenocarcinoma
after curative resection. Oncotarget. 7:54463–54473.
2016.PubMed/NCBI
|
|
53
|
Li Z, Yamada S, Inenaga S, Imamura T, Wu
Y, Wang KY, Shimajiri S, Nakano R, Izumi H, Kohno K and Sasaguri Y:
Polypeptide N-acetylgalactosaminyltransferase 6 expression in
pancreatic cancer is an independent prognostic factor indicating
better overall survival. Br J Cancer. 104:1882–1889. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kitada S, Yamada S, Kuma A, Ouchi S,
Tasaki T, Nabeshima A, Noguchi H, Wang KY, Shimajiri S, Nakano R,
et al: Polypeptide N-acetylgalactosaminyl transferase 3
independently predicts high-grade tumours and poor prognosis in
patients with renal cell carcinomas. Br J Cancer. 109:472–481.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang ZQ, Bachvarova M, Morin C, Plante M,
Gregoire J, Renaud MC, Sebastianelli A and Bachvarov D: Role of the
polypeptide N-acetylgalactosaminyltransferase 3 in ovarian cancer
progression: Possible implications in abnormal mucin
O-glycosylation. Oncotarget. 5:544–560. 2014.PubMed/NCBI
|
|
56
|
Onitsuka K, Shibao K, Nakayama Y, Minagawa
N, Hirata K, Izumi H, Matsuo K, Nagata N, Kitazato K, Kohno K and
Itoh H: Prognostic significance of
UDP-N-acetyl-alpha-D-galactosamine: polypeptide
N-acetylgalactosaminyltransferase-3 (GalNAc-T3) expression in
patients with gastric carcinoma. Cancer Sci. 94:32–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gill DJ, Chia J, Senewiratne J and Bard F:
Regulation of O-glycosylation through Golgi-to-ER relocation of
initiation enzymes. J Cell Biol. 189:843–858. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Punt CJ, Koopman M and Vermeulen L: From
tumour heterogeneity to advances in precision treatment of
colorectal cancer. Nat Rev Clin Oncol. 14:235–246. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mahasneh A, Al-Shaheri F and Jamal E:
Molecular biomarkers for an early diagnosis, effective treatment
and prognosis of colorectal cancer: Current updates. Exp Mol
Pathol. 102:475–483. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sinicrope FA, Okamoto K, Kasi PM and
Kawakami H: Molecular biomarkers in the personalized treatment of
colorectal cancer. Clin Gastroenterol Hepatol. 14:651–658. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Dienstmann R, Salazar R and Tabernero J:
Personalizing colon cancer adjuvant therapy: Selecting optimal
treatments for individual patients. J Clin Oncol. 33:1787–1796.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lavrsen K, Dabelsteen S, Vakhrushev SY,
Levann AMR, Haue AD, Dylander A, Mandel U, Hansen L, Frödin M,
Bennett EP and Wandall HH: De novo expression of human polypeptide
N-acetylgalactosaminyltransferase 6 (GalNAc-T6) in colon
adenocarcinoma inhibits the differentiation of colonic epithelium.
J Biol Chem. 293:1298–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Andre T, de Gramont A, Vernerey D,
Chibaudel B, Bonnetain F, Tijeras-Raballand A, Scriva A, Hickish T,
Tabernero J, Van Laethem JL, et al: Adjuvant fluorouracil,
leucovorin, and oxaliplatin in stage II to III colon cancer:
Updated 10-year survival and outcomes according to BRAF mutation
and mismatch repair status of the MOSAIC study. J Clin Oncol.
33:4176–4187. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Verhoeff SR, van Erning FN, Lemmens VE, de
Wilt JH and Pruijt JF: Adjuvant chemotherapy is not associated with
improved survival for all high-risk factors in stage II colon
cancer. Int J Cancer. 139:187–193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Guinney J, Dienstmann R, Wang X, de
Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda
G, Angelino P, et al: The consensus molecular subtypes of
colorectal cancer. Nat Med. 21:1350–1356. 2015. View Article : Google Scholar : PubMed/NCBI
|