Introduction
Breast cancer is one of the most common malignant
tumors in women. In many areas, breast cancer is the most common
malignancy in females. In China, the incidence of breast cancer is
increasing annually. Because the development and progression of
breast cancer are complex processes involving many genes, the
underlying mechanism of breast cancer remains unclear (1,2).
Single-stranded RNAs consisting of 19 to 25
nucleotides are known as microRNAs (miRNAs). These RNAs are
regulatory molecules that modulate the expression of functional
genes, play an important role in many biological processes, and are
expressed in a tissue- and time-specific manner. miRNAs regulate
gene expression at the post-transcriptional level mainly via
completely complementary or partially complementary binding to the
3′ UTR of the target gene, resulting in degradation or
translational repression of the target gene (2–4). Studies
have shown that a variety of miRNAs are involved in malignant
transformation processes, such as malignant proliferation,
metastasis and recurrence, and apoptosis inhibition. Studies have
reported that miRNA inhibitors can reduce the high miR-21
expression in breast cancer cells, inhibiting proliferation and
migration; that miR-373 can promote the metastasis of breast cancer
cells; that miR-520 plays a role in promoting the development of
breast cancer; and that miR-126 and miR-335 are able to inhibit
breast cancer to a certain extent (5–7). Recent
studies have demonstrated that miR-202 is associated with several
types of cancer, miR-202 has a lower expression in several of
cancers, and however, the expression and function of miR-202 have
not been investigated in breast cancer (8).
KRAS belongs to the RAS family; it is located on the
short arm of chromosome 12 and is 35 kb long. KRAS is a downstream
molecule of the EGF receptor. It possesses four exons and one 5′
end non-coding exon, encoding a 189-aa RAS protein. This gene
mainly functions in signaling pathways involving membrane receptors
and cAMP. KRAS has the strongest influence on malignant cancer in
the RAS family (9–12).
In our study, we found abnormal expression of
miR-202 and KRAS in breast cancer. We also found that miR-202 may
affect the proliferation and metastasis of breast cancer cells by
regulating KRAS. Our results provide a new target for the diagnosis
and treatment of breast cancer.
Materials and methods
Samples
Samples were collected from 30 patients (women aged
49–68 years) from May 2011 to 2013 at Zhongnan Hospital of Wuhan
University (Wuhan, China). All patients reviewed the methods and
significance of the study and provided written informed consent.
The project was approved by the ethics committee for medical
science research of Zhongnan Hospital of Wuhan University. All
cancer tissues were pathologically confirmed to be breast
cancer.
MicroRNA microarray analysis
A total of 500 ng of RNA was subjected to Agilent
miRNA microarray analysis (Guangzhou RiboBio Co., Ltd., Guangzhou,
China). Differences between groups were examined for statistical
significance with an unpaired Student's t test.
Cell culture
293, MCF7 and `-MB-231 cells were cultured in DMEM
supplemented with 10% FCS (both Invitrogen; Thermo Fisher
Scientific Inc., Waltham, MA, USA), 100 IU/ml penicillin and 100
mg/ml streptomycin (Shanghai Shenggong Biology Engineering
Technology Service, Ltd., Shanghai, China) at 37°C in a humidified
atmosphere containing 5% CO2.
MTT assays
Cells were plated in 96-well plates at a density of
1×104 cells per well; 24 h later, the cells were
transfected or treated as indicated. At the end of the incubation,
cellular proliferation was measured by MTT assays. The optical
density at 490 nm was measured using a microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Metastasis assay
A total of 1×105 cells were seeded onto
the upper part of a transwell chamber (BD Bioscience, Franklin
Lakes, NJ, USA) containing a gelatin-coated polycarbonate membrane
filter (pore size: 8 mm) for migration and invasion assays.
Serum-free medium was added to the upper well, and medium
containing 10% FBS was added to the lower well. After 24 h of
incubation at 37°C with 5% CO2, the filters were stained
with crystal violet (Amresco, LLC., Solon, OH, USA). Five random
fields were counted per chamber by using an inverted
microscope.
Real-time PCR
Total RNA was isolated with TRIzol (Invitrogen;
Thermo Fisher Scientific Inc.) according to the manufacturer's
protocol. Complementary DNA was synthesized by reverse
transcription of total RNA using an RT reaction kit (Promega Corp.,
Madison, WI, USA). Real-time PCR was performed according to the
manufacturer's instructions. SYBR Premix Ex Taq (TaKaRa Bio, Inc.,
Otsu, Japan) was used as a DNA-specific fluorescent dye. The primer
sequences in Table I were
synthesized.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Name | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| KRAS |
TACCAFCAACTCCAACGG |
GAACCCAAGGCATCTCCA |
| GAPDH |
AGAAGGCTGGGGCTCATTTG |
AGGGGCCATCCACAGTCTTC |
| microRNA-202 |
CTCCAGAGAGAUAGUAGAGCCT |
CTCAACCAATCACCTGGCACAGA |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
All reactions were repeated at least three times.
Gene expression levels were calculated relative to GAPDH or U6 with
Stratagene Mx 3000P software.
Western blot analysis
Total proteins were extracted using
radioimmunoprecipitation (RIPA) assay buffer (Wlaterson, Barcelona,
Spain) and were quantified using a bicinchoninic acid (BCA) protein
assay kit (Wlaterson). Equal amounts of protein (60 µg) were boiled
at 100°C for 5 min, separated on 12% sodium dodecyl sulfate
polyacrylamide gels (SDS-PAGE), and electrotransferred to
polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA,
USA). The membranes were blocked with 10% skim milk (w/v) at room
temperature for 2 h. Target proteins were probed with specific
antibodies against KRAS and GAPDH (Santa Cruz, Biotechnology, Inc.,
Dallas, TX, USA). The membranes were stripped and re-probed with an
antibody against GAPDH to verify equal loading.
Construction of stable
siRNA-expressing cell lines
To stably silence KRAS, cells were transfected with
the pRS-si-KRAS plasmid (Shanghai GeneChem Co., Ltd., Shanghai,
China) and were selected with G418 (400 µg/ml). After 3 weeks,
stable cells were selected, cultured and amplified.
Statistical analysis
All experiments were repeated at least three times.
A log-rank test was used to evaluate the correlation between
possible prognostic factors. Statistical significance between two
groups of data was evaluated by Student's t test (two-tailed).
One-way ANOVA and Dunnett's post hoc test were used for comparisons
among multiple groups. Statistical analysis was performed using
GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA,
USA). P<0.05 was considered to indicate statistically
significant difference.
Results
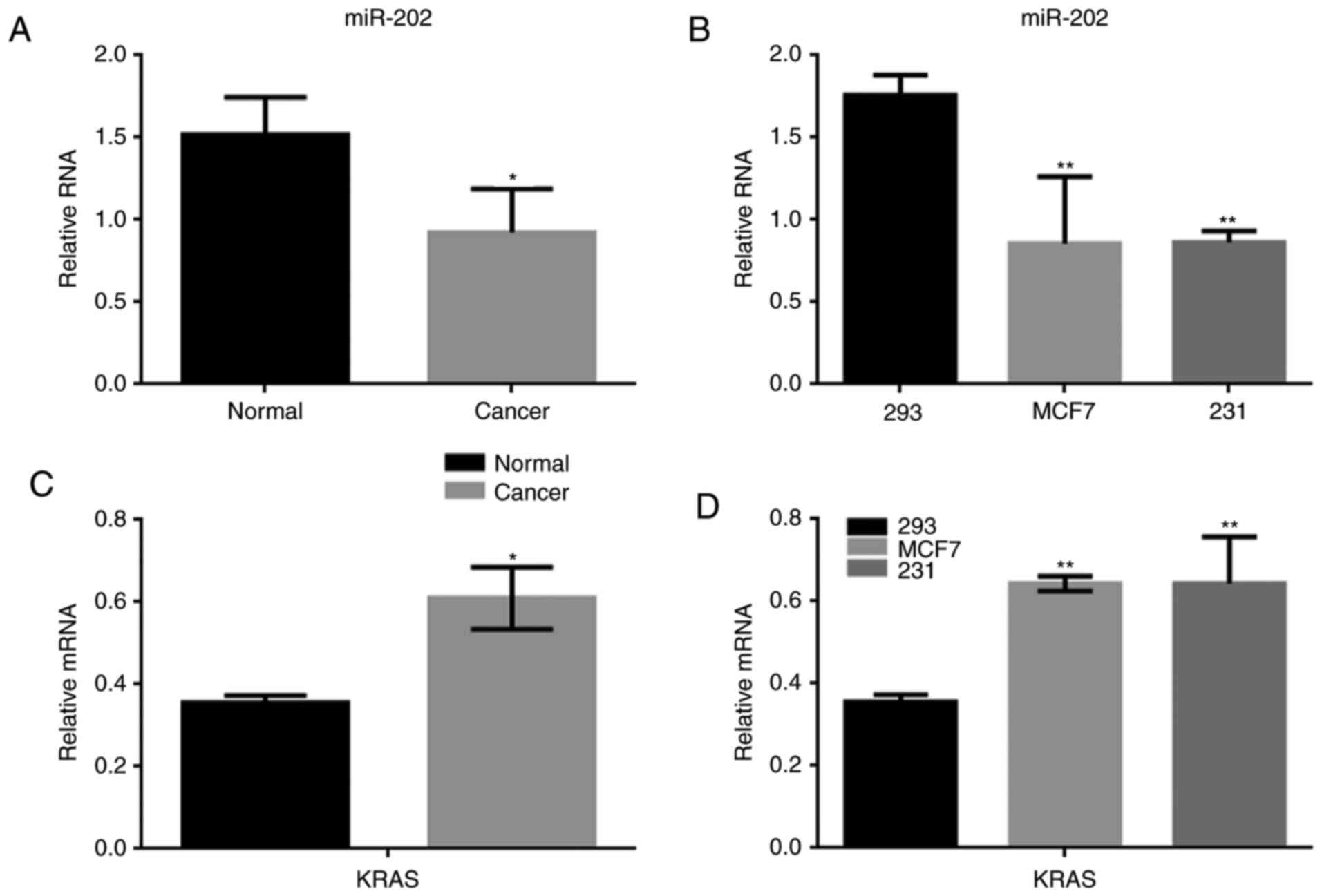
The expression of miR-202 and KRAS in
breast cancer
Many miRNAs were abnormally expressed in breast
cancer tissue. We found that the difference in expression of
miR-202 between breast cancer and normal tissues was the largest of
the miRNAs identified in our study (Table II). To determine the significance of
miR-202 and KRAS expression in breast cancer, the expression of
miR-202 was detected using real-time PCR in both breast cancer
tissues and in human cell lines, including 293, MCF7 and MDA-MB-231
cells. miR-202 expression was significantly lower in breast cancer
tissues than in adjacent tissues (Fig.
1A). We found the expression of miR-202 in MCF7 cells and
MDA-MB-231 cells was significantly lower than in normal 293 cells
(Fig. 1B). Meanwhile, KRAS expression
was obviously up regulated in the 30 breast cancer samples
analyzed. Similar results were also observed MCF7 and MDA-MB-231
cells (Fig. 1C, D).
 | Table II.Differentially expressed miRNAs in
breast cancer. |
Table II.
Differentially expressed miRNAs in
breast cancer.
| miRNA | Fold change | Trend |
|---|
| miR-202 | 38.92 | Down |
| miR-34 | 11.02 | Down |
| miR-29 | 8.43 | Down |
| miR-342 | 2.98 | Down |
| miR-21 | 17.98 | Up |
| miR-269 | 12.20 | Up |
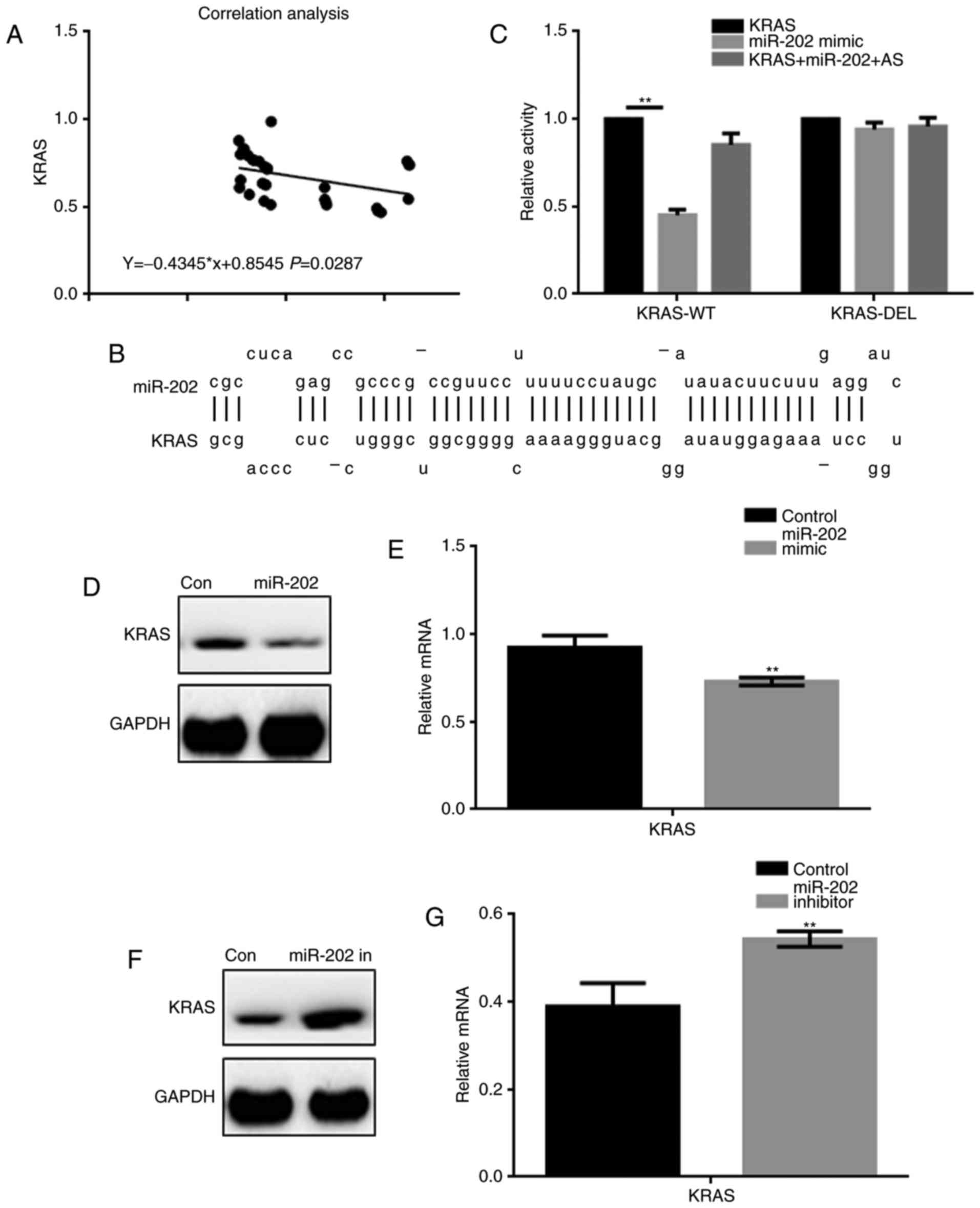
miR-202 inhibits KRAS expression in
breast cancer cells
A correlation analysis showed that miR-202
expression was negatively correlated with KRAS expression in breast
cancer (Fig. 2A). The miRDB tool
indicated that miR-202 could bind to the 3′ UTR of KRAS (Fig. 2B). A luciferase reporter assay was
used to determine whether KRAS was a potential target gene of
miR-202 (Fig. 2C). Co-transfection of
KRAS-WT and miR-202 resulted in lower luciferase activity in 293
cells, while co-expression studies with either KRAS-DEL or miR-202
AS (antisense) showed no change in luciferase activity. Further,
upon transfection of a miR-202 mimic, downregulation of KRAS
expression was observed at both the protein and mRNA levels.
Inhibiting miR-202 by transfecting cells with a miR-202 inhibitor
resulted in upregulation of KRAS (Fig.
2D-G).
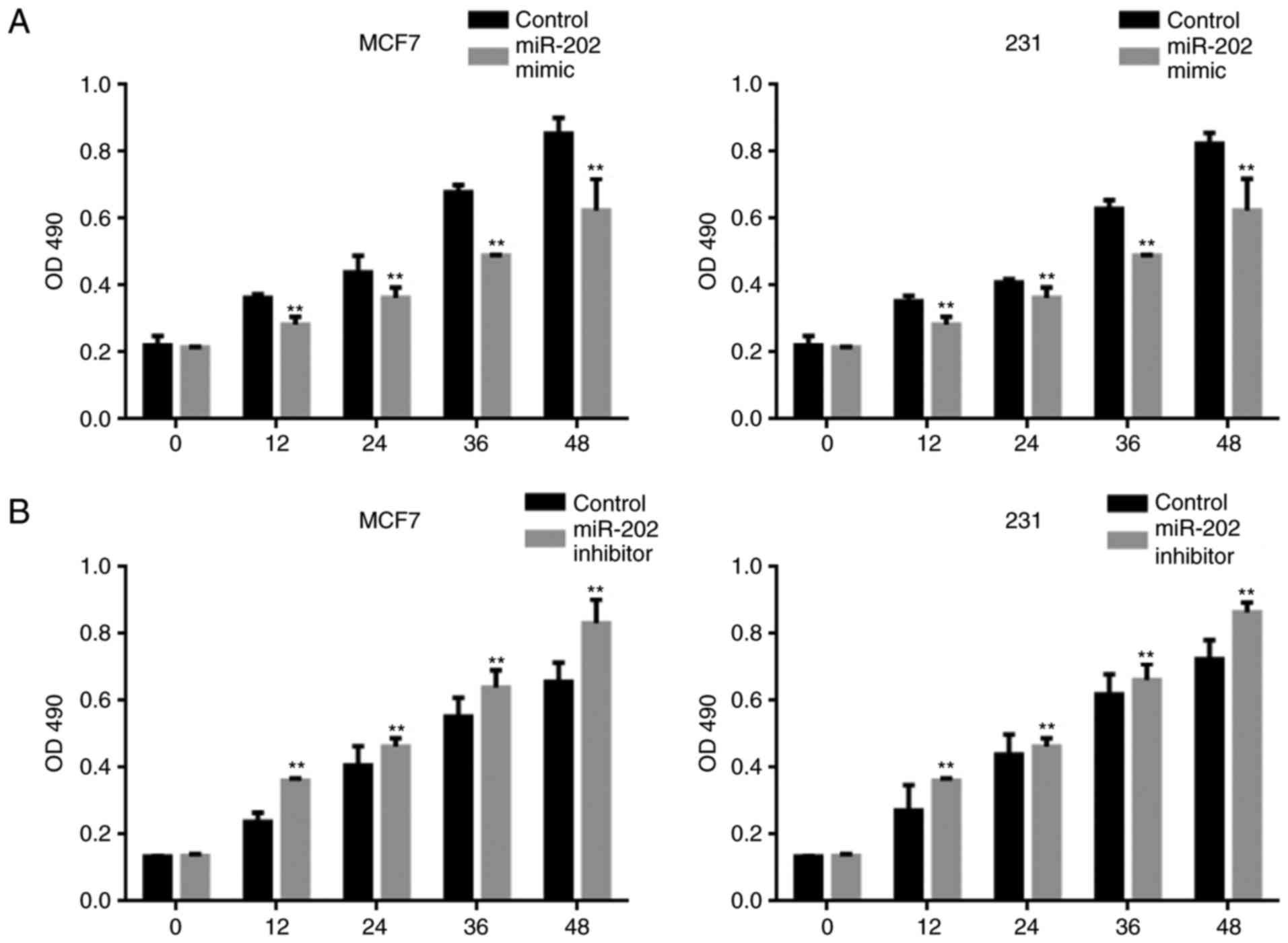
miR-202 inhibits the proliferation of
breast cancer cells
An MTT assay was used to examine the effect of
miR-202 on cell proliferation. Overexpression of miR-202
significantly blocked the proliferation of both MCF7 and MDA-MB-231
cells (Fig. 3A). By contrast, a lower
miR-202 expression level promoted cell proliferation (Fig. 3B).
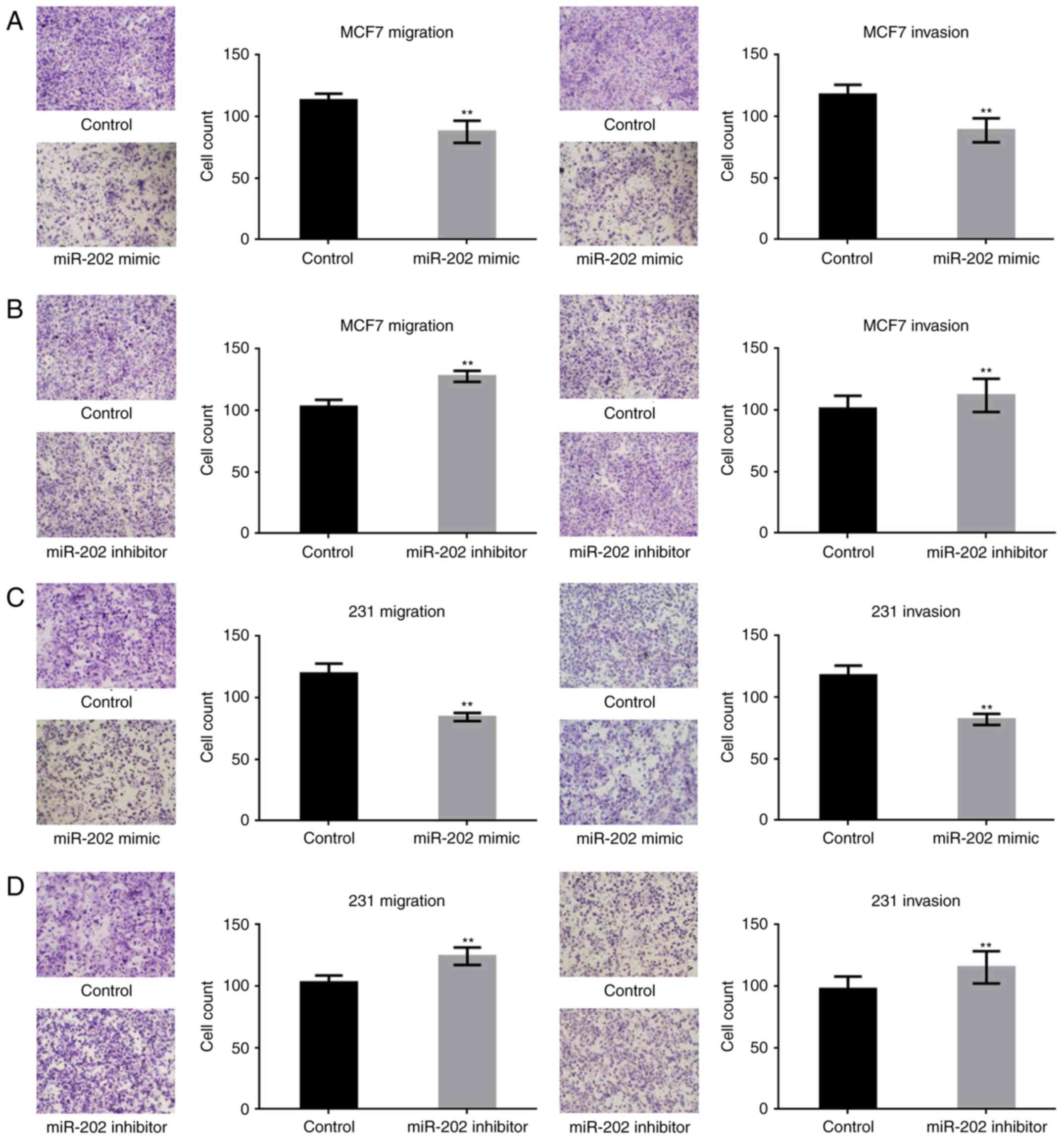
miR-202 inhibits the metastasis of
breast cancer cells
To study the effect of miR-202 on the metastasis of
breast cancer cells, transwell assays (with or without Matrigel)
were performed. miR-202 significantly suppressed the migration and
invasion potential of breast cancer cells (Fig. 4A-D). The above results indicated that
miR-202 could inhibit the growth and metastasis of breast cells,
possibly by targeting KRAS.
miR-202 inhibits the proliferation and
migration of breast cancer cells by targeting KRAS
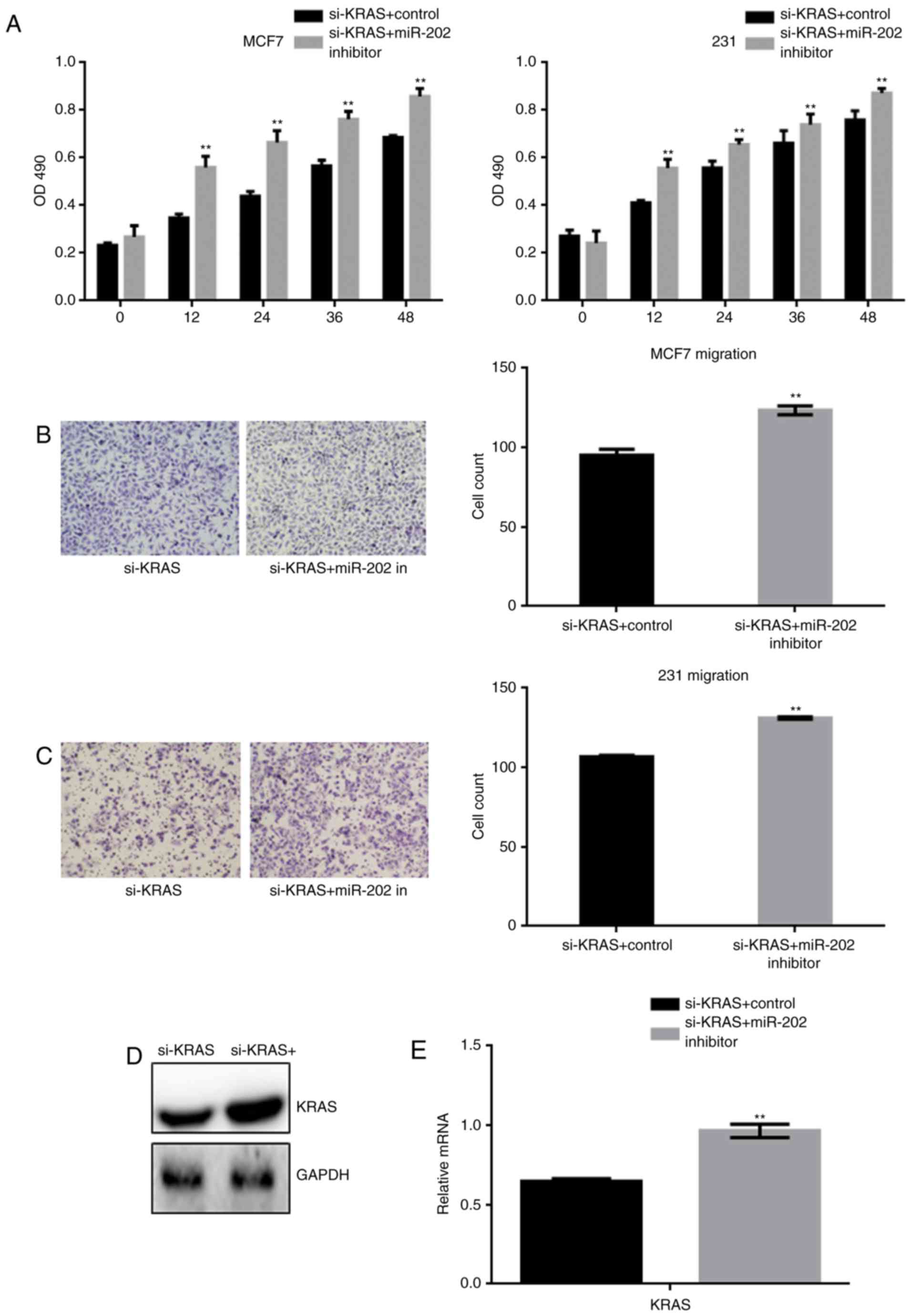
After generating stable si-KRAS-expressing cells,
the cells were transfected with a miR-202 inhibitor or a control
inhibitor. The effect of miR-202 inhibition on cell proliferation
and migration was detected by MTT and transwell assays (Fig. 5A-C). The results indicated that
miR-202 inhibition restored the cell proliferation and migration
that were blocked by si-KRAS. Western blot and real time PCR showed
that miR-202 inhibition resulted in re-expression of the silenced
KRAS (Fig. 5D-E).
The above results indicated that miR-202 could
inhibit the growth and metastasis of breast cancer cells by
targeting KRAS.
Discussion
Breast cancer is one of the most common malignant
tumors in the world and seriously affects women's physical and
mental health. The incidence of breast cancer has been rising since
the 1970s (13). Despite marked
improvement in the prognosis of breast cancer patients after
surgery and adjuvant therapy, breast cancer still has a high rate
of recurrence and mortality. In recent years, with the development
of targeted therapies for breast cancer, mortality has declined;
however, the mortality rate of breast cancer remains high, and
breast cancer is still a major threat to women's health. Therefore,
it is important to elucidate the pathogenesis of breast cancer and
develop new treatment strategies (14,15).
miRNAs are a group of non-protein coding, short,
single-stranded RNAs that are considered to be promising molecular
markers and therapeutic targets in several cancers. miR-202 has
been identified as a potential tumor suppressor in various cancers.
A previous study revealed that miR-202 upregulation induced novel
anticancer effects upregulation in NSCLC and indicated that STAT3
may be a molecular target of miR-202. In human ESCCs, miR-202
regulated apoptosis by targeting HSF2 via mechanism involving
caspase-3. A study also identified that miR-202 plays an important
role in regulating cell proliferation, migration and invasion of
cervical cancer (CC) by directly targeting cyclin D1, and miR-202
may represent a potential therapeutic target for patients with CC
(16–22). In addition, a previous study showed
that an increased serum concentration of miR-202 may be a strong
independent prognostic factor for breast cancer patients (20). Mutations in KRAS lead to sustained
activation of the RAS protein, independent of EGFR/HER receptor
activation, and eventually cause abnormalities in the RAS signaling
pathway. KRAS gene mutations can affect cell growth, proliferation,
migration and differentiation and lead to malignant transformation
of cells (23–25).
In our study, we found lower expression of miR-202
in breast cancer cells and tissues, indicating that miR-202 may
have an inhibitory effect on breast cancer. We identified several
potential reasons why miR-202 may have an inhibitory effect on
breast cancer. First, we found that miR-202 is negatively
correlated with KRAS and can regulate the activity of KRAS to some
extent. We also found that overexpression of miR-202 inhibited the
proliferation of breast cancer cells. Conversely, when the
expression of miR-202 was upregulated, the proliferative capacity
of breast cancer cells decreased. Afterwards, we demonstrated that
overexpression of miR-202 inhibited the migration and invasion of
breast cancer cells. Via a recovery experiment, we also found that
miR-202 inhibition could restore cell proliferation and migration
blocked by KRAS silencing. In follow-up experiments, the authors
intend to further study the effect of miR-202 on breast cancer
in vivo.
In summary, our data clarified that miR-202 could
inhibit the growth and metastasis of breast cancer cells by
targeting KRAS. Although further in vivo tests are needed to
confirm our results, our data provide an experimental basis for the
treatment of breast cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JT conceived and designed the study, acquired the
specimens and data, directed the drafting of manuscript and revised
it, and provided the financial support. SG designed the study and
took responsibility for the experiments. CC performed the
metastasis assay, QD performed the statistical analysis. JC
performed the western blot analysis.
Ethics approval and consent to
participate
All patients provided written informed consent. The
project was approved by the Ethics Committee for Medical Science
Research of Zhongnan Hospital of Wuhan University.
Consent for publication
All patients provided written informed consent for
the publication of their data.
Competing interests
The authors confirm that they have no competing
interests.
References
|
1
|
Rizzo S, Cangemi A, Galvano A, Fanale D,
Buscemi S, Ciaccio M, Russo A, Castorina S and Bazan V: Analysis of
miRNA expression profile induced by short term starvation in breast
cancer cells treated with doxorubicin. Oncotarget. 8:71924–71932.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mandujano-Tinoco EA, Garcia-Venzor A,
Muñoz-Galindo L, Lizarraga-Sanchez F, Favela-Orozco A,
Chavez-Gutierrez E, Krötzsch E, Salgado RM, Melendez-Zajgla J and
Maldonado V: miRNA expression profile in multicellular breast
cancer spheroids. Biochim Biophys Acta. 1864:1642–1655. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li P, Xu T, Zhou X, Liao L, Pang G, Luo W,
Han L, Zhang J, Luo X, Xie X and Zhu K: Downregulation of miRNA-141
in breast cancer cells is associated with cell migration and
invasion: Involvement of ANP32E targeting. Cancer Med. 6:662–672.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lykov AP, Kabakov AV, Kazakov OV,
Bondarenko NA, Poveshchenko OV, Raiter TV, Poveshchenko AF,
Strunkin DN and Konenkov VI: Levels of miRNA and hormones in
thoracic duct lymph in rats with experimental breast cancer induced
by N-methyl-N-nitrosourea. Bull Exp Biol Med. 162:387–390. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhardwaj A, Singh H, Rajapakshe K,
Tachibana K, Ganesan N, Pan Y, Gunaratne PH, Coarfa C and Bedrosian
I: Regulation of miRNA-29c and its downstream pathways in
preneoplastic progression of triple-negative breast cancer.
Oncotarget. 8:19645–19660. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Danza K, Summa S, Pinto R, Pilato B,
Palumbo O, Carella M, Popescu O, Digennaro M, Lacalamita R and
Tommasi S: TGFbeta and miRNA regulation in familial and sporadic
breast cancer. Oncotarget. 8:50715–50723. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fang H, Xie J, Zhang M, Zhao Z, Wan Y and
Yao Y: miRNA-21 promotes proliferation and invasion of
triple-negative breast cancer cells through targeting PTEN. Am J
Transl Res. 9:953–961. 2017.PubMed/NCBI
|
|
8
|
Zhang L, Xu J, Yang G, Li H and Guo X:
miR-202 inhibits cell proliferation, migration, and invasion by
targeting EGFR in human bladder cancer. Oncol Res. Jan 3–2018.(Epub
ahead of print). View Article : Google Scholar
|
|
9
|
Kim RK, Suh Y, Yoo KC, Cui YH, Kim H, Kim
MJ, Kim Gyu I and Lee SJ: Activation of KRAS promotes the
mesenchymal features of basal-type breast cancer. Exp Mol Med.
47:e1372015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim Y, Kim J, Lee HD, Jeong J, Lee W and
Lee KA: Spectrum of EGFR gene copy number changes and KRAS gene
mutation status in Korean triple negative breast cancer patients.
PLoS One. 8:e790142013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McVeigh TP, Jung SY, Kerin MJ, Salzman DW,
Nallur S, Nemec AA, Dookwah M, Sadofsky J, Paranjape T, Kelly O, et
al: Estrogen withdrawal, increased breast cancer risk and the
KRAS-variant. Cell Cycle. 14:2091–2099. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Su X, Zhang L, Li H, Cheng P, Zhu Y, Liu
Z, Zhao Y, Xu H, Li D, Gao H and Zhang T: MicroRNA-134 targets KRAS
to suppress breast cancer cell proliferation, migration and
invasion. Oncol Lett. 13:1932–1938. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zeng Y, Zhou Z, Fan M, Gong T, Zhang Z and
Sun X: PEGylated cationic vectors containing a protease-sensitive
peptide as a miRNA delivery system for treating breast cancer. Mol
Pharm. 14:81–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang J, Le TD, Liu L and Li J: Inferring
miRNA sponge co-regulation of protein-protein interactions in human
breast cancer. BMC Bioinformatics. 18:2432017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang L, Yang X, Lv Y, Xin X, Qin C, Han
X, Yang L, He W and Yin L: Cytosolic co-delivery of miRNA-34a and
docetaxel with core-shell nanocarriers via caveolae-mediated
pathway for the treatment of metastatic breast cancer. Sci Rep.
7:461862017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao Z, Lv B, Zhang L, Zhao N and Lv Y:
miR-202 functions as a tumor suppressor in non-small cell lung
cancer by targeting STAT3. Mol Med Rep. 16:2281–2289. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mody HR, Hung SW, Pathak RK, Griffin J,
Cruz-Monserrate Z and Govindarajan R: miR-202 diminishes TGFβ
receptors and attenuates TGFβ1-Induced EMT in pancreatic cancer.
Mol Cancer Res. 15:1029–1039. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Chen Z, Li MJ, Guo HY and Jing
NC: Long non-coding RNA metastasis-associated lung adenocarcinoma
transcript 1 regulates the expression of Gli2 by miR-202 to
strengthen gastric cancer progression. Biomed Pharmacother.
85:264–271. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yi Y, Li H, Lv Q, Wu K and Zhang W, Zhang
J, Zhu D, Liu Q and Zhang W: miR-202 inhibits the progression of
human cervical cancer through inhibition of cyclin D1. Oncotarget.
7:72067–72075. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Farhana L, Dawson MI and Fontana JA: Down
regulation of miR-202 modulates Mxd1 and Sin3A repressor complexes
to induce apoptosis of pancreatic cancer cells. Cancer Biol Ther.
16:115–124. 2016. View Article : Google Scholar
|
|
21
|
Joosse SA, Müller V, Steinbach B, Pantel K
and Schwarzenbach H: Circulating cell-free cancer-testis MAGE-A
RNA, BORIS RNA, let-7b and miR-202 in the blood of patients with
breast cancer and benign breast diseases. Br J Cancer. 111:909–917.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao Y, Li C, Wang M, Su L, Qu Y, Li J, Yu
B, Yan M, Yu Y, Liu B and Zhu Z: Decrease of miR-202-3p expression,
a novel tumor suppressor, in gastric cancer. PLoS One.
8:e697562013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cejalvo JM, Pérez-Fidalgo JA, Ribas G,
Burgués O, Mongort C, Alonso E, Ibarrola-Villava M, Bermejo B,
Martínez MT, Cervantes A and Lluch A: Clinical implications of
routine genomic mutation sequencing in PIK3CA/AKT1 and
KRAS/NRAS/BRAF in metastatic breast cancer. Breast Cancer Res
Treat. 160:69–77. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ustinova M, Daneberga Z, Bērziņa D,
Nakazawa-Miklaševiča M, Maksimenko J, Gardovskis J and Miklaševičs
E: Impact of KRAS variant rs61764370 on breast cancer morbidity.
Exp Oncol. 37:292–294. 2015.PubMed/NCBI
|
|
25
|
Huang X, Yang Y, Guo Y, Cao ZL, Cui ZW, Hu
TC and Gao LB: Association of a let-7 KRAS rs712 polymorphism with
the risk of breast cancer. Genet Mol Res. 14:16913–16920. 2015.
View Article : Google Scholar : PubMed/NCBI
|