Introduction
Renal cancer is a metabolic disease that starts in
the cells of the kidney (1) and is
responsible for ~3% of all malignancies in adults (2). Globally, 250,000 new cases of kidney
cancer are diagnosed each year (3).
Renal cell carcinoma (RCC) accounts for ~90% of all renal cancer
cases (4,5), and is the most common form of adult
kidney cancer, possessing unique genetic and histological features.
Radical nephrectomy (RN) is effective in curing early and local
RCC, however, one-third of all patients present with metastatic
disease at diagnosis (6).
Furthermore, 20–40% of patients with RCC who undergo surgical
nephrectomy will develop metastasis. Despite previous developments
in therapy, there remains no effective treatment for patients with
advanced stages of the disease, and RCC is generally resistant to
standard chemotherapy and radiotherapy. The 5-year survival rate of
patients with metastatic RCC is <10% (1,7). Important
prognostic factors of RCC include histological subtype, nuclear
grade, tumour size and evidence of metastatic disease at
presentation (8). In order to further
our understanding on the prognosis of RCC and in order to develop
novel biological therapeutic methods, the identification of
molecular markers, which are useful in assisting in the management
of the patients, and novel molecular targets for adjuvant
therapies, is necessary.
Lactate dehydrogenase (LDH) is a tetrameric enzyme,
belonging to the 2-hydroxy acid oxidoreductase family (9); it is a metabolic enzyme that catalyses
the interconversion of pyruvate and lactate during the processes of
glycolysis and gluconeogenesis depending on nutrient availability.
Previous studies have revealed that pre-treatment serum LDH is a
statistically significant prognostic factor in breast, renal, lung
and gastric cancer (10–13). LDH is under the translational control
of the hypoxia-inducible factor and MYC proto-oncogene, BHLH
transcription factor (MYC), and is thus regulated by key oncogenic
processes. Previous studies have revealed that pre-treatment serum
LDH is a significant prognostic factor in high-risk patients with
metastatic RCC (14,15). In addition, the activation of these
oncogenic pathways resulted in high serum LDH levels, which were
associated with drug resistance (14).
The human genome contains four LDH genes: LDHA,
LDHB, LDHC and LDHD (16).
The LDH family may increase the rate of the simultaneous
interconversion of pyruvate to lactate and nicotinamide adenine
dinucleotide (NAD)H to NAD+ by 14 orders of magnitude (17,18).
Numerous genes and proteins associated with apoptosis or tumour
survival have been reported to be associated with LDH activity
(19,20). However, the majority of previous
studies on renal cancer focused on serum LDH, and few studies have
analysed specifically which LDH gene serves a key function in
RCC.
Materials and methods
LDH expression data
LDH expression and clinical data from The Cancer
Genome Atlas (TCGA) database were sourced from the Cancer Genomics
Browser of University of California Santa Cruz (https://genome-cancer.ucsc.edu/). A total of 6
members of the LDH family are included in the database, including
LDHA, LDHB, LDHC, LDHD, LDH A like 6A (LDHAL6A) and
LDHAL6B. In total, 509 primary clear cell RCC (ccRCC)
tumours from patients with detailed LDH expression data obtained
between January 1998 and December 2013 were selected from the
updated TCGA database according to parameters defined in a previous
study (21). Patients with fully
characterized tumours, intact overall survival (OS) data, complete
RNAseq information and those without pre-treatment were included.
Data on clinicopathological characteristics, including age, sex,
tumour size, Tumour-Node-Metastasis (TNM) stage (22), Fuhrman grade (23), AJCC renal cancer stage (24), laterality, haemoglobin level, white
blood cell count, platelet level and OS time were collected. A
follow-up of the patients was completed, with a median length of
1,063 days. During the follow-up, 347 patients succumbed.
Patient enrollment
From the Fudan University Shanghai Cancer Centre
(FUSCC) cohort, a total of 192 patients with ccRCC who underwent RN
or nephron-sparing nephrectomy between 2007 and 2011 were
retrospectively enrolled. All the tissue samples were collected
during surgery and stored at −70°C in the tissue bank of the FUSCC.
The pathological subtypes were determined by two pathologists, who
were genitourinary specialists. Data on the clinicopathological
characteristics, including sex and tumour size, were collected.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA extraction, RT and RT-qPCR analysis were
performed on the FUSCC cohort. In the FUSCC cohort, total RNA was
isolated from the 192 ccRCC samples using TRIzol®
reagent (cat no. 15596–026; Invitrogen, Thermo Fisher Scientific,
Inc., Waltham, MA, USA). A PrimeScript RT reagent kit (cat no.
K1622; Thermo Fisher Scientific, Inc.) was used according to the
manufacturer's protocol to synthesize first-strand cDNA from 1 µg
total RNA isolated from renal carcinoma cells. Serial dilutions of
cDNA were amplified by qPCR using gene-specific primers. The most
concentrated sample contained cDNA derived from 1 ng total RNA.
Next, SYBR Green Real-Time PCR assays (Thermo Fisher Scientific,
Inc.) were performed using an ABI 7900HT (Applied Biosystems,
Thermo Fisher Scientific, Inc.). The expression level of RNA was
normalized to the level of β-actin. The primers for RT-qPCR
analysis were synthesized by Sangon Biotech Co., Ltd. (Shanghai,
China), and the sequences used were as follows: LDHD
forward, 5′-CAAAGCCAGGGAGGGGAAGAG-3′ and reverse,
5′-CGTAGTCAGGGAACTTGTGGG-3′: LDHAL6B forward,
5′-TTCCGAGAAGCCCGTTCATC-3′ and reverse, 5′-GTGAAAGGGCTGCCATGTTG-3′:
and β-actin forward, 5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse,
5′-GGGCACGAAGGCTCATCATT-3′. The efficiency of amplification of the
target gene (LDH family members) and the internal control
(β-actin) was examined using qPCR and TaqMan detection.
Thermocycling conditions were as follows: i) Initial denaturation
at 94°C for 4 min; ii) 30 cycles of denaturation at 94°C for 30
sec, annealing at 55°C for 25 sec and extension at 72°C for 45 sec;
and iii) a final extension at 72°C for 5 min (25). Quantification was performed using the
2−ΔΔCq method (25). TNM,
tumor grade, tumor stage, serum LDH level and tumor position were
obtained from electronic records. Patients received regular
follow-ups via telephone call or in the clinic once every 3 months.
Events, including tumor recurrence, progression, metastasis and
mortality were recorded.
Statistical analysis
The disease-free survival time was calculated from
the date of diagnosis until the date of mortality from any cause or
first recurrence. The OS time was defined as the time of diagnosis
until the date of the last follow-up or mortality from any cause.
Patients without recurrence events or mortality were recorded as
censored at the time of the last follow-up. Statistical analysis
was performed using Stata software (version 12.0; StataCorp LP,
College Station, TX, USA). All statistical tests were two-tailed,
and P<0.05 was considered to indicate a statistically
significant difference. Survival curves were constructed using
Kaplan-Meier curves and a log-rank test in order to assess the
differences between the groups. Adjusted odds ratios with 95%
confidence intervals (CIs) were calculated using Cox proportional
hazards models. Univariate and multivariate Cox proportional
hazards analysis assessed the expression of LDH family members and
OS for patients with ccRCC in the TCGA cohort. Genes known to be
associated with OS were further studied. Multivariate logistic
regression was performed in order to further study factors that may
affect the expression of LDHs. Student's t-test or Wilcoxon's
signed rank test were used for 70 paired patients in order to
reveal potential differences in the expression of LDH family
members between patients with ccRCC and a control population.
Spearman's correlation analysis was used to determine the
correlation between LDH levels and the expression of
LDHD.
Results
Clinical characteristics of patients
with ccRCC
In the TCGA cohort, the median age of the 509
patients with ccRCC was 61 years (range, 26–90 years). A total of
328 (64.4%) patients were male and 181 (35.6%) patients were
female. The median follow-up time of this cohort was 35.4 months.
TNM, tumor size, tumor grade, tumor stage, laterality, white blood
cell count, hemoglobin level and platelet level are presented in
Table I.
 | Table I.Clinicopathological characteristics
of patients in the TCGA and FUSCC cohorts. |
Table I.
Clinicopathological characteristics
of patients in the TCGA and FUSCC cohorts.
| Variables | TCGA cohort | FUSCC cohort |
|---|
| Total number of
patients, n (%) | 509 | 192 |
| Median age (range),
years | 61 (26–90) | 55.5 (17–84) |
| Sex, n (%) |
|
|
|
Male | 328 (64.44) | 131 (68.23) |
|
Female | 181 (35.56) | 61 (31.77) |
| Fuhrman grade, n
(%) |
|
|
| 1 and
2 | 234 (45.97) | 79 (41.15) |
| 3 and
4 | 271 (53.24) | 113 (58.85) |
| Gx | 4 (0.79) | 0 (0.00) |
| Mean longest
dimension (range), cm | 5.30 (0–20.0) | 5.01 (1–16.0) |
| T, n (%) |
|
|
| T1 | 258 (50.69) | 129 (67.19) |
| T2 | 63 (12.38) | 29 (15.10) |
| T3 | 178 (34.97) | 27 (14.06) |
| T4 | 10 (1.96) | 7 (3.64) |
| N, n (%) |
|
|
| N0 | 228 (44.79) | 181 (94.27) |
| N1 | 18 (3.54) | 4 (2.08) |
| Nx | 263 (51.67) | 7 (3.64) |
| M, n (%) |
|
|
| M0 | 406 (79.76) | 184 (95.80) |
| M1 | 78 (15.32) | 7 (3.60) |
| Mx | 25 (4.91) | 1 (0.50) |
| Stage, n (%) |
|
|
| I | 253 (49.71) | 130 (67.71) |
| II | 51 (10.02) | 30 (15.62) |
|
III | 125 (24.56) | 23 (11.98) |
| IV | 80 (15.72) | 9 (4.69) |
| Laterality, n
(%) |
|
|
|
Left | 239 (46.95) | 90 (46.87) |
|
Right | 269 (52.85) | 94 (48.95) |
|
Bilateral | 1 (0.20) | 8 (41.67) |
| Median hemoglobin
(range), g/l |
| 139 (76–398) |
|
Elevated, n (%) | 5 (0.98) | 20 (10.42) |
| Normal,
n (%) | 175 (34.38) | 86 (44.79) |
| Low, n
(%) | 251 (49.31) | 16 (8.33) |
|
Undefined, n (%) | 78 (15.32) | 70 (36.46) |
| Median white blood
cell count (range), 109/l |
| 6.2 (3.1–18.4) |
|
Elevated, n (%) | 160 (31.43) | 3 (1.56) |
| Normal,
n (%) | 251 (49.31) | 100 (52.08) |
| Low, n
(%) | 8 (1.57) | 17 (8.85) |
|
Undefined, n (%) | 89 (17.49) | 72 (37.5) |
| Median platelet
level (range), 109/l |
| 219 (97–450) |
|
Elevated, n (%) | 35 (6.88) | 4 (2.08) |
| Normal,
n (%) | 342 (67.19) | 103 (53.65) |
| Low, n
(%) | 45 (8.84) | 2 (1.04) |
|
Undefined, n (%) | 86 (16.90) | 83 (43.23) |
| Median follow-up
time (range), days | 1,063
(2–3,668) | 1,412
(117–2,245) |
| Median serum LDH
level (range), U/la |
| 155 (67–761) |
In the FUSCC cohort, the median age of the 192
patients with ccRCC was 55.5 years (range, 17–84 years). A total of
47 patients succumbed during the follow-up, and the median
follow-up time of this cohort was 47.1 months. A total of 131
(68.2%) of patients were male and 61 (31.8%) of patients were
female. TNM, tumor size, tumor grade, tumor stage and laterality
are additionally presented in Table
I.
LDHD and LDHAL6B are independent
prognostic factors for OS
In the TCGA cohort, a univariate Cox proportional
hazards model was performed to assess the factors predicting OS.
Age, TNM stage, Fuhrman grade, haemoglobin level, white blood cell
(WBC) count, platelet (PLT) count, and LDHC, LDHD and
LDHAL6B expression were significantly associated with the OS
of the patients with ccRCC (all P<0.05; Table II). A multivariate Cox analysis
performed following adjustment for all the potential prognostic
factors, which included age, tumor stage, Fuhrman grade,
laterality, WBC count, PLT count, hemoglobin content, LDHD,
LDHC and LDHAL6B expression indicated that age (HR,
1.036; 95% CI, 1.020–1.053; P<0.0001), tumor stage (HR, 1.603;
95% CI, 1.317–1.949; P<0.0001), laterality (HR, 0.664; 95% CI,
0.467–0.944; P=0.023), LDHD expression (HR, 0.872; 95% CI,
0.764–0.994; P=0.040) and LDHAL6B expression (HR, 1.285; 95%
CI, 1.048–1.576; P=0.016) were the only independent predictors of
OS (all P<0.05, Table II).
 | Table II.Univariate and multivariate Cox
proportional hazard analysis of LDHD expression and overall
survival for patients with clear cell renal cell carcinoma in the
TCGA and FUSCC cohort. |
Table II.
Univariate and multivariate Cox
proportional hazard analysis of LDHD expression and overall
survival for patients with clear cell renal cell carcinoma in the
TCGA and FUSCC cohort.
|
| Univariate
(TCGA) | Multivariate
(TCGA) | Univariate
(FUSCC) | Multivariate
(FUSCC) |
|---|
|
|
|
|
|
|
|---|
| Variables | HR | (95% CI) | P-value | HR | (95% CI) | P-value | HR | (95% CI) | P-value | HR | (95% CI) | P-value |
|---|
| Age | 1.028 | (1.015–1.042) | <0.0001 | 1.036 | (1.020–1.053) | <0.0001 | 1.001 | (0.997–1.025) | 0.932 | 0.996 | (0.961–1.034) | 0.858 |
| Sex | 1.073 | (0.781–1.473) | 0.665 |
|
|
|
|
|
|
|
|
|
| T | 1.964 | (1.658–2.325) | <0.0001 |
|
|
|
|
|
|
|
|
|
| N | 2.799 | (1.486–5.274) | 0.001 |
|
|
|
|
|
|
|
|
|
| M | 4.448 | (3.221–6.141) | <0.0001 |
|
|
|
|
|
|
|
|
|
| AJCC stage | 1.944 | (1.695–2.229) | <0.0001 | 1.603 | (1.317–1.949) | <0.0001 | 2.065 | (1.597–2.671) | <0.0001 | 2.325 | (1.538–3.514) | <0.0001 |
| Fuhrman grade | 2.350 | (1.899–2.908) | <0.0001 | 1.230 | (0.920–1.644) | 0.162 | 2.007 | (1.309–3.077) | 0.001 | 0.959 | (0.529–1.738) | 0.891 |
| Hb | 0.584 | (0.415–0.823) | 0.002 | 0.915 | (0.624–1.342) | 0.651 | 1.003 | (0.995–1.011) | 0.534 | 1.008 | (0.985–1.032) | 0.493 |
| WBC | 0.652 | (0.471–0.902) | 0.010 | 1.014 | (0.694–1.483) | 0.942 | 0.994 | (0.986–1.002) | 0.142 | 0.985 | (0.972–1.000) | 0.043 |
| PLT | 1.702 | (1.145–2.529) | 0.008 | 1.086 | (0.748–1.579) | 0.664 | 1.004 | (1.000–1.008) | 0.061 | 1.005 | (0.999–1.010) | 0.083 |
| Tumour size | 1.174 | (0.946–1.459) | 0.146 |
|
|
|
|
|
|
|
|
|
| Laterality | 0.669 | (0.491–0.913) | 0.011 | 0.664 | (0.467–0.944) | 0.023 | 1.075 | (0.606–1.909) | 0.803 | 1.197 | (0.544–2.635) | 0.655 |
| LDHA | 0.894 | (0.690–1.158) | 0.396 |
|
|
|
|
|
|
|
|
|
| LDHAL6B | 1.552 | (1.286–1.874) | <0.0001 | 1.285 | (1.048–1.576) | 0.016 | 1.010 | (0.963–1.059) | 0.680 | 1.066 | (0.994–1.144) | 0.073 |
| LDHAL6A | 0.895 | (0.752–1.065) | 0.212 |
|
|
|
|
|
|
|
|
|
| LDHB | 1.087 | (0.813–1.453) | 0.574 |
|
|
|
|
|
|
|
|
|
| LDHC | 1.093 | (1.002–1.193) | 0.046 | 1.073 | (0.978–1.177) | 0.137 |
|
|
|
|
|
|
| LDHD | 0.765 | (0.682–0.858) | <0.0001 | 0.872 | (0.764–0.994) | 0.040 | 0.944 | (0.897–0.994) | 0.029 | 0.899 | (0.826–0.979) | 0.014 |
Expression quantities of LDHD and
LDHAL6B are associated with prognosis and OS
As LDHD and LDHAL6B expression were
the only independent predictors of OS in multivariate Cox analysis,
they were selected for analysis. LDHD expression and
LDHAL6B expression were revealed to be normally distributed,
and were thus considered as categorical variables according to the
median expression level (divided into low and high expression
groups according to the median values of LDHD and
LDHAL6B of 7.24 and 1.49, respectively). As a result, it was
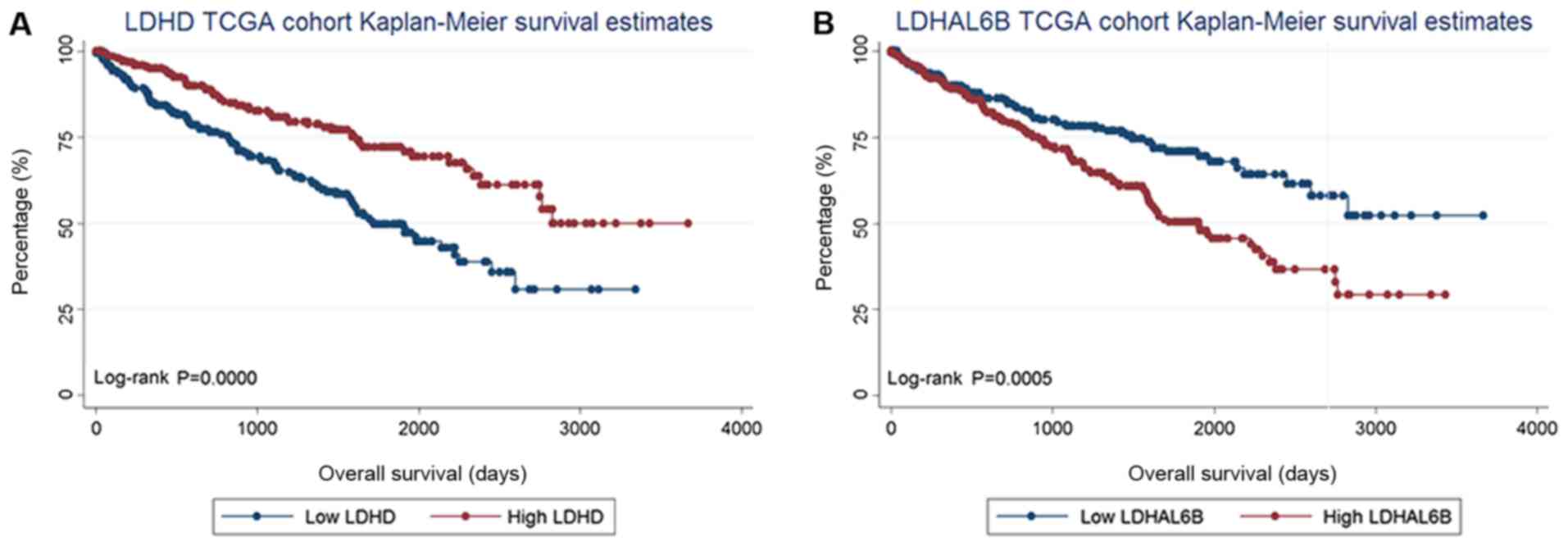
revealed that low LDHD expression (P<0.0001) was
associated with a poor prognosis for OS, whereas a low level of
LDHAL6B expression (P=0.0005) was associated with an
improved prognosis for OS, compared with their low expression
counterparts (Fig. 1). LDHD
and LDHAL6B expression were considered to be categorical
variables according to the median expression level. A log-rank test
was performed in order to compare the survival curves between the
different serum LDH levels.
In order to further understand the factors that may
affect the expression of LDHD and LDHAL6B, a
multivariate logistic regression analysis was performed. Tumor
pathological T stage was revealed to be significantly associated
with LDHD (P=0.003), whereas haemoglobin (P=0.003) was
significantly associated with LDHAL6B expression (Table III).
 | Table III.Multivariate logistic regression
analysis of factors that may affect the expression of LDHD
and LDHAL6B in The Cancer Genome Atlas cohort with clear cell renal
cell carcinoma. |
Table III.
Multivariate logistic regression
analysis of factors that may affect the expression of LDHD
and LDHAL6B in The Cancer Genome Atlas cohort with clear cell renal
cell carcinoma.
|
| LDHD | LDHAL6B |
|---|
|
|
|
|
|---|
| Variables | OR | (95% CI) | P-value | OR | (95% CI) | P-value |
|---|
| Age | 1.013 | (0.987–1.040) | 0.314 | 1.0114 | (0.986–1.038) | 0.382 |
| T | 0.565 | (0.389–0.819) | 0.003 | 1.0560 | (0.734–1.520) | 0.771 |
| N | 0.965 | (0.255–3.650) | 0.958 | 0.7600 | (0.216–2.671) | 0.669 |
| M | 1.652 | (0.695–3.926) | 0.256 | 0.6730 | (0.286–1.581) | 0.363 |
| Fuhrman grade | 0.788 | (0.499–1.243) | 0.305 | 1.0480 | (0.665–1.649) | 0.841 |
| Hb | 1.320 | (0.700–2.488) | 0.391 | 0.3800 | (0.202–0.717) | 0.003 |
|
WBC | 1.048 | (0.564–1.948) | 0.881 | 0.6880 | (0.371–1.276) | 0.236 |
|
PLT | 1.065 | (0.564–2.011) | 0.847 | 1.4330 | (0.756–2.718) | 0.271 |
The expression of LDH members in 70 paired patients
from the TCGA database was then analysed to understand the
difference in the expression of LDH family members between patients
with ccRCC and a control group. A paired Student's t-test was used
if the deviations of LDH expression between couples fitted a normal
distribution, and the Wilcoxon signed rank test was employed for
those that did not fit a normal distribution. In the paired
Student's t-tests, the expression of LDHD (P<0.0001) and
LDHAL6A (P<0.0001) were significantly different between
the patients with ccRCC and the paired controls, whereas the
expression of LDHAL6B (P=0.375) was not significantly
different. In the Wilcoxon's signed rank test analysis, the
expression of LDHA (P<0.0001), LDHB (P<0.0001)
and LDHC (P<0.0001) was significantly different between
patients with ccRCC and the paired controls (Table IV).
 | Table IV.Expression of LDH family members in
70 paired patients in The Cancer Genome Atlas cohort. |
Table IV.
Expression of LDH family members in
70 paired patients in The Cancer Genome Atlas cohort.
| Variable | P-value | Statistical
method | (95% CI) |
|---|
| LDHA | <0.0001 | Wilcoxon's rank rum
test |
|
| LDHAL6B | 0.3750 | Paired Student's
t-test | (−0.103–0.270) |
| LDHAL6A | <0.0001 | Paired Student's
t-test | (0.689–1.227) |
| LDHB | <0.0001 | Wilcoxon's rank sum
test |
|
| LDHC | <0.0001 | Wilcoxon's rank sum
test |
|
| LDHD | <0.0001 | Paired Student's
t-test | (2.269–3.168) |
LDHD expression is a prognostic factor
for OS in the FUSCC cohort
In the FUSCC cohort, LDHD and LDHAL6B
expression was validated. The expression of LDHD and
LDHAL6B was considered as categorical variables according to
the median expression level (low and high expression groups). As
the expression level of genes was based on the relative values of
the PCR results, patients were grouped by Δ-Cq (cycle threshold).
Δ-Cq=Cq(target genes)-Cq(reference genes). The median Δ-Cq values
of LDHD and LDHAL6B were 5.93 and 1.77, respectively.
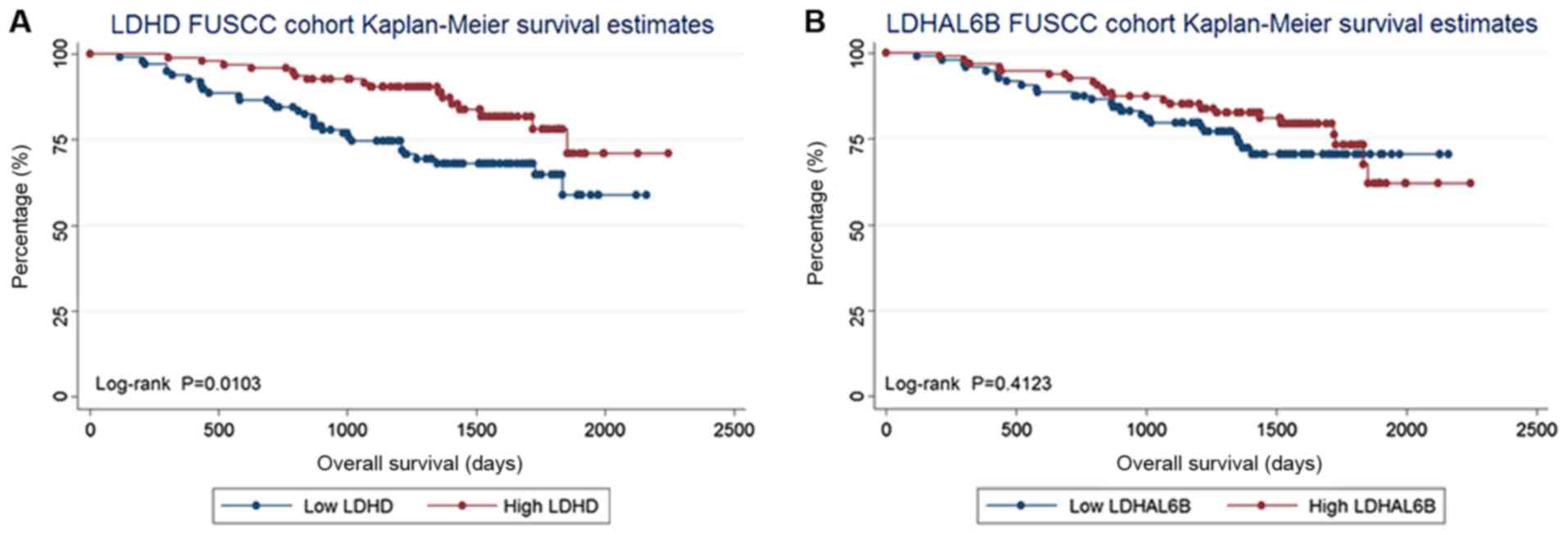
As a result, low LDHD expression was associated with a poor
prognosis for OS (log-rank test, P=0.010), whereas the expression
of LDHAL6B (log-rank test, P=0.412) was not associated with
OS. The Kaplan-Meier curves are presented in Fig. 2.
In order to further understand the factors that may
affect the expression of LDHD and LDHAL6B in the
FUSCC cohort, a multivariate logistic regression analysis using the
same parameters was performed. It was revealed that in the FUSCC
cohort, tumor pathological T stage was significantly associated
with the expression of LDHD (P=0.012), whereas age was
significantly associated with the expression of LDHAL6B
(P=0.043; Table V).
 | Table V.Multivariate logistic regression
analysis of factors that may affect the expression of LDHD
and LDHAL6B in the FUSCC cohort with ccRCC. |
Table V.
Multivariate logistic regression
analysis of factors that may affect the expression of LDHD
and LDHAL6B in the FUSCC cohort with ccRCC.
|
| LDHD | LDHAL6B |
|---|
|
|
|
|
|---|
| Variables | OR | (95% CI) | P-value | OR | (95% CI) | P-value |
|---|
| Age | 0.999 | (0.962–1.038) | 0.971 | 0.962 | (0.926–0.999) | 0.043 |
| T | 0.519 | (0.312–0.864) | 0.012 | 0.959 | (0.604–1.523) | 0.859 |
| N | 11.272 |
(0.856–148.417) | 0.065 | 2.651 | (0.223–31.479) | 0.440 |
| M | 1.056 | (0.069–16.042) | 0.969 | 0.521 | (0.6038–7.080) | 0.624 |
| Fuhrman grade | 0.587 | (0.295–1.169) | 0.130 | 0.579 | (0.300–1.117) | 0.103 |
| Hb | 0.986 | (0.958–1.013) | 0.305 | 1.000 | (0.974–1.026) | 0.978 |
|
WBC | 0.997 | (0.988–1.007) | 0.578 | 0.995 | (0.984–1.006) | 0.373 |
|
PLT | 1.003 | (0.997–1.009) | 0.300 | 1.001 | (0.995–1.007) | 0.733 |
Expression level of LDHD influences
the serum LDH level
As there was no data concerning the serum LDH level
in the TCGA database, the present study tested the serum LDH of
patients from the FUSCC cohort when they were diagnosed with a
kidney tumor in order to understand whether the expression of
LDHD is associated with serum LDH. In this further analysis
of the FUSCC cohort, the 192 patients were divided into two groups
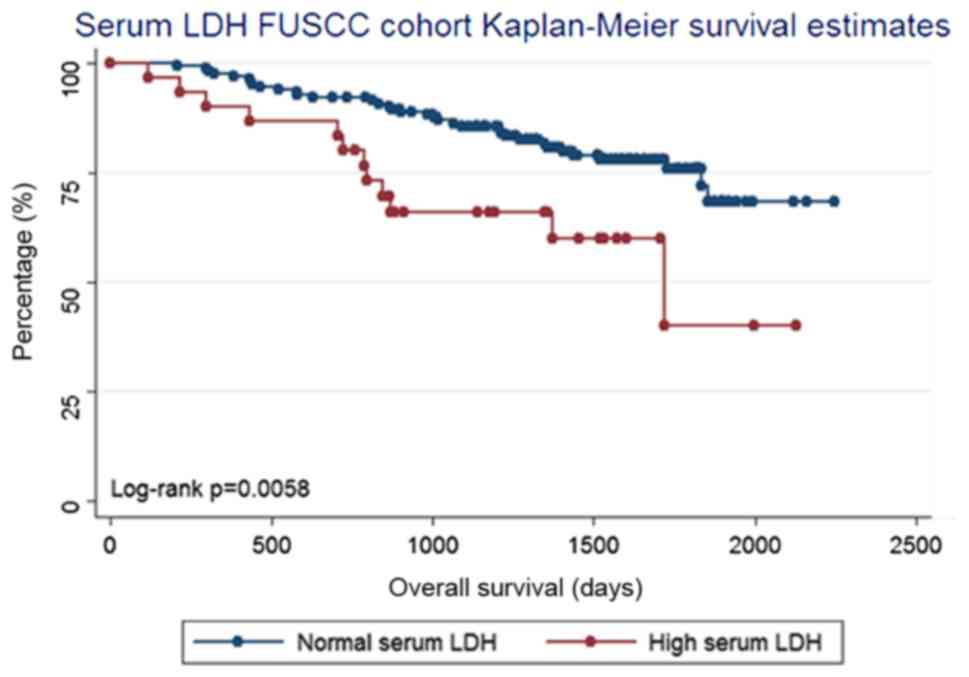
according to their serum LDH level. Log-rank testing revealed that
a serum LDH level higher than the upper limit of normal (215 U/l)
was associated with a poor prognosis for OS (log-rank test,
P=0.006; Fig. 3). This result is
concurrent with those of previous studies (11,14,21,26).
Furthermore, Spearman's correlation analysis (data not shown)
revealed that the serum LDH level has a negative correlation with
the expression of LDHD (P=0.028). Despite the fact that the
analysis of serum LDH is a protein test and the analysis of
LDHD is a gene test, it may be concluded that the expression
of LDHD is associated with the levels of serum LDH.
Expression of LDHD and LDHAL6B is not
associated with recurrence-free survival (RFS) and cancer-specific
survival (CSS) in the FUSCC cohort
In order to understand if the expression of
LDHD and LDHAL6B were associated with RFS and CSS,
LDHD and LDHAL6B expression in the FUSCC cohort was
considered as a categorical variable according to the median
expression level, and divided into high and low expression groups
as aforementioned. RFS was measured from the date of renal
resection until the date of radiographic detection of recurrence or
the last follow-up. CSS was measured from the date of renal
resection until the the date of mortality due to renal cancer or
the last follow-up. A total of 18 patients were diagnosed with
radiographic recurrence and 10 patients succumbed to renal cancer
during the follow-up. As a result, it was revealed that the
expression of LDHD was not associated with RFS (log-rank
test, P=0.887) or CSS (log-rank test, P=0.133). LDHAL6B
expression was also not associated with RFS (log-rank test,
P=0.364) and CSS (log rank test, P=0.430) (data not shown).
Discussion
In the present study, it was demonstrated that
certain LDH gene family members were associated with the OS of
patients with ccRCC. Members of this family, particularly
LDHD and LDHAL6B, were independent prognostic factors
for the OS of patients with ccRCC in the present study.
The human genome has four LDH genes: LDHA, LDHB,
LDHC, and LDHD. Of these genes (26), LDHB and LDHC are L
isomers and LDHD is a D isomer (27).
LDHD metabolism has been demonstrated to occur in
mitochondria, but its function in cancer is unclear. Previous
studies have demonstrated that in prostate cancer cells, the
metabolism of D-lactate inside mitochondria via LDHD and the
metabolic activity of LDHD in tumor cells were higher
compared with that in normal cells (28,29).
Furthermore, the LDHD protein level and activity are higher in
cancerous cells compared with that in normal prostate cells
(30). LDHD is a unique gene;
in Sporolactobacillus inulinus strain CASD, LDHD may use
NADH and NADPH, but preferentially uses NADPH as a coenzyme, which
is different from the coenzyme utilization of other LDHs (31,32).
A feature of tumor cells is their reliance upon
fermentative glycolysis, a common phenomenon coined ‘the Warburg
effect’ (33). LDH isoenzymes are
metabolically regulated, and are linked to glycolysis and the
Warburg effect (34). Previous
studies have revealed that the level of pre-treatment serum LDH is
a significant prognostic factor in numerous types of cancer,
including RCC (35–37). LDH is under the translational control
of hypoxia-inducible factor and MYC, and is thus regulated by key
oncogenic processes (33,38). However, in the field of oncology,
little is known concerning the subtypes of the LDH family. The few
previous studies focused on this topic have been mainly confined to
the cellular level. RCC is a disease that is closely associated
with metabolism (39,40). The LDH gene family serves an important
function in ccRCC. LDHA may possess multiple additional
functions in non-neoplastic and neoplastic tissues (41). There are numerous genes and proteins,
including tumor protein p53, vascular endothelial growth factor and
2′-deoxynucleoside 5′-phosphate N-hydrolase 1, amongst others,
reported to be associated with LDHA activity (41). The expression of LDHA is
associated with the prognosis of patients with brain tumors. A
previous study revealed that LDHA metabolic activity in
brain tumor cells was stronger compared with normal cells (41). However, the clinical association
between LDHD expression and tumors is not clear. There are
also few studies focused on renal cancer.
The present study revealed that LDHD may
serve an important function in the prognosis of patients with
ccRCC. It was revealed that the expression of LDHD and
LDHAL6B were independent prognostic factors for the OS of
patients with ccRCC. The downregulated expression of LDHD
was associated with a poor prognosis and a shorter OS. The
expression of LDHD is associated with the prognosis of
patients with ccRCC. In addition, the tumor stage was significantly
associated with the expression of LDHD. In the statistical
analysis of 70 paired patients, the expression of LDHD was
also significantly different between patients with ccRCC and paired
controls. Furthermore, the serum LDH level was significantly
associated with the expression of LDHD. The results of the
present study confirmed that LDHD is a useful biomarker for
patients with ccRCC. In addition, the expression of LDHD was
associated with patient prognosis. To the best of our knowledge, no
previous study has clarified the function of LDHD in renal
tumors. Although Girgis et al (42) had previously assessed the expression
of LDHA at the protein level via the use of
immunohistochemistry in 385 patients and indicated that LDHA
upregulation may be a predictor of a poor prognosis in patients
with ccRCC, in the present study, LDHA expression was not
significantly associated with the OS of patients with ccRCC in the
TCGA cohort or the FUSCC cohort.
LDHD is a subtype of LDH and serves an important
function in renal cancer. The integration of LDHD expression
has the potential to be a useful biomarker in the identification of
poor prognosis in patients with ccRCC. Although it has potential as
a novel renal cancer biomarker, its specific mechanism remains
unknown. To date, LDHD, which is produced in the methylglyoxal (MG)
pathway, is presumed to be released by cancer cells (43). The MG pathway produces LDHD as a final
product that is largely modified in cancer cells, with a specific
function in the glyoxalase systems, serving to eliminate the
cytotoxic MG mainly derived from glycolysis (44). The rationally engineered LDHD may
efficiently use NADH and NADPH as cofactors. Additionally, the
mitochondrial metabolism of LDHD in cancer cells is more active
compared with that of normal cells (45). Renal cancer is a disease that is
associated with metabolism and obesity. LDHD may serve a key
function in the progression and development of ccRCC.
There are a number of limitations to the present
study: Firstly, the patients included were from the FUSCC, with
good follow up, but patients from other centres were not included.
Secondly, all the tissue specimens in the present study were
sourced from patients who were suitable candidates for surgery, and
the same results may not apply to patients who are not suitable
candidates for surgery.
The present study has indicated at the association
between ccRCC outcome and LDHD, however, the underlying
mechanism remains poorly understood. The present study may have
opened a threshold to a novel aspect of ccRCC biomarkers or
therapeutic targets. Further studies are required.
In conclusion, LDHD was identified as an
independent prognostic factor for the OS of patients with ccRCC.
Low LDHD expression was associated with a poor prognosis for
OS, and tumor grade was significantly associated with LDHD
expression. LDHD may function as a tool to reveal further
genes associated with prognosis in ccRCC.
Acknowledgements
The present study was supported by the International
Cooperation and Exchange of Science and Technology Commission of
the Shanghai Municipality (grant no. 12410709300), the Guide
Project of Science and Technology Commission of Shanghai
Municipality (grant no. 124119a7300), and the Outstanding Young
Talent Training Plan of Shanghai Municipal Commission of Health and
Family Planning (grant no. XYQ2013102).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Linehan WM, Srinivasan R and Schmidt LS:
The genetic basis of kidney cancer: A metabolic disease. Nat Rev
Urol. 7:277–285. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Simard EP, Ward EM, Siegel R and Jemal A:
Cancers with increasing incidence trends in the United States: 1999
through 2008. CA Cancer J Clin. 62:118–128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ridge CA, Pua BB and Madoff DC:
Epidemiology and staging of renal cell carcinoma. Semin Intervent
Radiol. 31:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liang L, Li L, Zeng J, Gao Y, Chen YL,
Wang ZQ, Wang XY, Chang LS and He D: Inhibitory effect of silibinin
on EGFR signal-induced renal cell carcinoma progression via
suppression of the EGFR/MMP-9 signaling pathway. Oncol Rep.
28:999–1005. 2012.PubMed/NCBI
|
|
7
|
Maher ER: Genomics and epigenomics of
renal cell carcinoma. Semin Cancer Biol. 23:10–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ficarra V, Galfano A, Novara G, Iafrate M,
Brunelli M, Secco S, Cavalleri S, Martignoni G and Artibani W: Risk
stratification and prognostication of renal cell carcinoma. World J
Urol. 26:115–125. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Burgner JW II and Ray WJ Jr: On the origin
of the lactate dehydrogenase induced rate effect. Biochemistry.
23:3636–3648. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brown JE, Cook RJ, Lipton A and Coleman
RE: Serum lactate dehydrogenase is prognostic for survival in
patients with bone metastases from breast cancer: A retrospective
analysis in bisphosphonate-treated patients. Clin Cancer Res.
18:6348–6355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen J, Chen Z, Zhuang Q, Fan M, Ding T,
Lu H and He X: Prognostic value of serum lactate dehydrogenase in
renal cell carcinoma: A systematic review and meta-analysis. PLoS
One. 11:e01664822016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen Y, Zhang H, Xu A, Li N, Liu J, Liu C,
Lv D, Wu S, Huang L, Yang S, et al: Elevation of serum l-lactate
dehydrogenase B correlated with the clinical stage of lung cancer.
Lung cancer. 54:95–102. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang ZX, Yang LP, Qiu MZ, Wang ZQ, Zhou
YX, Wang F, Zhang DS, Wang FH, Li YH and Xu RH: Prognostic value of
preoperative serum lactate dehydrogenase levels for resectable
gastric cancer and prognostic nomograms. Oncotarget. 7:39945–39956.
2016.PubMed/NCBI
|
|
14
|
Armstrong AJ, George DJ and Halabi S:
Serum lactate dehydrogenase predicts for overall survival benefit
in patients with metastatic renal cell carcinoma treated with
inhibition of mammalian target of rapamycin. J Clin Oncol.
30:3402–3407. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cetin B, Afsar B, Deger SM, Gonul II,
Gumusay O, Ozet A, Benekli M, Coskun U and Buyukberber S:
Association between hemoglobin, calcium, and lactate dehydrogenase
variability and mortality among metastatic renal cell carcinoma.
Int Urol Nephrol. 46:1081–1087. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Adeva-Andany M, López-Ojén M,
Funcasta-Calderón R, Ameneiros-Rodríguez E, Donapetry-García C,
Vila-Altesor M and Rodríguez-Seijas J: Comprehensive review on
lactate metabolism in human health. Mitochondrion. 17:76–100. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fan J, Hitosugi T, Chung TW, Xie J, Ge Q,
Gu TL, Polakiewicz RD, Chen GZ, Boggon TJ, Lonial S, et al:
Tyrosine phosphorylation of lactate dehydrogenase A is important
for NADH/NAD(+) redox homeostasis in cancer cell. Mol Cell Biol.
31:4938–4950. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Markert CL, Shaklee JB and Whitt GS:
Evolution of a gene. Multiple genes for LDH isozymes provide a
model of the evolution of gene structure, function and regulation.
Science. 189:102–114. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun Q, Chen X, Ma J, Peng H, Wang F, Zha
X, Wang Y, Jing Y, Yang H, Chen R, et al: Mammalian target of
rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is
critical for aerobic glycolysis and tumor growth. Proc Natl Acad
Sci USA. 108:4129–4134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zha X, Wang F, Wang Y, He S, Jing Y, Wu X
and Zhang H: Lactate dehydrogenase B is critical for hyperactive
mTOR-mediated tumorigenesis. Cancer Res. 71:13–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hoffmann R, Franzke A, Buer J, Sel S,
Oevermann K, Duensing A, Probst M, Duensing S, Kirchner H, Ganser A
and Atzpodien J: Prognostic impact of in vivo soluble cell adhesion
molecules in metastatic renal cell carcinoma. Br J Cancer.
79:1742–1745. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sobin LH and Fleming ID: TNM
classification of malignant tumors, fifth edition (1997). Union
Internationale Contre le Cancer and the American Joint Committee on
Cancer. Cancer. 80:1803–1804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rioux-Leclercq N: The Fuhrman grading
system for kidney cancer prognosis. Prog Urole. 16(4 Suppl): FMC):.
S5–S8. 2006.(In French).
|
|
24
|
Kim SP, Alt AL, Weight CJ, Costello BA,
Cheville JC, Lohse C, Allmer C and Leibovich BC: Independent
validation of the 2010 American Joint Committee on Cancer TNM
classification for renal cell carcinoma: Results from a large,
single institution cohort. J Urol. 185:2035–2039. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gallo M, Sapio L, Spina A, Naviglio D,
Calogero A and Naviglio S: Lactic dehydrogenase and cancer: An
overview. Front Biosci (Landmark Ed). 20:1234–1249. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gleason FH and Nolan RA: D(−)-lactate
dehydrogenase in lower fungi. Science. 152:1272–1273. 1966.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rojo EE, Guiard B, Neupert W and Stuart
RA: Sorting of D-lactate dehydrogenase to the inner membrane of
mitochondria. Analysis of topogenic signal and energetic
requirements. J Biol Chem. 273:8040–8047. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lapierre L, Germond JE, Ott A, Delley M
and Mollet B: D-Lactate dehydrogenase gene (ldhD) inactivation and
resulting metabolic effects in the Lactobacillus johnsonii strains
La1 and N312. Appl Environ Microbiol. 65:4002–4007. 1999.PubMed/NCBI
|
|
30
|
de Bari L, Moro L and Passarella S:
Prostate cancer cells metabolize d-lactate inside mitochondria via
a D-lactate dehydrogenase which is more active and highly expressed
than in normal cells. FEBS Lett. 587:467–473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pagala VR, Park J, Reed DW and Hartzell
PL: Cellular localization of D-lactate dehydrogenase and NADH
oxidase from Archaeoglobus fulgidus. Archaea. 1:95–104. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bernard N, Johnsen K, Holbrook JJ and
Delcour J: D175 discriminates between NADH and NADPH in the
coenzyme binding site of Lactobacillus delbrueckii subsp.
bulgaricus D-lactate dehydrogenase. Biochem Biophys Res Commun.
208:895–900. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim JW and Dang CV: Cancer's molecular
sweet tooth and the Warburg effect. Cancer Res. 66:8927–8930. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Saedeleer CJ, Copetti T, Porporato PE,
Verrax J, Feron O and Sonveaux P: Lactate activates HIF-1 in
oxidative but not in Warburg-phenotype human tumor cells. PLoS One.
7:e465712012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
von Eyben FE, Madsen EL, Liu F, Amato R
and Fritsche H: Serum lactate dehydrogenase isoenzyme 1 as a
prognostic predictor for metastatic testicular germ cell tumours.
Br J Cancer. 83:1256–1259. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Halabi S, Small EJ, Kantoff PW, Kattan MW,
Kaplan EB, Dawson NA, Levine EG, Blumenstein BA and Vogelzang NJ:
Prognostic model for predicting survival in men with
hormone-refractory metastatic prostate cancer. J Clin Oncol.
21:1232–1237. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Agarwala SS, Keilholz U, Gilles E,
Bedikian AY, Wu J, Kay R, Stein CA, Itri LM, Suciu S and Eggermont
AM: LDH correlation with survival in advanced melanoma from two
large, randomised trials (Oblimersen GM301 and EORTC 18951). Eur J
Cancer. 45:1807–1814. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim JW and Dang CV: Multifaceted roles of
glycolytic enzymes. Trends Biochem Sci. 30:142–150. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cho E, Giovannucci EL and Joh HK:
Nutrients related to one-carbon metabolism and risk of renal cell
cancer. Cancer Causes Control. 24:373–382. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang OC, Maxwell PH and Pollard PJ: Renal
cell carcinoma: Translational aspects of metabolism and therapeutic
consequences. Kidney Int. 84:667–681. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Serganova I, Rizwan A, Ni X, Thakur SB,
Vider J, Russell J, Blasberg R and Koutcher JA: Metabolic imaging:
A link between lactate dehydrogenase A, lactate, and tumor
phenotype. Clin Cancer Res. 17:6250–6261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Girgis H, Masui O, White NM, Scorilas A,
Rotondo F, Seivwright A, Gabril M, Filter ER, Girgis AH, Bjarnason
GA, et al: Lactate dehydrogenase A is a potential prognostic marker
in clear cell renal cell carcinoma. Mol Cancer. 13:1012014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Santel T, Pflug G, Hemdan NY, Schäfer A,
Hollenbach M, Buchold M, Hintersdorf A, Lindner I, Otto A, Bigl M,
et al: Curcumin inhibits glyoxalase 1: A possible link to its
anti-inflammatory and anti-tumor activity. PLoS One. 3:e35082008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rulli A, Carli L, Romani R, Baroni T,
Giovannini E, Rosi G and Talesa V: Expression of glyoxalase I and
II in normal and breast cancer tissues. Breast Cancer Res Treat.
66:67–72. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Meng H, Liu P, Sun H, Cai Z, Zhou J, Lin J
and Li Y: Engineering a d-lactate dehydrogenase that can
super-efficiently utilize NADPH and NADH as cofactors. Sci Rep.
6:248872016. View Article : Google Scholar : PubMed/NCBI
|