Introduction
The development of ameloblastoma (AB) may be similar
to tooth germ development, as it is likely derived from the
epithelial lining of the dental follicle, the epithelial residue of
the tooth plate and the basal cells of the oral mucosa (1).
From C. elegans to humans, WNT genes
are conserved across many species. Previous studies have
demonstrated that WNT1 is a key signal molecule for controlling
cell growth and proliferation. It transfers regulatory information
between cells, and serves an important role in stem cell
differentiation and neural development. Kumamoto and Ooya (2) reported that WNT pathway abnormalities
induce tumor occurrence, as WNT1 activation phosphorylates
β-catenin in the nucleus to activate the classic WNT pathways,
inducing cell proliferation and inhibiting apoptosis to generate
anomalous growth.
As the effect of the WNT signaling pathway in the
process of tooth development is well characterized, and as multiple
associated factors are expressed during different developmental
periods of the tooth germ (3), we
hypothesized that this pathway may be associated with the
development of AB. Therefore, immunohistochemistry, western
blotting and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) were used to examine WNT1 expression in AB, with
the aim of providing a basis for diagnosing and treating AB.
Materials and methods
Tissue samples
Immunohistochemical specimens were obtained from
paraffin blocks archived at the Department of Oral Pathology,
Stomatological Hospital of China Medical University (Shenyang,
China), between June 2009 and April 2013. The study protocol was
approved by the Medical Ethics Committee of Stomatological Hospital
of China Medical University. The study was performed in accordance
with the Declaration of Helsinki. All specimens were analyzed
subsequent to obtaining verbal informed patient consent, This was
not a retrospective study. There were 80 cases of AB (Table I) with no chemotherapy or radiotherapy
administered prior to tissue extraction, 10 of keratocystic
odontogenic tumor (KCOT) and 10 samples of normal oral mucosa
(NOM). There were 40 male and 40 female patients with AB; the age
range was 18–79 years and the median age was 49 years. All
specimens were classified according to the 2005 World Health
Organization (WHO) criteria (4). The
areas the tumor tissue was extracted from included the mandible (65
cases), maxilla (12 cases) and gingiva (3 cases). The histological
types were as follows: 76.3% (61/80) solid/polycystic, 1.6% (13/80)
unicystic, 3.8% (3/80) desmoplastic and 3.8% (3/80) peripheral. Of
the solid type, 42 cases were typed as follicular, 5 as plexiform,
7 as follicular/plexiform, 3 as acanthomatous, 2 as basal cell and
2 as keratinous.
 | Table I.Histopathological classification of
the 80 AB tumor samples used in immunohistochemistry. |
Table I.
Histopathological classification of
the 80 AB tumor samples used in immunohistochemistry.
| AB type | Cases, n |
|---|
| Solid/polycystic | 61 |
|
Follicular | 42 |
|
Plexiform | 5 |
|
Follicular/plexiform | 7 |
|
Acanthomatus | 3 |
| Basal
cell | 2 |
|
Keratinous | 2 |
| Unicystic | 13 |
| Desmoplastic | 3 |
| Peripheral | 3 |
The fresh tissue samples obtained between January
2004 and December 2009 for western blotting and RT-qPCR were
surgical resection specimens from the Department of Oral and
Maxillofacial Surgery, Stomatological Hospital of China Medical
University. The samples included 30 cases of AB (Table II), 5 cases of KCOT, and 5 NOM
samples. In addition, 3 cases of tooth germ tissue were abortuses
from the Department of Obstetrics of the People's Liberation Army
no. 202 Hospital (Shenyang, China). The samples were cryopreserved
at −86°C and were classified according to the 2005 WHO criteria.
There were 16 male and 14 female patients with AB; the age range
was 16–72 years, and the median age of which was 44 years. The
areas involved included the mandible (24 cases), maxilla (3 cases),
gingiva (1 case), left parapharyngeal space palate (1 case) and
left cheek (1 case). The histological type was as follows: 83.3%
(25/30) were solid/polycystic, 3.3% (1/30) unicystic, 3.3% (1/30)
desmoplastic and 10% (3/30) peripheral. Of the solid type, 14 were
follicular, 5 plexiform, 3 acanthomatous, 2 basal cell and 1
follicular/plexiform.
 | Table II.Histopathological classification of
the 30 AB tumor samples used for western blotting and reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Histopathological classification of
the 30 AB tumor samples used for western blotting and reverse
transcription-quantitative polymerase chain reaction.
| AB type | Cases, n |
|---|
| Solid/polycystic | 25 |
|
Follicular | 14 |
|
Plexiform | 5 |
|
Follicular/plexiform | 1 |
|
Acanthoma | 3 |
| Basal
cell | 2 |
| Unicystic | 1 |
|
Desmoplastic | 3 |
|
Peripheral | 1 |
Immunohistochemistry
Sections (5 µm) were obtained from the
paraffin-embedded tissues of the first group. Following the
inactivation of endogenous peroxidase by quenching with
H2O2, sections were treated overnight with a
goat anti-WNT1 polyclonal primary antibody (dilution, 1:400; Abcam,
Cambridge, MA, USA; cat no. ab15251) at 4°C. The
streptavidin-peroxidase (SP) method was used; an SP kit (Zymed;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) contained
biotinylated secondary antibodies that were incubated for 10 min at
37°C; sections were washed three times with PBS buffer between
steps. One drop (38 µl) of liquid Diaminobenzidine (DAB, Maxim
Biotech, Inc., Fuzhou, China) chromogen was added to 1 ml of stable
DAB substrate buffer (cat no. ab64238; Abcam, Cambridge, MA, USA)
in the mixing vial. This DAB chromogen reagent was added at room
temperature for 5 to 15 min until the color turned brown. Samples
in which the primary antibody had been replaced with PBS buffer
were used as the negative control.
Immunohistochemical reactivity for WNT1 was
evaluated based on brown staining in the cytoplasm. The evaluation
considered (A) the intensity of staining: Colorless, 0; light
yellow, 2; brown-yellow, 2; brown, 3; and (B) the proportion of
stained tumor cells: <25%, 1; 25–75%, 2; >75%, 3. The score
for each case was derived as A × B, and cases were classified
according to their score: 0–1, negative (−); 2–4, weakly positive
(+); 4–7, moderately positive (++); and >7, strong positive
(+++), as previously described (5).
Western blot
Pre-made protein cracking fluid (20 mmol/l Tris-HCL
pH 7.4, 150 mmol/l NaCl, 1% Triton X-100, 0.1% SDS. 1% deoxycholic
acid socium, 1 mmol/l EDTA, 1 mmol/l PMSF, 1 µg/ml aprotinin, 1
mmol/l Na3VO4) was used to extract proteins
and extracted proteins were separated using SDS-PAGE (12%
separation polyacrylamide gel and 4% polyacrylamide concentration
glue). The amount of protein per lane was 38.8 µg. The coomassie
brilliant blue method (6) was used
for protein quantification. The proteins were transferred to a
nitrocellulose membrane (Biomol; Enzo Life Sciences, Inc.,
Farmingdale, NY, USA) and blocked with Tween-20 in Tris-buffered
saline containing 5% non-fat dry milk overnight at 4°C. The goat
anti-human WNT1 polyclonal antibody (dilution, 1:400; cat no.
ab15251; Abcam) was used as the primary antibody; the secondary
antibody was alkaline phosphatase-labeled anti-goat IgG (dilution,
1:1,000; cat no. A0208; Seikagaku Corporation, Tokyo, Japan).
β-tubulin was used as the internal control antibody (dilution,
1:1,000; cat no. A01030; Abbkine Scientific Co., Ltd., Wuhan,
China). The 5-bromo-4-chloro-3-indolyl phosphate/nitro blue
tetrazolium Alkaline Phosphatase Color Development kit was used for
visualization (cat no. C3206; Beyotime Institute of Biotechnology,
Jiangsu, China). AlphaView 2.0 gel imaging analysis software
(ProteinSimple; Bio-Techne, Minneapolis, MN, USA) was used for
quantitative analysis.
RT-qPCR
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.)
was used to extract total RNA, and cDNA was synthesized with
reverse transcription using the PrimeScript One Step RT-PCR kit
Ver.2 (cat no. RR055A; Takara Biotechnology Co., Ltd., Dalian,
China). PCR amplification of the cDNA was performed in 20 µl
mixtures according to the protocol of the SYBR Premix Ex Taq™ II
kit (cat no. RR820A; Takara Biotechnology Co., Ltd.). The
thermocycling conditions were as follows: Initial denaturation
stage at 95°C for 30 sec, followed by 40 cycles of denaturation at
95°C for 5 sec, and then primer annealing and template
amplification at 60°C for 34 sec. Primer3 software was used to
design the PCR primers, which were synthesized by Beijing Genomics
Institute (Beijing, China) (Table
III). β-actin (ACTB) was used as an internal control.
qPCR results were analyzed using Rotor gene Real-Time Analysis
software 2.3.1 (Corbett Life Science; Qiagen GmbH, Hilden,
Germany). The comparative Cq (2−ΔΔCq) method was used to
determine the relative quantitative result (7).
 | Table III.WNT1 and ACTB
primers. |
Table III.
WNT1 and ACTB
primers.
| Gene | Primers | Sequence | Product size, bp |
|---|
| WNT1 | Upstream |
5′-CGGGCAACAACCAAAGTC-3′ | 107 |
|
| Downstream |
5′-GCAGCAGCGTAGCAGAAAC-3′ |
|
| ACTB | Upstream |
5′-AGTTGCGTTACACCCTTTC-3′ | 492 |
|
| Downstream |
5′-TGTCACCTTCACCGTTCC-3′ |
|
Statistical analysis
Data are presented as the mean ± standard error of
the mean. All experiments were conducted with a minimum of three
repeats. Statistical analysis was performed using SPSS 17.0
software. χ2 tests and one-way analysis of variance
followed by Fisher's least significant difference tests were used
for the comparisons. P<0.05 was considered to represent a
statistically significant difference.
Results
WNT1 protein expression is increased
in AB, as determined with immunohistochemistry
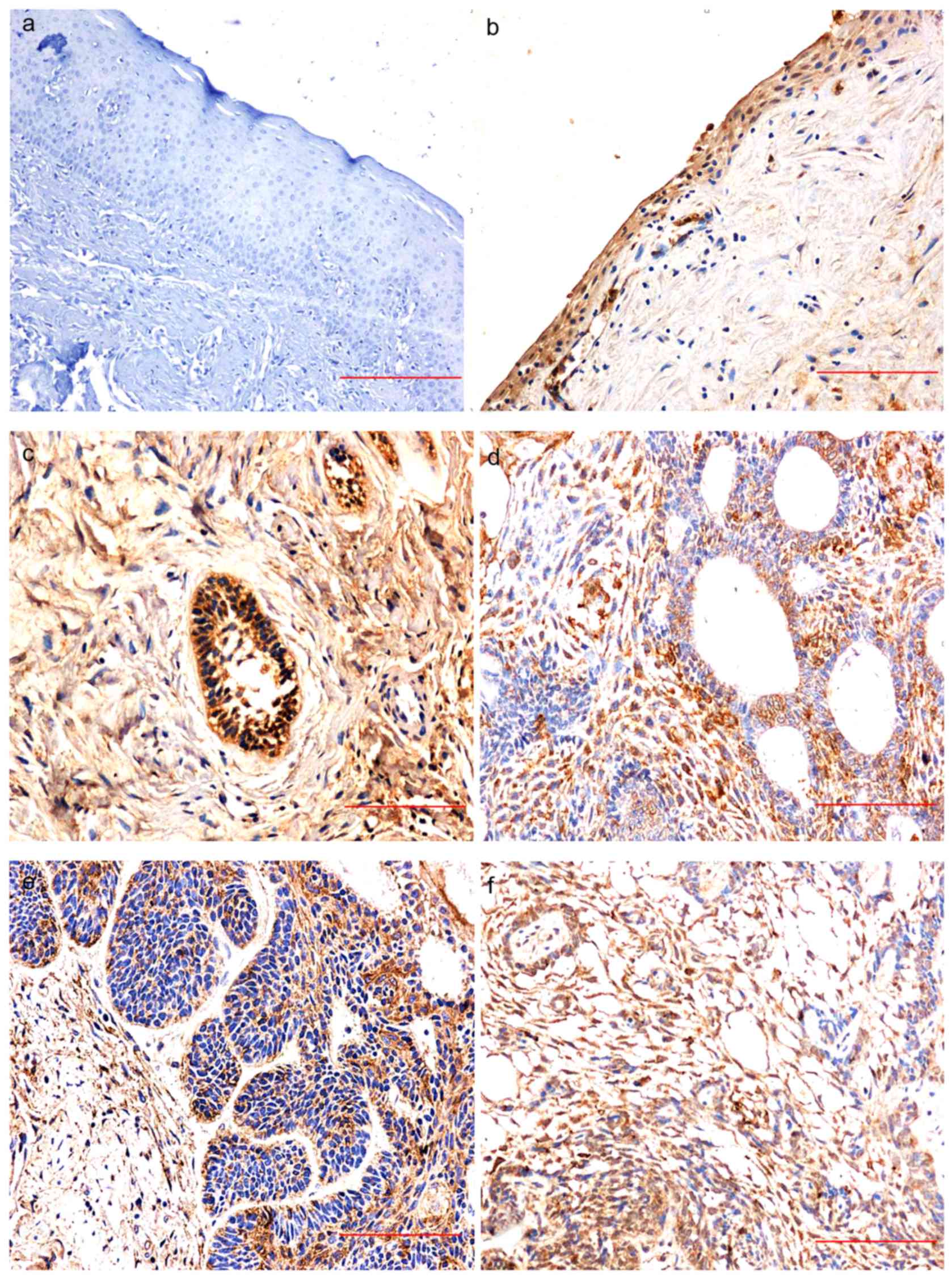
WNT1 expression in NOM was negative for all samples
(Fig. 1A) while there was positive
WNT1 expression in 70% of KCOT samples (7/10); WNT1 was expressed
in the cytoplasm of the lining epithelial cells, and staining was
generally weak-to-moderately positive (Fig. 1B). There was positive WNT1 expression
in 89% of AB samples (71/80); staining was predominantly moderately
or strongly positive in the outer periphery of the columnar or
cuboidal cells of the dental epithelium, the cytoplasm of the
central stellate reticular layer, and the inflammatory interstitial
lymphocytes (Fig. 1C-F).
As shown by Tables
IV–VI, WNT1 expression in AB was
significantly higher than in NOM (P<0.001), but not
significantly different from in KCOT (P>0.05). There was no
significant difference between AB types (P>0.05).
 | Table IV.Difference in WNT1 expression between
AB and NOM as determined by immunohistochemistry. |
Table IV.
Difference in WNT1 expression between
AB and NOM as determined by immunohistochemistry.
|
|
| WNT1 expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Tissue type | n | Positive | Negative | χ2 | P-value |
|---|
| AB | 80 | 71 | 9 | 36.88 | <0.001 |
| NOM | 10 | 0 | 10 |
|
|
 | Table VI.Difference in WNT1 expression between
different types of AB as determined by immunohistochemistry. |
Table VI.
Difference in WNT1 expression between
different types of AB as determined by immunohistochemistry.
|
|
| WNT1
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| AB type | n | Positive | Negative | χ2 | P-value |
|---|
|
Solid/polycystic | 61 | 55 | 6 | 2.073 | 0.56 |
| Unicystic | 13 | 11 | 2 |
|
|
| Desmoplastic | 3 | 2 | 1 |
|
|
| Peripheral | 3 | 3 | 0 |
|
|
WNT1 protein expression is increased
in AB, as determined with western blotting
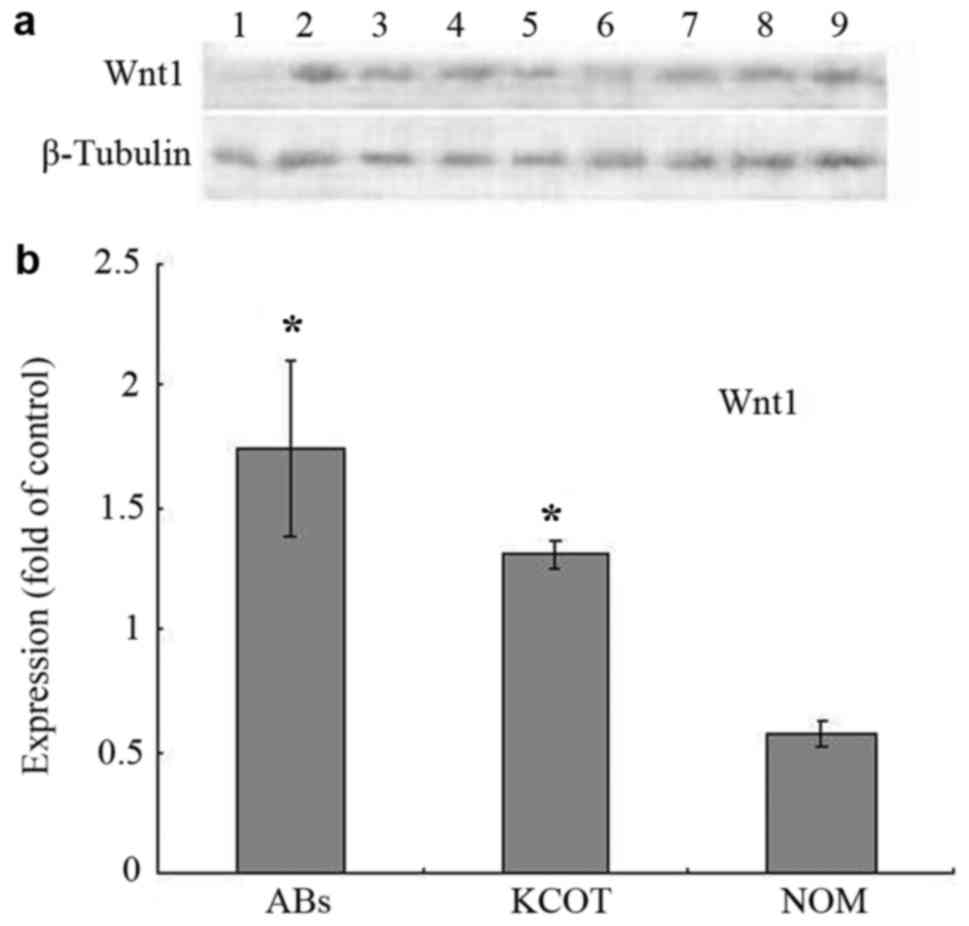
The WNT1 protein expression level in each sample was
determined with western blotting (Fig.
2A); expression levels in AB, KCOT and NOM were 1.74±0.36,
1.31±0.06, and 0.57±0.05 (Fig. 2B).
WNT1 expression in AB was significantly higher than in NOM
(P<0.05), but not significantly different from in KCOT
(P>0.05).
WNT1 mRNA expression is increased in
AB, as determined with RT-qPCR
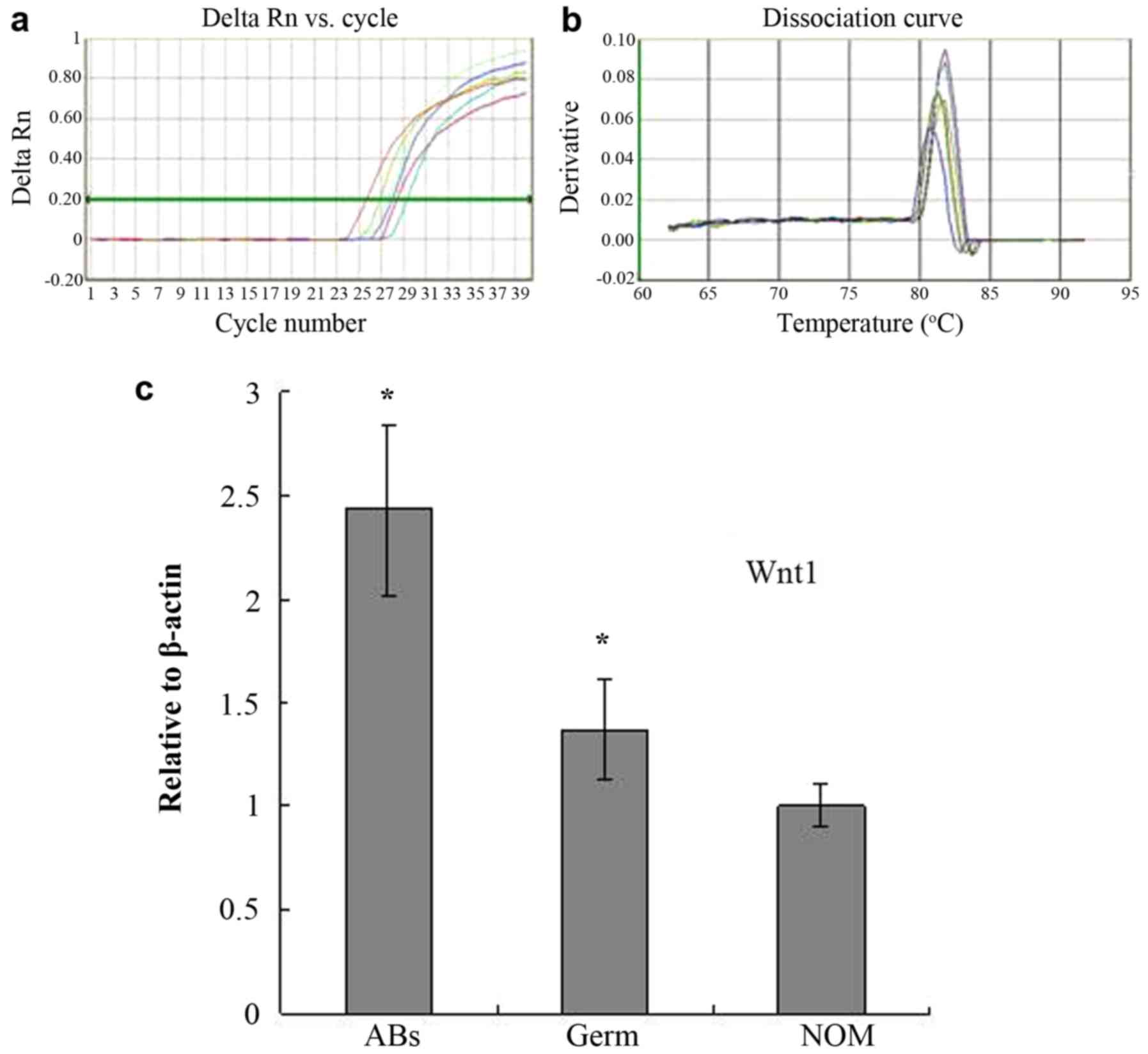
RT-qPCR was used to assess the WNT1 mRNA
expression level in 30 AB, 3 tooth germ and 5 NOM tissue samples
(Fig. 3A and B). The WNT1 PCR
product melting curve peaks of all samples were at 83°C, the
dissolution temperature was uniform and the peaks were sharp,
indicating that the product was specific. It was determined that
the expression of WNT1 was increased 2.43-fold in AB
compared with NOM tissue samples and 1.77-fold in AB compared with
tooth germ tissue samples (both P<0.05; Fig. 3C).
Discussion
The WNT signaling pathway is named for its
initiating protein, WNT1; WNT1 is the first member of the
19-member WNT gene family and encodes a 370-amino acid
protein (8). Howe et al
(9) reported that WNT1 induces
overexpression of downstream target genes by activating the
WNT/β-catenin signaling pathway, resulting in excessive cell
growth. The diverse biological effects of WNT signaling include the
regulation of cell proliferation, morphogenesis, and cell survival
and death processes; abnormal activation of WNT signaling is
associated with a variety of developmental disorders, degenerative
diseases, types of cancer e.g. colon cancer (10), retinopathy (11), limbless malformations (12), and bone and cartilage diseases,
including arthritis (13). WNT1 is a
tumor-associated protein which is upregulated in breast cancer,
hepatocellular carcinoma, oral carcinoma, colorectal cancer and
other types of epithelial malignancy; it may be involved in the
occurrence of these tumors (14–17). Joeng
et al demonstrated that WNT1 effectively reduced β-catenin
expression in the cytoplasm, and induced apoptosis by recovering
the function of WNT inhibitory factor-1 (WIF-1) in colon cancer
cells, increasing WNT siRNA or using anti-WNT monoclonal antibodies
(18).
In the present study, WNT1 expression was positive
in 71 cases of AB; of these, expression was strongly positive in 60
cases (84.5%) and weakly positive in 11 cases (15.5%), suggesting
heterogeneity in WNT1 expression intensity in AB. In addition, WNT1
was expressed only in the cytoplasm of tumor cells, and not in the
nucleus. WNT1 was expressed in the peripheral cells of tumor cell
nests and the stellate reticular cell layer. We hypothesize that
positive WNT1 expression in tumor-adjacent tissues may be
associated with the invasiveness of AB. Positive WNT1 expression
was also detected in cells surrounding the epithelial islands of
follicular and acanthoma AB, reflecting the importance of WNT1
signaling in cell proliferation (19). However, it is difficult to compare the
difference in WNT1 content using only immunohistochemical
staining.
Western blotting demonstrated that WNT1 levels in
the tumor tissue were significantly higher than in NOM tissue,
suggesting that WNT1 expression was enhanced in AB. RT-qPCR was
used to determine that WNT1 mRNA levels in 30 cases of AB
were 2.43 times higher than that in NOM tissue, suggesting that
WNT1 expression is enhanced at mRNA level. As mRNA levels directly
reflect gene expression, they support the hypothesis that
WNT1 gene activation is associated with AB. The mechanisms
for the activation of the WNT1 gene in AB has yet to be
determined.
The WNT gene was designated as such as it
contains a virus integration site; such integration sites are also
present upstream of the WNT1 gene promoter (20). Papillary ductal carcinoma of the
breast is potentially induced by human papilloma virus infection,
and high WNT1 mRNA expression has been detected in tumors of
this type (14). Integrating a viral
gene into the viral integration site upstream of the WNT1
gene promoter induces mammary tumor formation (20), suggesting that the activation of the
WNT1 gene in cancer may be associated with the transfection
of viral genes. As AB development due to viral infection has never
been reported, further studies are warranted to determine whether
there are mutations at the WNT1 gene regulatory site.
Immunohistochemical results of this study showed
that the expression level of WNT1 was significantly higher in the
AB than in normal oral mucosa tissues, combined with western
blotting and RT-qPCR results suggest that WNT1 and the occurrence
of ABs and its aggressive has a high correlation. Through the study
of biological behavior of AB and the relationship between WNT1
expression, WNT1 could also be the diagnosis and evaluation of
prognosis of effective index of AB. For this reason, the WNT1 as a
target genes to treat AB is helpful.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation Project of China (grants nos. 30672332
and 81072197).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GW performed the immunohistochemical, western blot
analysis and RT-qPCR experiments and was a major contributor in
writing the manuscript. MZ made substantial contributions to
conception and design. YC and JJ collected and organized the data
of 80 patients with ameloblastoma. XG and TW performed the
statistical analysis of data. All authors reviewed the final
manuscript and approved it for publication.
Ethics approval and consent to
participate
The study protocol was approved by the Medical
Ethics Committee of Stomatological Hospital of China Medical
University. The study was performed in accordance with the
Declaration of Helsinki. Informed verbal patient consent was
obtained for the use of all specimens.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kharaishvili G, Simkova D, Makharoblidze
E, Trtkova K, Kolar Z and Bouchal J: Wnt signaling in prostate
development and carcinogenesis. Biomed Pap Med Fac Univ Palacky
Olomouc Czech Repub. 155:11–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kumamoto H and Ooya K: Immunohistochemical
detection of beta-catenin and adenomatous polyposis coli in
ameloblastomas. Oral Pathol Med. 34:401–406. 2005. View Article : Google Scholar
|
|
3
|
Peng L, Ye L, Dong G, Ren LB, Wang CL, Xu
P and Zhou XD: WNT5A inhibits human dental papilla cell
proliferation and migration. Biochem Biophys Res Commun.
390:1072–1078. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Barnes L, Eveson JW, Reichart P and
Sidransky D: Pathology and Genetics of Head and Neck Tumours. World
Health Organization Classification of Tumours. IARC Press; Lyon;
2006
|
|
5
|
Li TJ, Wu YT, Yu SF and Yu GF: Unicystic
ameloblastoma: A clinicopathologic study of 33 Chinese patients. Am
J Surg Pathol. 24:1385–1392. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grintzalis K, Georgiou CD and Schneider
YJ: An accurate and sensitive Coomassie brilliant blue G-250-based
assay for protein determination. Anal Biochem. 480:28–30. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reya T and Clevers H: Wnt signalling in
stem cells and cancer. Nature. 434:843–850. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Howe LR, Subbaramaiah K, Chung WJ,
Dannenberg AJ and Brown AM: Transcriptional activation of
cyclooxygenase-2 in Wnt-1 transformed mouse mammary epithelial
cells. Cancer Res. 59:1572–1577. 1999.PubMed/NCBI
|
|
10
|
Yoshida N, Kinugasa T, Ohshima K, Yuge K,
Ohchi T, Fujino S, Shiraiwa S, Katagiri M and Akagi Y: Analysis of
Wnt and β-catenin expression in advanced colorectal cancer.
Anticancer Res. 35:4403–4410. 2015.PubMed/NCBI
|
|
11
|
Wittkorn E, Sarkar A, Garcia K,
Kango-Singh M and Singh A: The Hippo pathway effector Yki
downregulates Wg signaling to promote retinal differentiation in
the Drosophila eye. Developmental. 142:2002–2013. 2015. View Article : Google Scholar
|
|
12
|
Liu B, Chen S, Cheng D, Jing W and Helms
JA: Primary cilia integrate hedgehog and Wnt signaling during tooth
development. J Dent Res. 93:475–482. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maasalu K, Nikopensius T, Kõks S, Nõukas
M, Kals M, Prans E, Zhytnik L, Metspalu A and Märtson A:
Whole-exome sequencing identifies de novo mutation in the COL1A1
gene to underlie the severe osteogenesis imperfecta. Hum Genomics.
9:62015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Katoh M: Expression and regulation of Wnt1
in human cancer: Upregulation of Wnt1 by β-estradiol in MCF-7
cells. Int J Oncol. 22:209–212. 2003.PubMed/NCBI
|
|
15
|
Jia J, Jia S, Ji K and Jiang WG: Wnt1
inducible signalling pathway protein-2 (WISP-2/CCN5): Roles and
regulation in human cancers (Review). Oncol Rep. 31:533–539. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choi AR, Park JR, Kim RJ, Kim SR, Cho SD,
Jung JY and Nam JS: Inhibition of Wnt1 expression reduces the
enrichment of cancer stem cells in a mouse model of breast cancer.
Biochem Biophys Res Commun. 425:436–442. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Keupp K, Beleggia F, Kayserili H, Barnes
AM, Steiner M, Semler O, Fischer B, Yigit G, Janda CY, Becker J, et
al: Mutations in WNT1 cause different forms of bone fragility. Am J
Hum Genet. 92:565–574. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Joeng KS, Lee YC, Jiang MM, Bertin TK,
Chen Y, Abraham AM, Ding H, Bi X, Ambrose CG and Lee BH: The
swaying mouse as a model of osteogenesis imperfecta caused by WNT1
mutations. Hum Mol Genet. 23:4035–4042. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kato K, Nakatani Y, Kanno H, Inayama Y,
Ijiri R, Nagahara N, Miyake T, Tanaka M, Ito Y, Aida N, et al:
Possible linkage between specific histological structures and
aberrant reactivation of Wnt pathway in adamantinomatous
craniopharyngioma. J Pathol. 203:814–821. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fuchs-Young R, Shirley SH, Lambertzl I,
Colby JK, Tian J, Johnston D, Gimenez-Conti IB, Donehower LA, Conti
CJ and Hursting SD: P53 genotype as a determinant of ER expression
and tamoxifen response in the MMTV-Wnt-1 model of mammary
carcinogenesis. Breast Cancer Res Treat. 130:399–408. 2011.
View Article : Google Scholar : PubMed/NCBI
|