Introduction
Branchial cleft cyst (BRCC) is generally accepted to
be a congenital cyst derived from the first and second branchial
clefts, particularly the second (1,2). BRCC
appears as a circumscribed nodular swelling on the side of the
neck, with neither an opening in the pharynx nor an external sinus,
whereas a branchial fistula communicates internally and/or
externally (1,2).
Human papillomaviruses (HPVs) are small
non-enveloped doubled-stranded DNA viruses that belong to the
Papillomaviridae family (3).
To date, more than 200 types of HPV have been identified in
clinical samples (3). HPVs are known
to infect the epithelium of the skin or mucosa. Mucosal HPVs are
divided into two types according to their association with
carcinoma: Low-risk and high-risk (4). The 14 types of high-risk HPV of most
concern are HPV −16, −18, −31, −33, −35, −39, −45, −51, −52, −56,
−58, −59, −66 and −68 (5). The
integration of high-risk HPV DNA into the host genome is a notable
step in malignant transformation in uterine cervical cancer
(6,7).
Viral integration into the host genome often leads to viral
E2 gene disruption. Since E2 controls the expression
of viral oncogenes E6 and E7 by repressing the viral
promoter, integration leads to overexpression of E6 and
E7 (8). The E6 oncoprotein is
able to disrupt the function of tumor protein p53 (hereinafter
referred to as p53), which has anticancer functions, by targeting
the p53 protein for ubiquitination and degradation. Furthermore,
the E7 oncoprotein binds and functionally inactivates the
retinoblastoma protein (pRb), which regulates the progression from
the G1 phase to the S phase of the cell cycle.
Functional inactivation of pRb by E7 is known to induce
upregulation of p16 (also known as cyclin-dependent kinase
inhibitor 2A) expression (9,10). These oncoproteins are expressed in
high-risk HPV infections, which indicates that they serve a role in
the initiation and/or progression of tumors and their interactions
may substantially enhance the efficiency of tumor cell
immortalization (11).
High-risk HPV DNA detected using the polymerase
chain reaction (PCR) and pyrosequencing was previously reported to
be present in BRCC: Of 19 BRCCs analyzed, 7 (36.8%) contained
HPV-16 and/or HPV-18 genomic DNA (12). However, HPV DNA could not be
identified using in situ hybridization (ISH) in these cases.
These disparate findings may be due to the differing analytic
sensitivities between PCR/pyrosequencing and ISH. Further
information, such as viral load and the integration of HPV, is
required to clarify the role of HPV infection in BRCC. The aim of
the present study was therefore to confirm the presence of HPV
infection in BRCC.
Materials and methods
Patient sample collection
The present study was approved by the Ethics
Committee of the University of the Ryukyus (Nakagami, Japan). All
patients provided written informed consent to participate prior to
surgery. Procedures were performed in accordance with the
Declaration of Helsinki. The participants in the present study
consisted of 6 patients (3 men and women; age range, 2–29 years)
with BRCC and 1 patient with HPV-associated oropharyngeal carcinoma
(OPC) with metastasis in the lymph node and cystic formation as the
HPV-positive control. Each patient had undergone surgical resection
and no other treatment, and pathologists had confirmed the
histological diagnoses. Formalin-fixed paraffin-embedded (FFPE; 10%
formalin natural buffer solution at 4°C for a 24-h incubation)
and/or fresh-frozen cyst tissues taken from these patients were
used for the detection of HPV infection. In addition, cyst fluid
from 1 patient was also examined to detect the HPV genome.
Culture of HPV-16-positive CaSki cell
line
To obtain HPV-16 positive DNA, an HPV-16-infected
cervical cancer cell line, the CaSki cell line, was purchased from
the European Collection of Cell Cultures (Salisbury, Wiltshire,
UK). The cell line has been tested and authenticated by DNA short
tandem repeat profiling by the Japanese Collection of Research
Bioresources Cell Bank (Osaka, Japan). The CaSki cells were
cultured at a density of 0.7×106 cells per 25
cm2 with 5 ml RPMI-1640 containing 10% fetal bovine
serum, 100 IU/ml penicillin and 100 µg/ml streptomycin. When the
culture was near confluence, the cells were detached by cell
scraper and placed into a 15-ml tube. The tube was centrifuged at
2,600 × g (4°C) for 5 min, and the supernatant was discarded and
pelleted. The cell pellet was stored at −80°C until DNA
extraction.
Detection of HPV DNA
The HPV types present in FFPE, fresh-frozen samples
and/or fresh-frozen CaSki cell line were analyzed. In the case of
FFPE samples, three 10-µm thick sections were deparaffinized in
xylene and rehydrated in 100% alcohol. Following air-drying,
genomic DNA was extracted using the Gentra Puregene Tissue kit
(Qiagen Sciences, Inc., Gaithersberg, MD, USA), as described
previously (13). To remove PCR
inhibitors (by removing formaldehyde cross-linking between DNA
fragments), DNA samples were heated at 100°C in 25 mM borate-NaOH
buffer (pH 11.0) for 30 min according to a previously published
method (14). Genomic DNA was then
re-purified by ethanol precipitation and dissolved in distilled
water. In the case of fresh-frozen samples, genomic DNA was
extracted as described previously (13). The presence and the integrity of the
DNA in all samples were examined by PCR amplification of the
β-globin gene using primers PC04 and GH20 (Table I), as previously described (13). Negative (water) and positive (DNA of
HPV-16-positive CaSki cell line) controls were included in each
amplification series.
 | Table I.Primers used in the present
study. |
Table I.
Primers used in the present
study.
| Primer name | Sequence |
|---|
| Cloning
primers |
|
|
GP5+ |
5′-TTTGTTACTGTGGTAGATACTAC-3′ |
|
GP6+ |
5′-GAAAAATAAACTGTAAATCATATTC-3′ |
|
MY09 |
5′-CGTCCMARRGGAWACTGATC-3′ |
|
MY11 |
5′-GCMCAGGGWCATAAYAATGG-3′ |
|
E2-forward |
5′-AACGAAGTATCCTCTCCTGAAATTATTAG-3′ |
|
E2-reverse |
5′-CCAAGGCGACGGCTTTG-3′ |
|
E6-forward |
5′-GAGAACTGCAATGTTTCAGGACC-3′ |
|
E6-reverse |
5′-TGTATAGTTGTTTGCAGCTCTGTGC-3′ |
|
E7-forward |
5′-CCGGACAGAGCCCATTACAA-3′ |
|
E7-reverse |
5′-CGAATGTCTACGTGTGTGCTTTG-3′ |
|
PC04 |
5′-CAACTTCATCCACGTTCACC-3′ |
|
GH20 |
5′-GAAGAGCCAAGGACAGGTAC-3′ |
| qPCR primers and
Taqman probe |
|
|
E2-forward |
5′-AACGAAGTATCCTCTCCTGAAATTATTAG-3′ |
|
E2-reverse |
5′-CCAAGGCGACGGCTTTG-3′ |
|
E2-probe |
5′-FAM-CACCCCGCCGCGACCCATATAMRA-3′ |
|
E6-forward |
5′-GAGAACTGCAATGTTTCAGGACC-3′ |
|
E6-reverse |
5′-TGTATAGTTGTTTGCAGCTCTGTGC-3′ |
|
E6-probe |
5′-FAM-CAGGAGCGACCCAGAAAGTTACCACAGTTTAMRA-3′ |
|
β-globin-F |
5′-TGGGTTTCTGATAGGCACTGACT-3′ |
|
β-globin-R |
5′-AACAGCATCAGGAGTGGACAGAT-3′ |
|
β-globin-probe |
5′-FAM-TCTACCCTTGGACCCAGAGGTTCTTTGAGTTAMRA-3′ |
The consensus primer sets GP5+/GP6+ and MY09/MY11
were used to analyze the presence of HPV DNA in PCR (Table I) as described previously (15). When no PCR amplification occurred with
GP5+/GP6+ or MY09/MY11 primers, 10-fold
diluted first PCR products were used as template DNA for
(auto-)nested PCR using the GP5+/GP6+ primer pair (16). PCR cycling was performed as follows:
initial denaturing at 95°C for 10 min; 30 cycles at 95°C for 30
sec, 58°C for 1 min, and 72°C for 30 sec, with a final extension at
72°C for 5 min. PCR products of expected size (GP5+/GP6+, 150 bp;
MY09/MY11, 450 bp) were purified and directly sequenced with an ABI
PRISM 3,130×l Genetic Analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. The sequences were then aligned and compared with those
of known HPV types in the GenBank database using BLAST (blast.ncbi.nlm.nih.gov/Blast.cgi).
DNA extracted from FFPE samples may not have been
amplified using GP5+/GP6+ or MY09/MY11 primers because of DNA
fragmentation during paraffin fixation, which may result in a
false-negative result (17). Since
the dominant HPV type is HPV-16 in head and neck carcinomas
(15), primer sets for the E2,
E6 and E7 regions of the HPV-16 sequence were designed
to produce small products to prevent a false-negative PCR (Table I). PCR amplification was performed
using the GoTaq® Green Master Mix (Promega Corporation,
Madison, WI, USA) according to the manufacturer's protocol. PCR was
performed as follows: Denaturation at 95°C for 15 min, followed by
50 cycles at 95°C for 15 sec and 60°C for 1 min, and finally 72°C
for 5 min. PCR fragments of expected size
(E2-forward/E2-reverse, 82 bp;
E6-forward/E6-reverse, 81 bp; E7-
forward/E7-reverse, 91 bp) were purified using the Wizard SV
GEL and PCR Clean-UP system (Promega Corporation) The purified PCR
products were cloned into a PGEM-T easy vector (Promega
Corporation) and sequenced using ABI PRISM 3,130×l Genetic
Analyzer, as DNA subcloning is an appropriate method for the
analysis of DNA sequences of PCR products of <100 bp, when
compared with direct sequencing. The sequences obtained were
analyzed as aforementioned.
Quantitative (q)PCR analysis of viral
load and physical status of HPV-16
To evaluate the viral load and physical status of
HPV-16, qPCR was performed as described previously (15). Briefly, primers and TaqMan probes
targeting the HPV-16 E2 and E6 open reading frames
were used (Table I). The primers and
probes recognize the E2 hinge region, which was deleted on
HPV-16 integration. Two standard curves for the E2 and
E6 genes were created by amplification of 10-fold serial
dilutions (102, 103, 104,
105 and 106 viral copies) of the plasmid
pB-actin containing the complete HPV-16 early region (Addgene,
Cambridge, MA, USA). Viral DNA load was assessed by calculating
E6 copy numbers. An external standard curve was created
using known serial dilutions (0.3, 3, 30 and 300 ng) of human
genomic placental DNA (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) for cellular DNA quantification and β-globin was amplified
as described previously (15,18). The amount of DNA was calculated by
plotting the Cq values against the logarithm of the standard curve.
The physical status of HPV-16 was assessed based on a previously
published method (7,15). The total E6 copy number in 50
ng cellular DNA was then determined (15). Ratios of E2 copy number/total
E6 of <1 indicate the presence of both integrated and
episomal forms. An E2/E6 ratio ≥1 indicates the
predominance of the episomal form, whereas a ratio of 0 indicates
the presence of the integrated form only.
ISH with HPV DNA probes
Biotinyl tyramide-based ISH was performed using the
GenPoint™ HPV biotinylated DNA probe and the GenPoint tyramide
signal amplification system for biotinylated probes according to
the manufacturer's protocol (Dako; Agilent Technologies, Inc.,
Santa Clara, CA, USA). The GenPoint HPV biotinylated DNA probe has
been found to react with HPV types 16, 18, 31, 33, 35, 39, 45, 51,
52, 56, 58, 59 and 68 in FFPE sections by ISH. Serial 4-µm-thick
sections of FFPE samples were deparaffinized in xylene and
rehydrated using a graded alcohol series. Target HPV DNA retrieval
was performed in 10 mM sodium citrate (at pH 6.0) at 95°C for 40
min. The slides were digested with proteinase K (10,000-fold
dilution with Tris-buffered saline; Dako; Agilent Technologies,
Inc.) for 10 min at room temperature. Endogenous peroxidases were
blocked with 0.3% H2O2 in methanol for 20
min. A drop of the HPV probe was added to the section and a
coverslip was applied. The probe and the target DNA were denatured
by incubating the slides at 92°C for 5 min. Following denaturation,
the slides were transferred to a humidified chamber for
hybridization at 37°C for 16 h. Next, coverslips were removed and
slides were bathed in TBS containing 0.05% Tween-20 (TBST).
Coverslips were then washed using GenPoint Detection system
stringent wash solution (Dako; Agilent Technologies, Inc.) at 48°C
for 30 min, followed by a rinse in TBST. Detection of the
hybridized probe was performed using the GenPoint Detection system
according to the manufacturer's protocol, with included primary
streptavidin-horseradish peroxidase (HRP), biotinyl tyramide,
secondary streptavidin-HRP and 3–3′-diaminobenzidine (DAB; Dako;
Agilent Technologies, Inc.). Slides were counterstained with
hematoxylin.
Immunohistochemistry for
p16INK4a
Immunohistochemistry for p16INK4a was
performed using the CINTec®p16 Histology kit (MTM
Laboratories; Roche Applied Science, Penzberg, Germany) (19). Serial 4-µm-thick sections of FFPE
samples were deparaffinized in xylene and rehydrated in a graded
alcohol series, followed by heating at 95–99°C for 10 min in kit
epitope-retrieval solution. Endogenous peroxidases were blocked
with kit peroxidase-blocking reagents at room temperature for 5
min. The sections were incubated for 30 min at room temperature
with primary monoclonal mouse anti-p16INK4a antibody
(ready-to-use antibody solution) from the aforementioned kit.
Following washes in PBS, slides were incubated at room temperature
for 30 min with HRP-conjugated goat anti-mouse secondary antibody
(MTM Laboratories; Roche Applied Science). Immunolabeling was
visualized following incubation in DAB at room temperature for 10
min. Stained slides were counterstained with hematoxylin to
visualize cyst structures for analysis under light microscopic
observation.
The scoring criteria for p16INK4A
immunoreactivity (p16INK4A expression) were defined for
the present study based on a previous scoring method (19): 0, no staining; 1, 1–10% of the tumor
cells positive; 2, 11–40% positive; 3, 40–70% positive; and 4,
>70% positive. The term ‘p16INK4A overexpression’ was
defined as a score of 3 or 4.
Results
Identification of HPV types and
analysis of the physical status of HPV-16
Table II presents the
results of PCR using primer sets GP5+/GP6+, MY09/MY11, E2-F/E2-R,
E6-F/E6-R and E7-F/E7-R. Primer sets GP5+/GP6+ and MY09/MY11 did
not amplify the HPV genome effectively, unlike the E2-F/E2-R and
E6-R/E6-R sets. PCR amplification, and the nucleotide sequences of
the PCR products generated, demonstrated that 4/6 patients with
BRCC were positive for HPV-16 (66.7%), as was the patient with OPC.
HPV-16 genomic DNA was also detected in the cyst fluid obtained
from patient 4 using PCR.
 | Table II.Sample conditions, PCR amplification,
HPV infection status, ISH signal status and p16INK4a
score in branchial cleft cyst. |
Table II.
Sample conditions, PCR amplification,
HPV infection status, ISH signal status and p16INK4a
score in branchial cleft cyst.
|
|
|
|
|
| PCR
amplification |
|
| Integration |
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
| Patient | Sex | Age, years | Disease | Sample | GP5+/GP6+ | MY09/MY11 | E2-F/E2-R | E6-F/E6-R | E7-F/E7-R | HPV type | Viral load
(copies/50 ng DNA) | E2/E6 | Status | ISH signal
status | p16 score |
|---|
| 1 | Male | 2 | Branchial cleft
cyst | FFPE | – | – | + | + | + | 16 |
1.65×106 | 1.03 | Episomal | D | 0 |
| 2 | Male | 29 | Branchial cleft
cyst | FFPE | + | + | + | + | + | 16 |
7.39×102 | 0.56 | Mixed | D, P | 1 |
| 3 | Female | 18 | Branchial cleft
cyst | FFPE | – | – | + | + | – | 16 |
2.21×104 | 0.03 | Mixed | D, P | 1 |
| 4 | Female | 6 | Branchial cleft
cyst | Fresh-frozen | – | – | + | + | Not tested | 16 |
9.21×103 | 0.03 | Mixed | D, P | 1 |
| 4 |
|
| Cyst fluid | Fresh-frozen | – | – | + | + | Not tested | 16 |
7.10×102 | 0.49 | Mixed | NA | NA |
| 5 | Male | 16 | Branchial cleft
cyst | FFPE | – | – | – | – | – | No |
|
|
| (−) | 1 |
| 6 | Female | 9 | Branchial cleft
cyst | FFPE | – | – | – | – | – | No |
|
|
| (−) | 1 |
| C | Male | 50 | Metastatic lymph
node from oropharyngeal cancer | Fresh-frozen | + | + | + | + | + | 16 |
1.06×106 | 0.92 | Mixed | D, P | 4 |
Viral load and physical status of
HPV-16
qPCR revealed HPV-16 loads (Table II). In the lymph node metastasis from
the OPC case, the HPV-16 load was 1.06×106 copies/50 ng
genomic DNA, whereas the loads in the four HPV-positive BRCCs
ranged between 7.39×102 and 1.65×106
copies/50 ng DNA. Cyst fluid obtained from patient 4 was subjected
to viral load analysis and was identified to have
7.10×102 copies/50 ng genomic DNA.
Since there were no marked differences between the
amplification efficiencies of E2 (88.1%) and E6
(89.7%) PCR, the qPCR conditions in the present study were deemed
suitable for evaluating integration. Analysis of the
E2/E6 ratio demonstrated that one patient with BRCC
(patient 1) had an episomal-type infection, whereas three
mixed-type (episomal- and integrated-type) infections were noted in
the three HPV-positive BRCCs (patients 2, 3 and 4) and in the lymph
node of the patient with OPC.
ISH for identification of HPV DNA
Positive HPV DNA signals were observed in cyst wall
tissues in 4/6 patients with BRCC (66.7%; Table II, Figs.
1–3). Positive ISH was only
observed in samples in which the HPV-16 genome was detected using
PCR. Diffuse reactions in the nuclei of the basal to granular
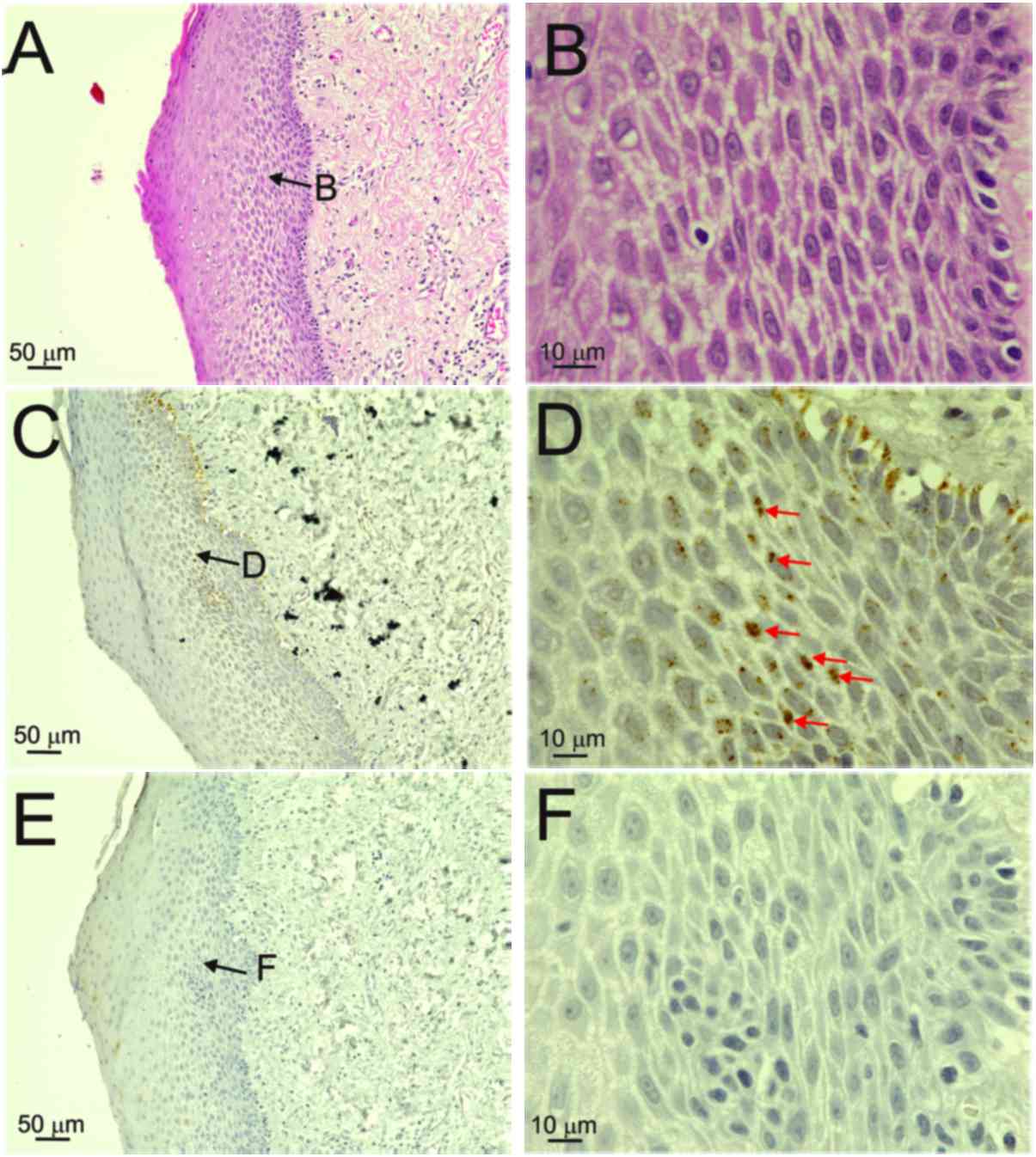
layers, particularly in prickle cells, were observed in patient 1,
who had an episomal-type HPV infection (Fig. 1). Fig.
1D shows diffuse ISH signals in the nuclei of prickle cells.
Although the basal cells displayed diffuse reactions in the nuclei,
positive reactions were also observed outside the nuclei
(false-positive reactions; Fig. 1D).
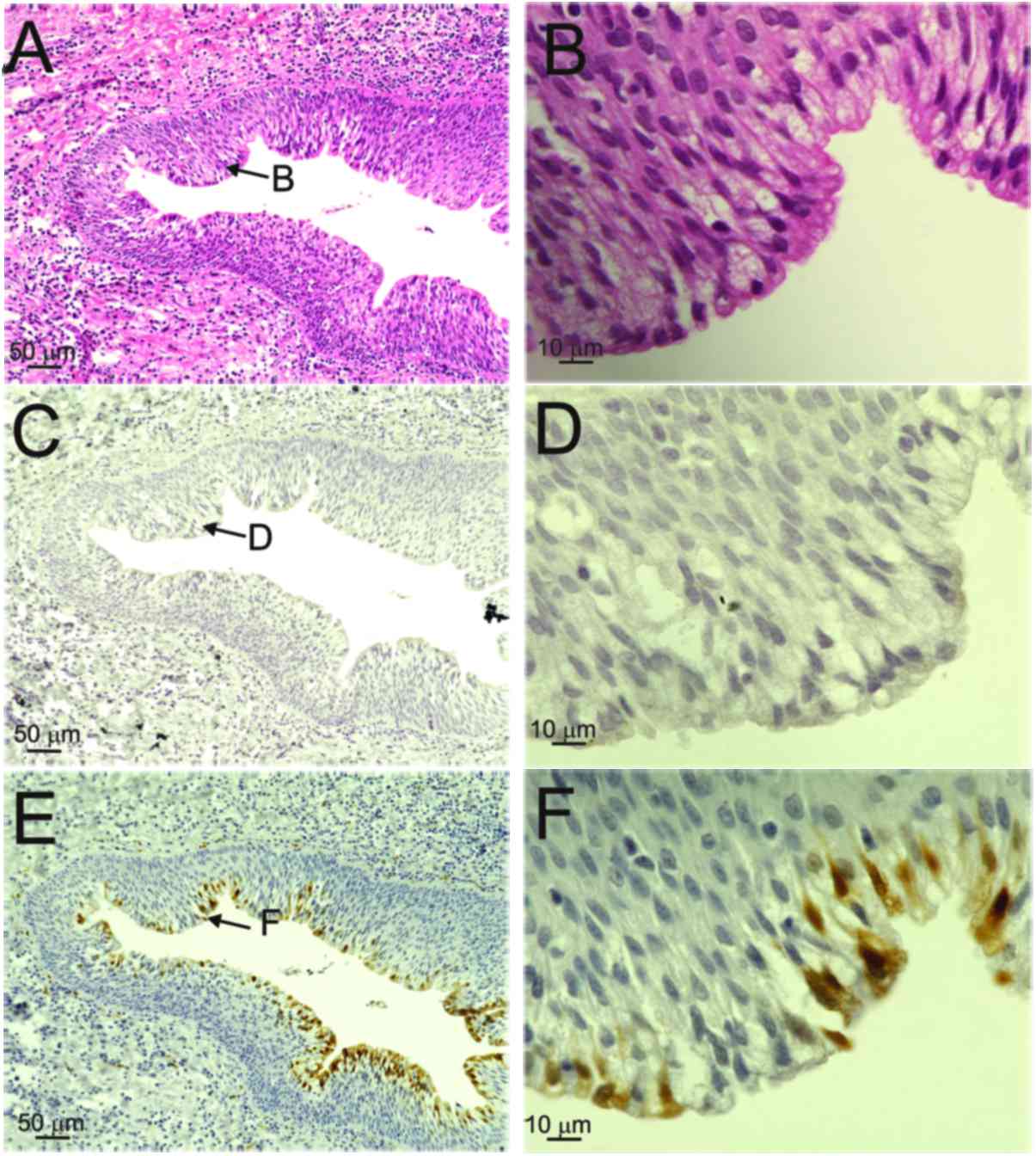
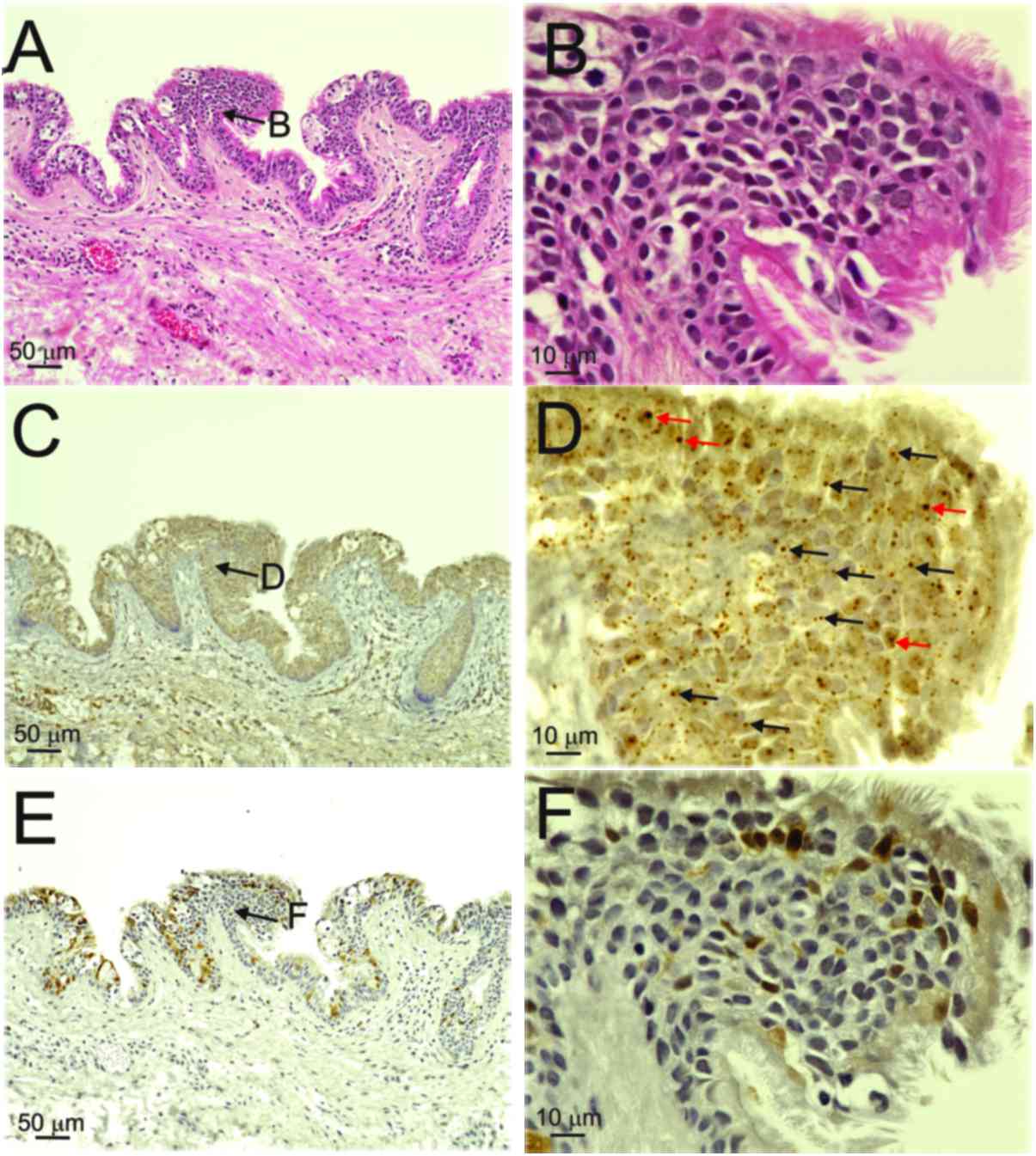
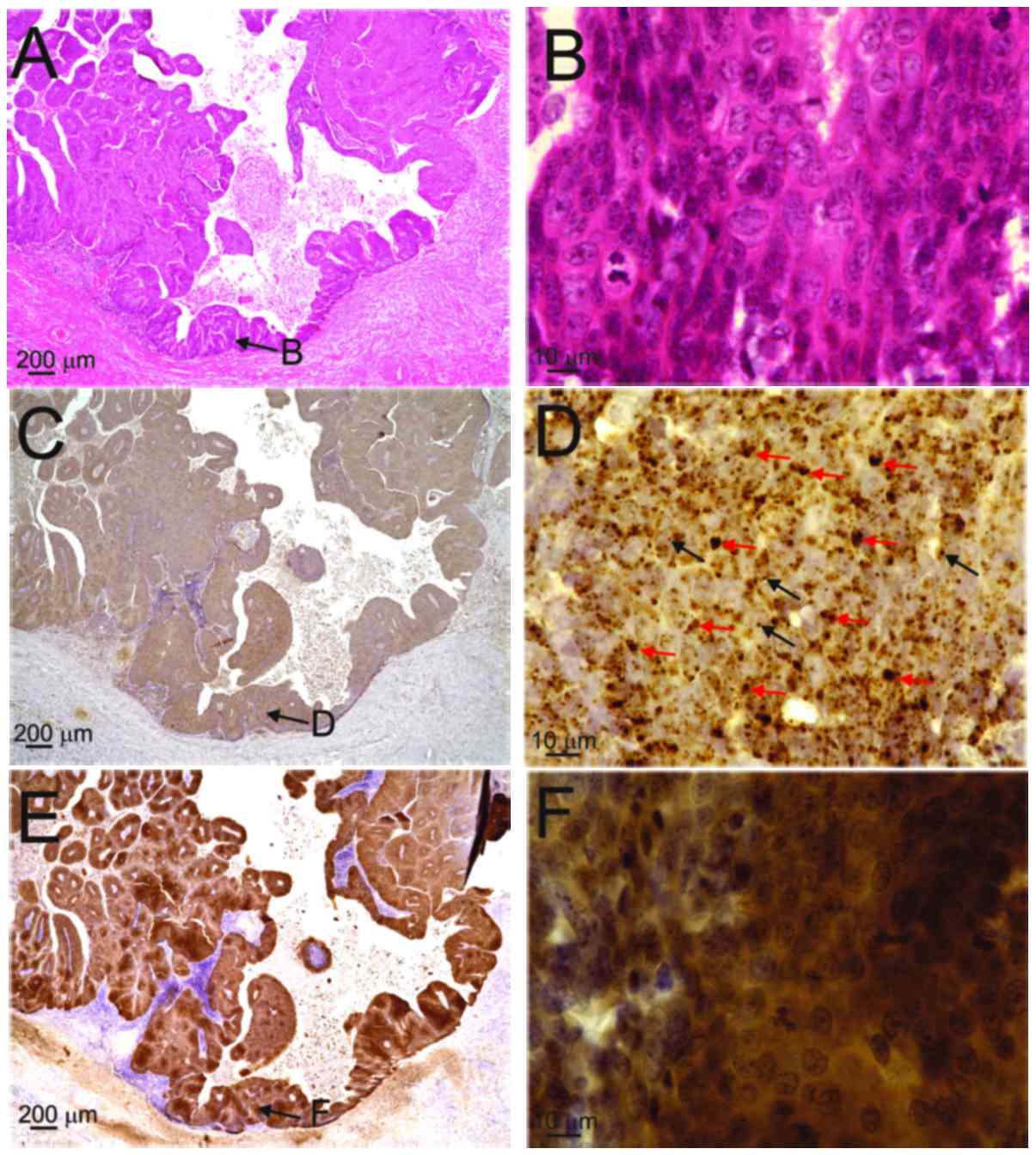
By contrast, both diffuse (red arrows) and punctate patterns (black
arrows) were observed in the nuclei of the basal-to-granular layers
in patients 2, 3 and 4, and in those of the OPC case (Table II, Figs.
2 and 4). However, there was no
positive ISH signal in patients 5 and 6, who did not have HPV
infection (Fig. 3C and D).
p16INK4a
immunoreactivity
The expression of p16INK4a was weak in
the nuclei and/or cytoplasm in the basal-to-keratinized layers of
the cyst wall (score 1) in the three patients with BRCC with
mixed-type infections (Table II,
Fig. 2E and F), whereas the OPC case
exhibited p16INK4a overexpression (Fig. 4E and F; score 4). The superficial
cyst-lining squamous cells of the cyst wall in specimens from
patients 5 and 6 (who did not have HPV infection) also exhibited
faint immunoreactivity (Fig. 3E and
F; score 1). On the other hand, the specimen from patient 1,
who had an episomal infection, exhibited no p16INK4a
immunoreactivity (Fig. 1E and F;
score 0). Since 5/6 patients with patients with BRCC exhibited a
weak p16INK4a reaction in the cyst wall, the expression
of p16INK4a was not associated with the presence of HPV
detected by PCR.
Discussion
In the present study, HPV-16 genome DNA was detected
in 4/6 samples from patients with BRCC using PCR with
HPV-16-specific primers. However, HPV-16 genome was detected in
only one using PCR using the consensus primers. Paraffin fixation
of samples induces DNA fragmentation and can occasionally result in
a false-negative PCR result (17).FFPE samples of BRCCs were examined by
PCR using consensus primers (GP5+/GP6+ and MY09/MY11), except in
the case of patient 4. A previous study concerning HPV infection in
BRCCs demonstrated that 7 of 19 BRCCs (36.8%) contained high-risk
HPV-16 and/or HPV-18 genomes (12).
In that study, FFPE samples were examined and the GP5+/GP6+ PCR
primer set was used to detect the HPV genome in the samples from
patients with BRCC (12). The
divergent results, which reveal a difference in the presence of HPV
between the present study and the previous study (12) may relate to the PCR method and sample
conditions (i.e., whether the sample was fresh-frozen or FFPE). In
a previous study of head and neck squamous cell carcinomas using
fresh-frozen samples (13), HPV-16
genome DNA was detected via PCR using consensus primers in 39/45
(86.7%) squamous cell carcinoma samples infected with high-risk
type HPV subtypes. Since HPV-16 is the major HPV type present in
head and neck carcinomas, specific primers for HPV-16 were used in
the present PCR analysis. ISH with the HPV DNA probes used in the
present study can react with HPV types 16, 18, 31, 33, 35, 39, 45,
51, 52, 56, 58, 59 and 68 in FFPE sections. The consistent results
between PCR and ISH experiments suggest that HPV-16 is a major type
of HPV infection in BRCC.
In a previous study (15), the viral load of HPV-16 in tonsillar
carcinomas ranged between 1.54×102 and
1.34×107 (median, 4.13×105), and was
significantly increased compared with that in non-tonsillar head
and neck carcinomas (1.20×101-3.89×106;
median, 9.7×101), detected using the same methods as
those used in the present study. The HPV-16-positive BRCC samples
in the present study had decreased viral loads
(7.10×102-1.65–106; median,
9.21×103) compared with the tonsillar carcinomas in the
previous study (15), and patients
with BRCC with integration of HPV genome had decreased viral loads
compared with the metastatic lymph node from the OPC case (Table II, control case).
The integration of DNA from high-risk HPV types into
the host genome is a major step in malignant transformation
(6,8).
In the present study, viral integration was observed in three of
four BRCC samples with HPV infection. The punctate and diffuse HPV
DNA ISH staining patterns corresponded to integrated- and
episomal-type infections, respectively (20). Therefore, the consistent results of
the physical status of HPV infection in ISH- and PCR-based HPV
detection in the present study indicated that persistent high-risk
type HPV infections may occur in patients with BRCC. In the present
study, immunoreactive p16INK4a was weakly expressed,
regardless of HPV infection (Table
II and Fig. 3). Pai et al
(12) reported that
p16INK4A expression in the superficial BRCC cyst-lining
cells may represent the process of cellular senescence prior to the
shedding of cells into the cyst lumen. Although the functional
inactivation of host pRb by HPV protein E7 results in the
overexpression of p16INK4a, weak p16INK4a
expression, despite the mixed integration of HPV-16, might
represent low E7 expression and a low BRCC viral load. These
results also suggest that the weak expression of
p16INK4a observed in the present study may not be an
indicator of HPV infection.
In the present study, none of the patients with BRCC
had fistulas, either externally or internally, but high-risk type
HPV genomic DNA was present. Further study is required to clarify
the route of HPV infection. A possible explanation for HPV
infection in BRCC might be the entry of the virus through the
branchial groove and/or pouch via amniotic fluid in the prenatal
period. There have been several reports demonstrating the presence
of HPV DNA in the amniotic fluid of pregnant women (21–25). The
presence of HPV DNA in the amniotic fluid and/or placenta increases
the risk of HPV infection in the neonate at birth (21,22).
Possible perinatal vertical HPV transmission is suspected, because
cesarean section delivery does not protect against mother-to-child
HPV transmission (25). These
previous studies suggest that neonates have early contact with HPV
prior to delivery. The most commonly detected HPV types in the
amniotic fluid are HPV-6, −11, −16 and −18 (26–29). The
most prevalent HPV type in asymptomatic oral infections is HPV-16,
followed by HPV-6 and −11 (30,31). In
the present study, the HPV DNA detected in the cyst wall and cyst
fluid of BRCC cases was the high-risk subtype HPV-16. These results
are consistent with reports concerning the HPV types observed in
amniotic fluid (21–25). Since the participants in the present
study were young (2–29 years), vertical infection was the most
likely route of HPV infection in these BRCC cases, although whether
the HPV genome is present in the amniotic fluid remains
unknown.
Although BRCC is usually treated with surgical
excision, several studies recommend sclerotherapy as the primary
treatment (32,33). The results of these studies indicate
that sclerotherapy for cystic neck lesions has a satisfactory
outcome, lower morbidity and fewer complications compared with
surgical treatment. However, imaging modalities and even aspiration
cytology cannot sufficiently differentiate between benign and
malignant cysts. Since lymph node metastasis from OPC may appear as
a cystic mass (20,34) and certain patients with BRCC exhibit
persistent high-risk-type HPV infection, as observed in the present
study, these issues should be taken into account when considering
sclerotherapy for BRCC. Since the HPV genome in the cystic fluid of
BRCC may be amplified by PCR in the present study, this method may
aid the detection of HPV infection.
In summary, high-risk-type HPV infection was
frequently observed in patients with BRCC. To the best of our
knowledge, the present study is the first to demonstrate that HPV
integration occurs in BRCC. Since a considerable number of patients
with BRCC exhibit persistent infection and integration of
high-risk-type HPV DNA, surgical treatment may be more beneficial
in these cases. Further studies are required to clarify the
clinical relevance of infection with high-risk HPV types to the
etiology and malignant transformation of BRCC.
Acknowledgements
The authors would like to thank the Ryukyu Society
for the Promotion of Oto-Rhino-Laryngology for providing assistance
with writing and for technical assistance.
Funding
This study was supported by a KAKENHI grant (no.
26462610) awarded by the Japan Society for the Promotion of
Science.
Availability of data and materials
All data generated and analyzed during this study
are included in this published article.
Authors' contributions
TI and TU contributed to the experimental studies,
data acquisition and preparation of the manuscript. ZD contributed
to the experiments and data acquisition. SK, HM, AK, SA, HH, YY and
AG contributed to the acquisition of samples and to surgical
treatment. MS contributed to the study design, supervision of
experiments and manuscript review. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the University of the Ryukyus (Nakagami, Japan). All
patients provided written informed consent to participate prior to
surgery. Procedures were performed in accordance with the
Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BRCC
|
branchial cleft cyst
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
HPV
|
human papillomavirus
|
|
ISH
|
in situ hybridization
|
|
OPC
|
oropharyngeal carcinoma
|
|
PCR
|
polymerase chain reaction
|
References
|
1
|
Waldhausen JH: Branchial cleft and arch
anomalies in children. Semin Pediatr Surg. 15:64–69. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Veivers D and Dent J: Lateral cervical
cysts: An Australian perspective. ANZ J Surg. 82:799–802. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bzhalava D, Eklund C and Dillner J:
International standardization and classification of human
papillomavirus types. Virology. 476:341–344. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clifford GM, Smith JS, Plummer M, Munoz N
and Franceschi S: Human papillomavirus types in invasive cervical
cancer worldwide: A meta-analysis. Br J Cancer. 88:63–73. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jacobs MV, Snijders PJ, van den Brule AJ,
Helmerhorst TJ, Meijer CJ and Walboomers JM: A general primer
GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid
detection of 14 high-risk and 6 low-risk human papillomavirus
genotypes in cervical scrapings. J Clin Microbiol. 35:791–795.
1997.PubMed/NCBI
|
|
6
|
Badaracco G, Venuti A, Sedati A and
Marcante ML: HPV16 and HPV18 in genital tumors: Significantly
different levels of viral integration and correlation to tumor
invasiveness. J Med Virol. 67:574–782. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peitsaro P, Johansson B and Syrjänen S:
Integrated human papilloma virus type 16 is frequently found in
cervical cancer precursers as demonstrated by a novel quantitative
real time PCR technique. J Clin Microbiol. 40:886–891. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arias-Pulido H, Peyton CL, Joste NE,
Vargas H and Wheeler CM: Human papillomavirus type 16 integration
in cervical carcinoma in situ and in invasive cervical cancer. J
Clin Microbiol. 44:1755–1762. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khleif SN, DeGregori J, Yee CL, Otterson
GA, Kaye FJ, Nevins JR and Howley PM: Inhibition of cyclin
D-CDK4/CDK6 activity is associated with an E2F-mediated induction
of cyclin kinase inhibitor activity. Proc Natl Acad Sci USA.
93:4350–4354. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Serrano M, Hannon GJ and Beach D: A new
regulatory motif in cell-cycle control causing specific inhibition
of cyclin D/CDK4. Nature. 366:704–707. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jeon S and Lambert PF: Integration of
human papillomavirus type 16 DNA into the human genome leads to
increased stability of E6 and E7 mRNAs: Implications for cervical
carcinogenesis. Proc Natl Acad Sci USA. 92:1654–1658. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pai RK, Erickson J, Pourmand N and Kong
CS: p16INK4A immunohistochemical staining may be helpful in
distinguishing branchial cleft cysts from cystic squamous cell
carcinomas originating in the oropharynx. Cancer. 117:108–119.
2009.PubMed/NCBI
|
|
13
|
Deng Z, Hasegawa M, Matayoshi S, Kiyuna A,
Yamashita Y, Maeda H and Suzuki M: Prevalence and clinical features
of human papillomavirus in head and neck squamous cell carcinoma in
Okinawa, southern Japan. Eur Arch Otorhinolaryngol. 268:1625–1631.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Taga M, Eguchi H, Shinohara T, Takahashi
K, Ito R, Yasui W, Nakachi K, Kusunoki Y and Hamatani K: Improved
PCR amplification for molecular analysis using DNA from long-term
preserved formalin-fixed, paraffin-embedded lung cancer tissue
specimens. Int J Clin Exp Pathol. 6:76–79. 2013.PubMed/NCBI
|
|
15
|
Deng Z, Hasegawa M, Kiyuna A, Matayoshi S,
Uehara T, Agena S, Yamashita Y, Ogawa K, Maeda H and Suzuki M:
Viral load, physical status, and E6/E7 mRNA expression of human
papillomavirus in head and neck squamous cell carcinoma. Head Neck.
35:800–808. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deng Z, Hasegawa M, Yamashita Y, Matayoshi
S, Kiyuna A, Agena S, Uehara T, Maeda H and Suzuki M: Prognostic
value of human papillomavirus and squamous cell carcinoma antigen
in head and neck squamous cell carcinoma. Cancer Sci.
103:2127–2134. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Specht K, Richter T, Müller U, Walch A,
Werner M and Höfler H: Quantitative gene expression analysis in
microdissected archival formalin-fixed and paraffin-embedded tumor
tissue. Am J Pathol. 158:419–429. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
van Duin M, Snijders PJF, Schrijnemakers
HFJ, Voorhorst FJ, Rozendaal L, Nobbenhuis MAE, van den Brule AJC,
Verheijen RHM, Helmerhorst TJ and Meijer CJ: Human papillomavirus
16 load in normal and abnormal cervical scrapes: An indicator of
CIN II/III and viral clearance. Int J Cancer. 98:590–595. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Deng Z, Hasegawa M, Aoki K, Matayoshi S,
Kiyuna A, Yamashita Y, Uehara T, Agena S, Maeda H, Xie M and Suzuki
M: A comprehensive evaluation of human papillomavirus positive
status and p16INK4aoverexpression as a prognostic
biomarker in head and neck squamous cell carcinoma. Int J Oncol.
45:67–76. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Evans MF, Mount SL, Beatty BG and Cooper
K: Biotinyl-tyramide-based in situ hybridization signal patterns
distinguish human papillomavirus type and grade of cervical
intraepithelial neoplasia. Mod Pathol. 15:1339–1347. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Armbruster-Moraes E, Ioshimoto LM, Leão E
and Zugaib M: Presence of human papillomavirus DNA in amniotic
fluids of pregnant women with cervical lesions. Gynecol Oncol.
54:152–158. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sarkola ME, Grénman SE, Rintala MA,
Syrjänen KJ and SyrjäNen SM: Human papillomavirus in the placenta
and umbilical cord blood. Acta Obstet Gynecol Scand. 87:1181–1188.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Syrjänen S: Current concepts on human
papillomavirus infections in children. APMIS. 118:494–509. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tseng CJ, Lin CY, Wang RL, Chen LJ, Chang
YL, Hsieh TT and Pao CC: Possible transplacental transmission of
human papillomaviruses. Am J Obstet Gynecol. 166:35–40. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Smith EM, Johnson SR, Cripe TP, Pignatari
S and Turek L: Perinatal vertical transmission of human
papillomavirus and subsequent development of respiratory tract
papillomatosis. Ann Otol Rhinol Laryngol. 100:479–483. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cason J and Mant CA: High-risk mucosal
human papillomavirus infections during infancy & childhood. J
Clinical Virology. 32(Suppl 1): S52–S58. 2005. View Article : Google Scholar
|
|
27
|
Fredericks DB, Balkin A, Daniel HW,
Schonrock J, Ward B and Frazer IH: Transmission of human
papillomaviruses from mother to child. Aust NZJ Obstet Gynaecol.
33:30–32. 1993. View Article : Google Scholar
|
|
28
|
Kaye JN, Cason J, Pakarian FB, Jewers RJ,
Kell B, Bible J, Raju KS and Best JM: Viral load as a determinant
for transmission of human papillomavirus type 16 from mother to
child. J Med Virol. 44:415–421. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pakarian F, Kaye J, Cason J, Kell B,
Jewers R, Derias NW, Raju KS and Best JM: Cancer associated human
papillomaviruses: Perinatal transmission and persistence. Br J
Gynaecol Obstet. 101:514–517. 1994. View Article : Google Scholar
|
|
30
|
Syrjänen S: HPV infections in children.
Papillomavirus Rep. 14:93–110. 2003. View Article : Google Scholar
|
|
31
|
Syrjänen S and Puranen M: Human
papillomavirus infections in children: The potential role of
maternal transmission. Crit Rev Oral Biol Med. 11:259–274. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim MG, Kim SG, Lee JH, Eun YG and Yeo SG:
The therapeutic effect of OK-432 (picibanil) sclerotherapy for
benign neck cysts. Laryngoscope. 118:2177–2181. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Knipping S, Goetze G, Neumann K and
Bloching M: Sclerotherapy of cervical cysts with Picibanil
(OK-432). European Archives of Oto-Rhino-Laryngology. 264:423–427.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yasui T, Morii E, Yamamoto Y, Yoshii T,
Takenaka Y, Nakahara S, Todo T and Inohara H: Human papillomavirus
and cystic node metastasis in oropharyngeal cancer and cancer of
unknown primary origin. PLoS One. 9:e953642014. View Article : Google Scholar : PubMed/NCBI
|