Introduction
Non-small cell lung cancer (NSCLC) is one of the
most frequent and lethal malignancies worldwide; the 5-year
survival rate is only 20% (1).
Previous studies have indicated an increasing trend of NSCLC
mortality in China in the previous 20 years, particularly in urban
areas and among older (≥60 years old) people (2,3). An
increasing number of studies have demonstrated the emerging roles
of long non-coding RNAs (lncRNAs) in the tumorigenesis of NSCLC and
have suggested their potential clinical applications as diagnostic
markers and therapeutic targets in NSCLC (4,5).
LncRNAs are a class of non-coding RNAs that are
>200 nucleotides in length (6).
Dysregulation of lncRNAs contributes to the development and
progression of several human diseases, notably lung cancer
(7). For example, Hox Transcript
Antisense RNA promotes proliferation, survival, invasion,
metastasis and drug resistance in lung cancer cells (8). Metastasis-associated lung adenocarcinoma
transcript 1 is an important target for anti-metastatic therapy in
NSCLC (9,10). Antisense Noncoding RNA in the
Inhibitors of cyclin dependent kinase 4 locus promotes NSCLC cell
proliferation and inhibits apoptosis (11). Additional characterization of other
lung cancer-associated lncRNAs will provide an improved
understanding of their potential roles as therapeutic targets.
Ubiquitin-like with PHD and ring finger domains 1
(UHRF1) is overexpressed in various types of cancer, and was
identified as a novel diagnostic marker of lung cancer (12). UHRF1, also known as ICBP90, was
identified as a functional determinant of growth regulation genes,
including p16INK4A and p14ARF, and the
expression of UHRF1 is only detectable in proliferating cells, not
in quiescent cells (13). Previous
studies identified that UHRF1 serves a central function in
epigenetic modulation during DNA duplication in the S phase
(14–16). Taniue et al (17) demonstrated that lncRNA UHRF1 protein
associated transcript (UPAT) stabilizes the UHRF1 protein by
interfering with its ubiquitination and degradation, and promotes
colon tumorigenesis (17).
The present study demonstrated that UPAT expression
was also upregulated in NSCLC tissues. Additionally, UPAT
significantly promoted cell growth and G1-S phase transition of
NSCLC cells. Furthermore, lncRNA UPAT suppressed the expressions of
Ras association domain-containing protein 1 (RASSF1) and
Cadherin-13 (CDH13) by increasing UHRF1 expression, thereby
promoting NSCLC cell proliferation. In conclusion, the data of the
present study suggested that the lncRNA UPAT promoted the
proliferation of NSCLC cells and may be a potential therapeutic
target of NSCLC.
Materials and methods
Tissue collection and ethics
statement
A total of 43 paired tumor tissues and matched
normal tissues (>2.0 cm distance from the tumor edge) were
collected from patients with NSCLC (age range, 33–85 years old;
mean age, 51.7 years old; 31 male and 12 female) who received
surgical treatment between August 2011 and September 2015 at The
Second Affiliated Hospital of Jiaxing University (Jiaxing, China).
All experiments were approved by the Research Ethics Committee of
Jiaxing University (Jiaxing, China). Written informed consent was
obtained from all patients.
Cell culture
The human lung epithelial BEAS-2B cell line and
NSCLC H1299, H1650, H358 and A549 cell lines were purchased from
Shanghai Institute of Biochemistry and Cell Biology, Shanghai
Institutes for Biological Sciences, Chinese Academy of Sciences
(Shanghai, China). The cells were cultured in RPMI-1640 medium
(Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) containing 10% fetal bovine serum (Life Technologies; Thermo
Fisher Scientific, Inc.), 100 IU/ml penicillin and 100 mg/ml
streptomycin, and maintained at 37°C in humidified air containing
5% CO2.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA of tissues and cells were extracted using
of TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Total RNA was reverse transcribed to cDNA
by using the PrimeScript RT Master Mix Kit (Takara Biotechnology
Co., Ltd., Tokyo, Japan). qPCR was carried out using SYBR Green
remix (Takara Biotechnology Co., Ltd.) using an ABI Step One
instrument (Thermo Fisher Scientific, Inc.) with the following
thermocycling conditions: 2 min at 94°C, followed by 40 cycles of
30 sec at 94°C, 30 sec at 60°C, 30 sec at 72°C, then 2 min at 72°C.
The primers sequences were obtained from PrimerBank (https://pga.mgh.harvard.edu/primerbank/;
date of access, November 15, 2011). The sequences were as follows:
UPAT forward, AACCAAGAGCCTGAAGACG, reverse, CTCACCTCCTTTCTCACTCC;
UHRF1 forward, GCCACCCAAAGTTCACATCTT and reverse,
TGTTGCTATGACATTGCAGTCC; RASSF1 forward, CCCCGCAGTGCTATTGCAT and
reverse, CACGAAGCGCACATTCTCTT; CDH13 forward,
AGTGTTCCATATCAATCAGCCAG and reverse, CCTTACAGTCACTGAAGGTCAAG; GAPDH
forward, TGTGGGCATCAATGGATTTGG and reverse, ACACCATGTATTCCGGGTCAAT.
The relative amount of mRNA was calculated using the
2−ΔΔCq method (18). Gene
expression was normalized by GAPDH. All data were obtained from
three individual experiments.
Transfection of NSCLC cells
UPAT and UHRF1 siRNAs were purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). The siRNA sequence of UHRF1
was AAACAGAUGGAGGACGGCCA, and the siRNA sequence of UPAT was
AGGAGGTGAGAGGGAATGT. A549 cells (1×105 cells/well) were
seeded in a 6-well culture plate containing complete medium 24 h
prior to transfection. The negative control scramble or UPAT siRNA
(50 pmol/well) or UHRF1 siRNA (50 pmol/well) were transfected with
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in
A549 cells according to the manufacturer's protocol. The
full-length complementary DNA of UPAT was synthesized and subcloned
into the pcDNA3 vector by Genewiz, Inc. (Suzhou, China), named
pcDNA3-UPAT. The empty pcDNA3 vector (8 µg) or pcDNA3-UPAT (8 µg)
were transfected with Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in H1299 cells according to the manufacturer's
protocol. At 24 h after transfection, the cells were treated and
harvested.
Western blotting
A549 and H1299 cells (1×107) were lysed
in radioimmunoprecipitation assay lysis buffer (Beyotime Institute
of Biotechnology, Haimen, China) and protein concentrations were
quantified using the BCA protein assay (Pierce; Thermo Fisher
Scientific, Inc.). Equal quantities (20 µg) of protein were
separated via SDS-PAGE (10%) and then transferred to polyvinylide
fluoride membranes. The membranes were blocked with 5% skimmed milk
in Tris-buffered saline with Tween-20 for 30 min at room
temperature. This was followed by an incubation at 4°C overnight
with primary antibodies: UHRF1 (sc-365392, 1:250 dilution; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA); RASSF1 (sc-18722, 1:200
dilution; Santa Cruz Biotechnology, Inc.); CDH13 (sc-166875, 1:300
dilution; Santa Cruz Biotechnology, Inc.); and GAPDH (sc-47724,
1:500 dilution; Santa Cruz Biotechnology, Inc.). The membranes were
washed and then incubated with goat anti-mouse horseradish
peroxidase-conjugated secondary antibody (sc-2005, 1:2,000
dilution; Santa Cruz Biotechnology, Inc.) at room temperature for 2
h. The membranes were then analyzed using ECL substrate (32106,
Thermo Fisher Scientific, Inc.) following the manufacturer's
protocol. GAPDH was used as an endogenous protein for
normalization. Protein bands were quantified using the Tanon-5200
Image Analyzer (Shanghai, China). Quantification of blots using
Image J software (NIH, USA).
CCK-8 assay
The proliferation of A549 and H1299 cells were
analyzed with a Cell Counting Kit-8 (CCK-8) (Dojindo Molecular
Technologies, Kumamoto, Japan). A549 and H1299 cells
(1×104 cells/well) were seeded in 96-well plates at 37°C
in humidified 5% CO2 atmosphere. Following transfection
treatment, 10 µl CCK-8 solution was added into each well and then
incubated for another 4 h at 37°C. CCK-8 reagent was added at 0,
24, 48 and 72 h according to the manufacturer's protocol. The
absorbance at 450 nm was measured using a microplate reader.
Cell cycle analysis
Following transfection treatment, A549 and H1299
cells were trypsinized and washed with cold PBS. The cells
(1×106) were fixed in pre-chilled 70% ethanol at −20°C
overnight. For measurement of DNA content, cells were stained with
propidium iodide solution (50 µg/ml propidium iodide, 100 µg/ml
RNase A, 0.05% Triton X-100 in PBS), and incubated at 37°C in the
dark for 30 min. DNA content analysis by flow cytometry with a
FACSCalibur system (BD Biosciences, Franklin Lakes, NJ, USA). The
percentage of the cell population in each phase was calculated with
the FlowJo FACS analysis software (version 7.0; Tree Star, Inc.,
Ashland, OR, USA).
Plasmid transfection and
dual-luciferase reporter assay
The sequence of human UHRF1 (Gene ID: 29128)
promoter (−2,000 bp) was obtained from the UCSC Genome Browser
(uc002mbo.3, Human GRCh37/hg19; http://genome.ucsc.edu/; date of access, February 13,
2017) and purchased from Genewiz, Inc. (Suzhou, China). The
sequence was cloned into the luciferase reporter vector pGL4
(Promega Corporation, Madison, WI, USA) to produce the pGL4-UHRF1
plasmid. The pGL4.74 vector was also purchased from Promega
Corporation. The siRNA sequence of siRNA sequence of UPAT is
AGGAGGTGAGAGGGAATGT. NSCLC A549 cells (1×105) were
seeded in a 6-well culture plate containing complete medium 24 h
prior to transfection. The negative control scramble or UPAT siRNA
(50 pmol/well) were transfected with Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) in A549 cells
according to the manufacturer's protocol. The empty pcDNA3 vector
(8 µg) or pcDNA3-UPAT (8 µg) were transfected with Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in A549 cells
according to the manufacturer's protocol. After 24 h
post-transfection, the A549 cells were transfected with pGL4-UHRF1
and pGL4.74 vectors using Lipofectamine® 3000 according
to manufacturer's protocol. Cells were cultured for 8 h after
plasmid transfection. Then, the luciferase activity was measured
using a Dual-Luciferase Reporter assay system (E1910; Promega
Corporation) according to the manufacturer's protocol. Data were
normalized to the activity of Renilla luciferase.
Statistical analysis
Data are presented as mean ± standard deviation of
three independent experiments. Differences between groups were
analyzed by GraphPad Prism 5 software (GraphPad Software, Inc., La
Jolla, CA, USA) with unpaired Student's t-test or one-way analysis
of variance with a Tukey-Kramer post hoc test. The Pearson's
Chi-square test was used to evaluate the difference between levels
of UPAT expression in specimens and clinicopathological
characteristics. P<0.05 was considered to indicate a
statistically significant difference.
Results
UPAT is upregulated in human NSCLC
tissues and cells
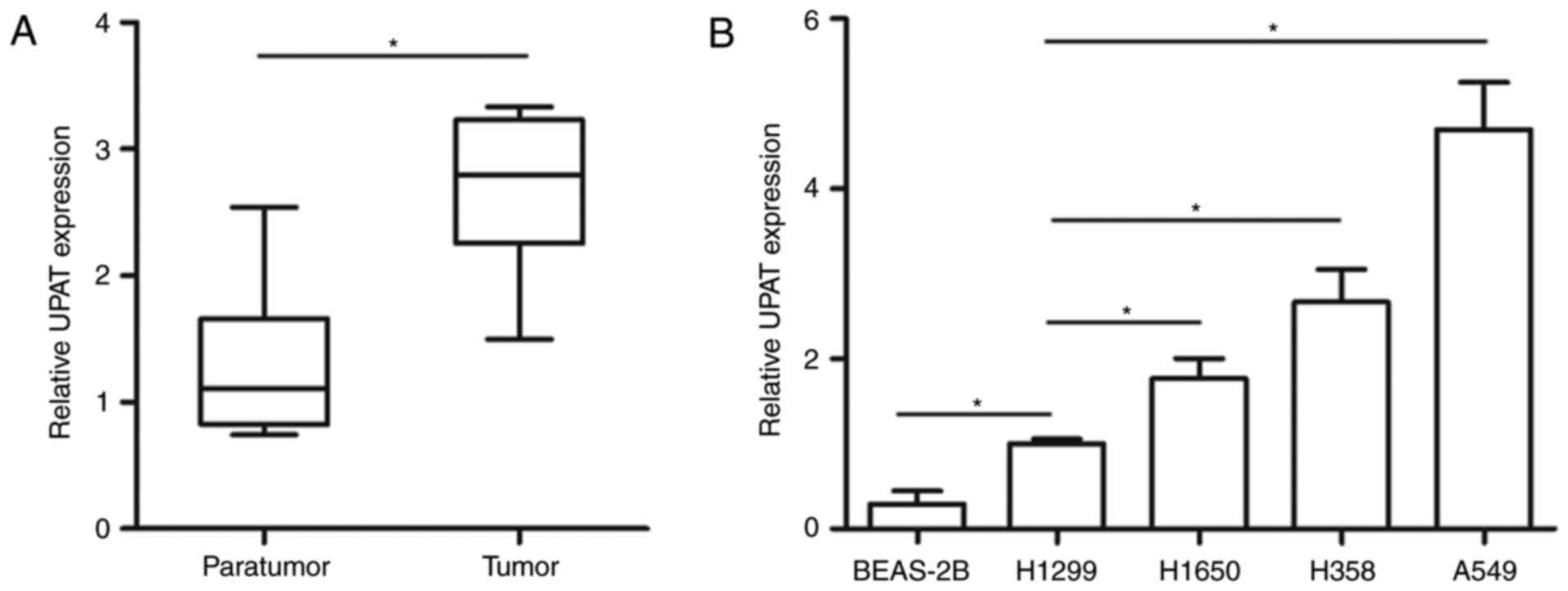
The expression of the UPAT in 43 human NSCLC tissues
and matched adjacent normal tissues was first examined using
RT-qPCR analysis. The results indicated that UPAT expression was
significantly upregulated in NSCLC tissues compared with the
control (Fig. 1A). The expression of
UPAT was also analyzed in normal human lung epithelial BEAS-2B cell
line and NSCLC H1299, H1650, H358 and A549 cell lines; the UPAT
level was significantly increased in NSCLC cells compared with its
expression in the BEAS-2B cells. In addition, the UPAT level was
significantly increased in the A549 cells (4.48-fold change)
compared with its expression in the H1299 cells (Fig. 1B). Furthermore, all samples from
patients with NSCLC were divided into two groups, including a UPAT
low-expression group (n=21) and a high-expression group (n=22). The
median (2.13, in tumor group) was used as cut-off value. Then, the
χ2 analysis was performed to explore the potential
associations between UPAT expression and clinical characteristics;
the data revealed that the expression level of UPAT was
significantly associated with tumor size, Tumor-Node-Mestastasis
stage and history of smoking, which suggests that the abnormal
expression of UPAT contributed to the development of NSCLC
(Table I).
 | Table I.Association between UPAT expression
and clinicopathological characteristics of patients with non-small
cell lung cancer (n=43). |
Table I.
Association between UPAT expression
and clinicopathological characteristics of patients with non-small
cell lung cancer (n=43).
| Characteristics | All patients | UPAT low expression
[< median]a(%) | UPAT high expression
[≥ median]a (%) | P-valueb |
|---|
| Total | 43 | 21 (48.8) | 22 (51.2) | – |
| Age, years |
|
|
| 0.85 |
|
<60 | 26 | 13 (50.0) | 13 (50.0) | – |
| ≥60 | 17 | 8
(47.1) | 9
(52.9) | – |
| Sex |
|
|
| 0.56 |
| Male | 31 | 16 (51.6) | 15 (48.4) | – |
|
Female | 12 | 5
(41.7) | 7
(58.3) | – |
| Tumor size, cm |
|
|
| 0.01c |
|
<3 | 18 | 13 (72.2) | 5
(27.8) | – |
| ≥3 | 25 | 8
(32.0) | 17 (68.0) | – |
| TNM stage |
|
|
| 0.04c |
|
I–II | 24 | 15 (62.5) | 9
(37.5) | – |
|
III–IV | 19 | 6
(31.6) | 13 (68.4) | – |
| Metastasis |
|
|
| 0.92 |
| No | 29 | 14 (48.3) | 15 (51.7) | – |
|
Yes | 14 | 7
(50.0) | 7
(50.0) | – |
| History of
smoking |
|
|
| 0.03c |
|
Never | 12 | 9
(75.0) | 3
(25.0) | – |
|
Ever | 31 | 12 (38.7) | 19 (61.3) | – |
UPAT promotes NSCLC cell proliferation
and inhibits G1 arrest in vitro
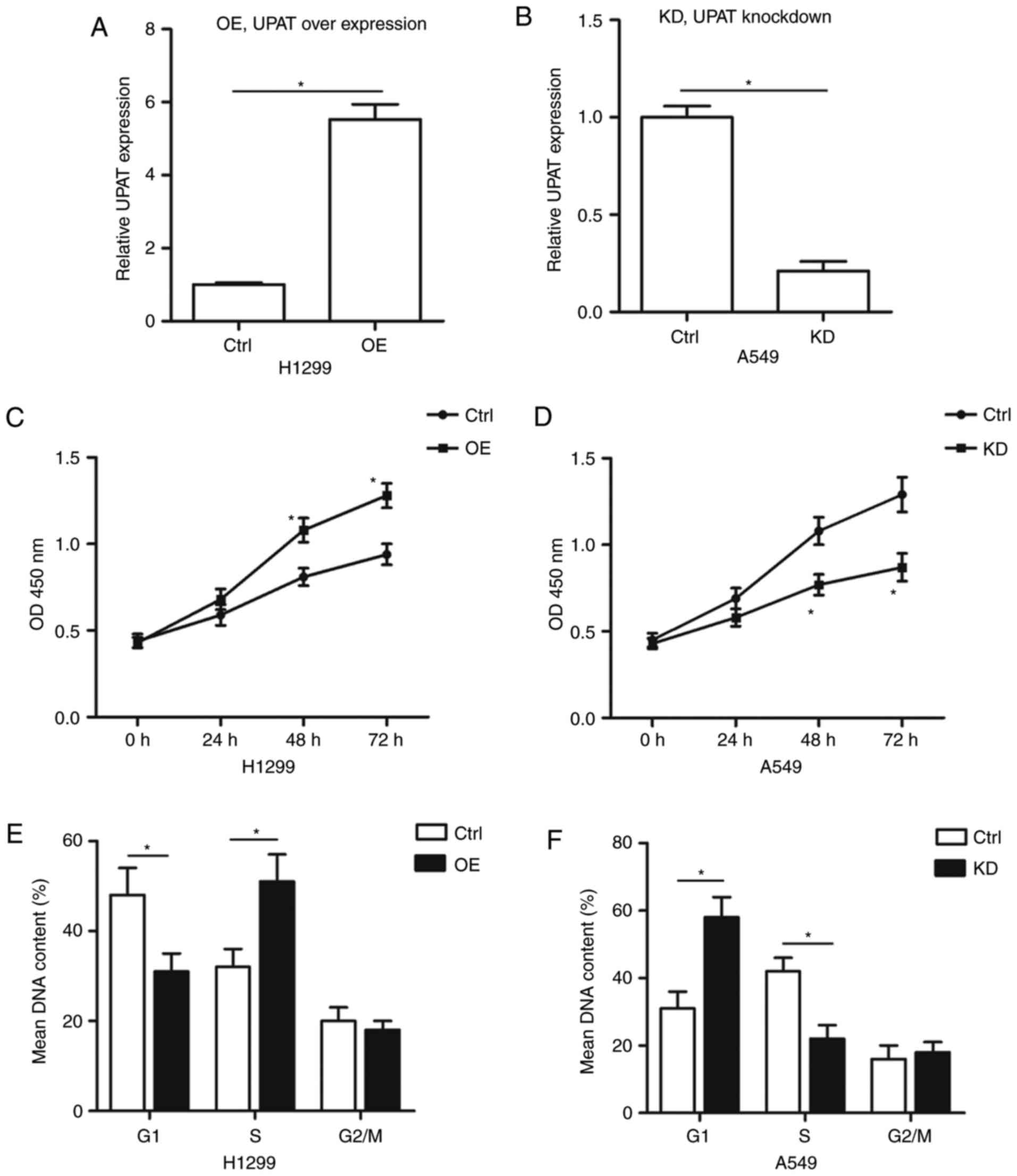
To explore the function of UPAT in NSCLC cells, the
A549 and H1299 cell lines were selected for additional studies, as
they exhibited the highest and lowest UPAT expression levels,
respectively (Fig. 1B). Then, the
overexpression plasmid and siRNA were used for transfection into
the H1299 and A549 cell lines, respectively. The qPCR analysis
indicated that the level of UPAT was increased in H1299 cells with
pcDNA3-UPAT transfection (Fig. 2A).
The level of UPAT was decreased in A549 cells with UPAT siRNA
transfection (Fig. 2B). Therefore,
these 2 cell lines were used in all additional experiments for UPAT
overexpression or silencing, respectively. To determine the effect
of UPAT on NSCLC cell proliferation, a CCK-8 assay was performed.
The results indicated that the cell proliferation of H1299 cells
transfected with pCDNA-UPAT was significantly increased compared
with the control groups (Fig. 2C). By
contrast, UPAT knockdown inhibited the A549 cell proliferation
(Fig. 2D). These data indicated that
UPAT markedly promoted the proliferation of NSCLC cells. The
effects of UPAT on the cell cycle were additionally investigated.
As indicated in Fig. 2E,
overexpressing UPAT decreased the number of H1299 cells in the G1
phase and increased the number of cells in the S phase compared
with the control. However, UPAT siRNA transfection caused a G1
phase cell-cycle arrest, with a significant increase in the
percentage of A549 cells in G1 stage (Fig. 2F). These results suggest that the
proliferation-inducing effects of UPAT are probably attributable to
the promotion of the G1-S phase transition.
UPAT inhibits RASSF1 and CDH13
expression
A previous study demonstrated that UPAT is
associated with UHRF1 and increases its protein level (17). Additionally, UHRF1 knockdown
contributes to lower methylation levels of RASSF1 and CDH13
promoters in A549 NSCLC cells (19).
To study the underlying molecular mechanisms and downstream targets
involved in the effect of UPAT on cell cycle and growth, the mRNA
levels of RASSF1 and CDH13, which are tumor suppressor genes
involved in cell cycle and proliferation and are also inhibited by
UHRF1 in NSCLC (20,21), were examined. The qPCR analysis
demonstrated that RASSF1 and CDH13 mRNA levels were decreased in
H1299 cells transfected with pCDNA-UPAT compared with the control
(Fig. 3A). By contrast, UPAT
knockdown increased the RASSF1 and CDH13 mRNA levels in A549 cells
(Fig. 3B). Similarly, RASSF1 and
CDH13 protein levels were decreased in H1299 cells transfected with
pCDNA-UPAT but increased in A549 cells transfected with UPAT siRNA
compared with the respective controls (Fig. 3C and D). Furthermore, it was also
confirmed that UPAT overexpression increased the protein level of
UHRF1 in the H1299 cells, but that UPAT knockdown decreased the
protein level of UHRF1 in the A549 cells (Fig. 3C and D). These results suggested that
UPAT may promote NSCLC cell proliferation partly through epigenetic
silencing of RASSF1 and CDH13 transcription.
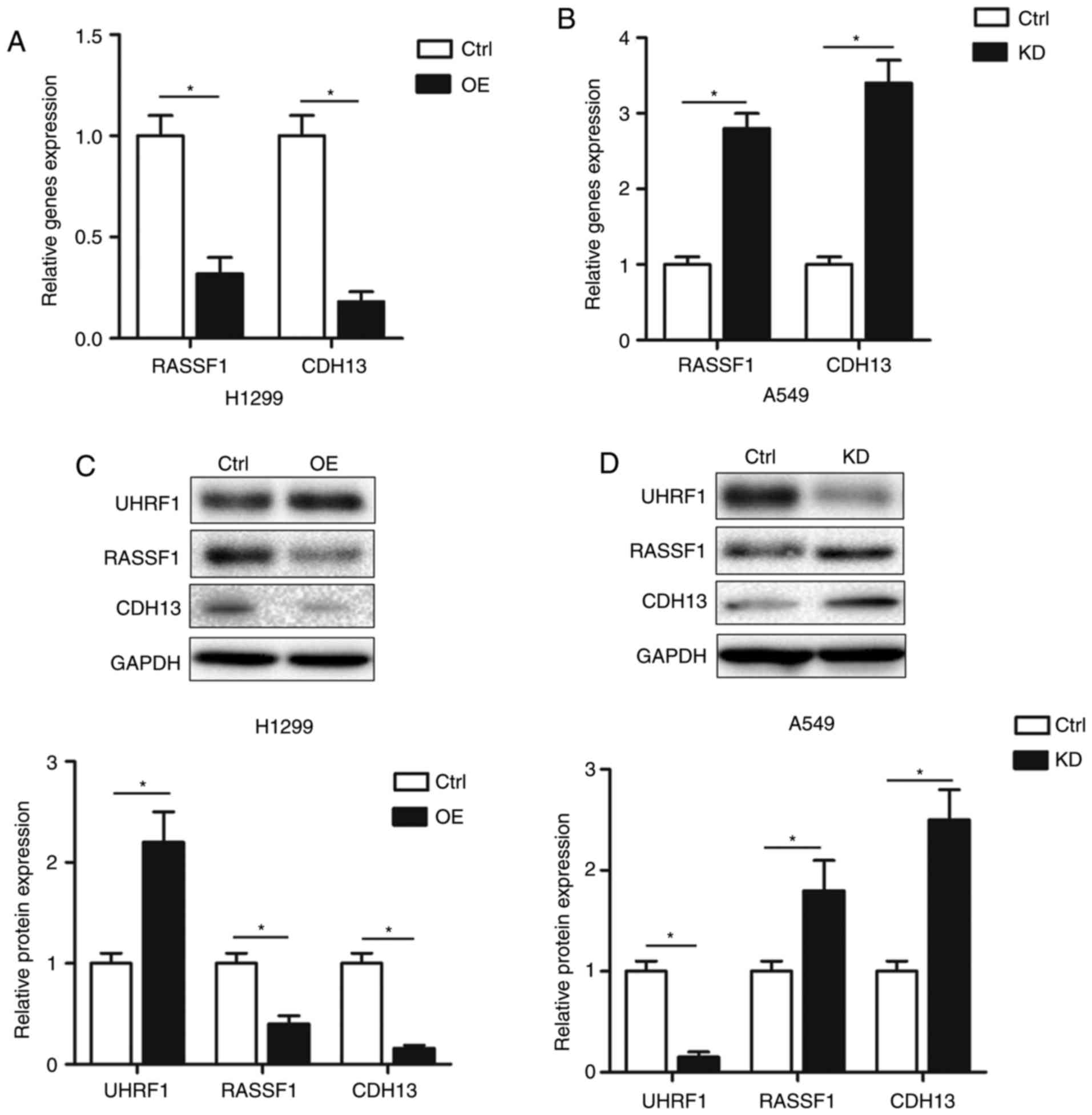 | Figure 3.UPAT inhibits RASSF1 and CDH13
expression. (A and B) Quantitative polymerase chain reaction assays
were used to examine the changes of RASSF1 and CDH13 mRNAs in (A)
H1299 cells transfected with pcDNA-UPAT (OE) and (B) A549 cells
transfected with UPAT siRNA (KD) or ctrl. GAPDH was used as an
internal control. Data are presented as mean ± SD. n=3. *P<0.05
by Student's t-test. Western blotting analysis of UHRF1, RASSF1 and
CDH13 protein levels in (C) H1299 cells transfected with pcDNA-UPAT
(OE) and (D) A549 cells transfected with UPAT siRNA (KD) or ctrl.
GAPDH was used as an internal control. Data are presented as mean ±
SD. n=3. *P<0.05 by Student's t-test. UHRF1, Ubiquitin-like with
PHD and RING finger domains 1; UPAT, UHRF1 protein associated
transcript; OE, overexpressed; KD, knockdown; SD, standard
deviation; si, small interfering; ctrl, control; RASSF1, Ras
association domain-containing protein 1; CDH13, Cadherin-13. |
UHRF1 overexpression is potentially
involved in the oncogenic function of UPAT
Luciferase vectors carrying the UHRF1 promoter
(−2,000 bp) in front of the firefly luciferase gene coding region
were constructed. As indicated in Fig.
4A, pCDNA-UPAT significantly increased the luciferase activity
of the UHRF1 promoter; concurrently, treatment with UPAT siRNA
suppressed the luciferase activity of UHRF1 promoter. To validate
whether UPAT regulated NSCLC cell proliferation by increasing UHRF1
expression, rescue experiments were performed. A549 cells were
co-transfected with pcDNA-UPAT and UHRF1 siRNA (Fig. 4B), and the CCK-8 and cell cycle assays
results indicated that the knockdown of UHRF1 partially decreased
the promotion of cell growth and G1-S phase transition resulting
from UPAT overexpression (Fig. 4C and
D). These results indicated that the effects of UPAT on NSCLC
cell proliferation partially depended on its effect of increasing
UHRF1 levels.
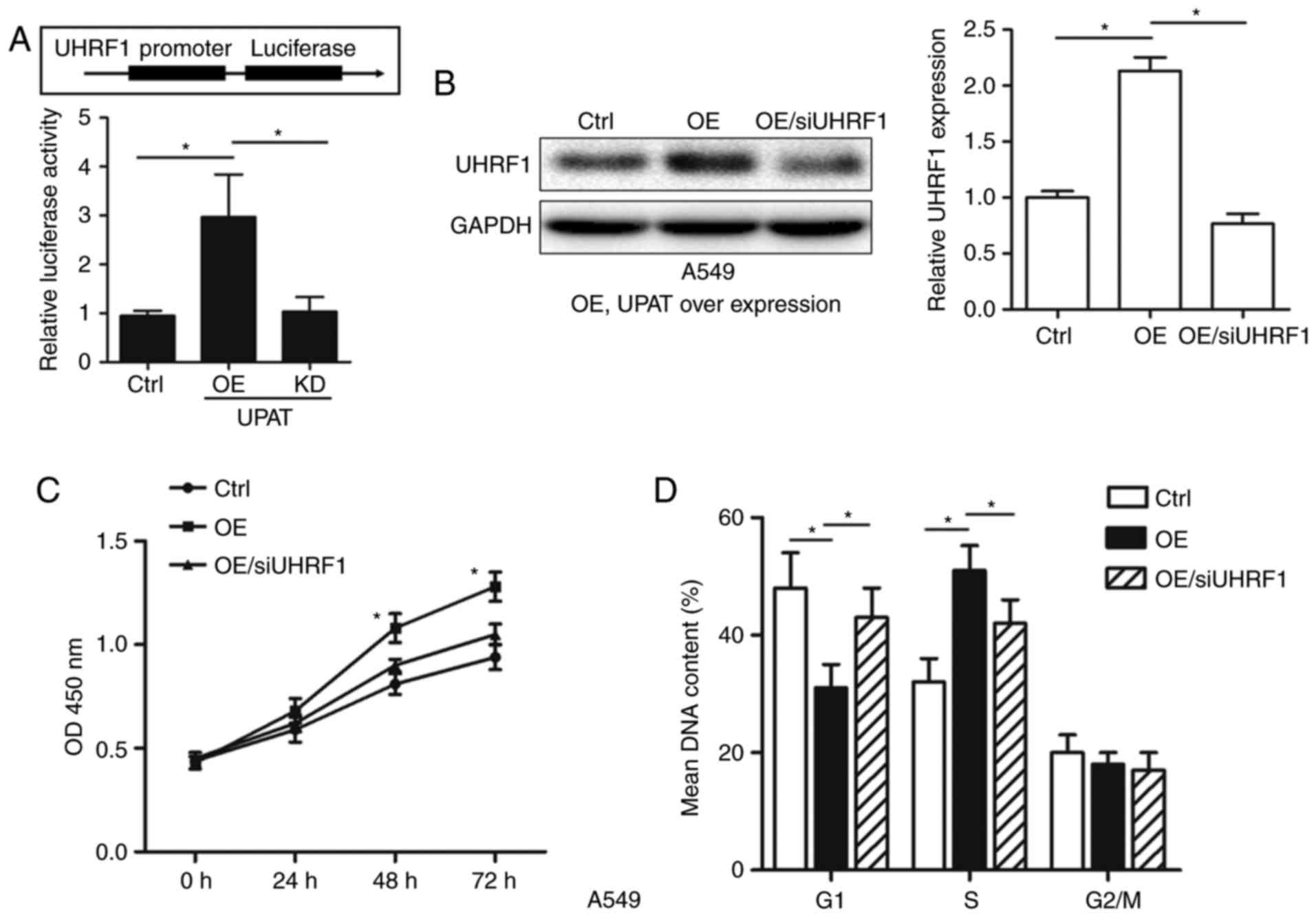 | Figure 4.UHRF1 overexpression is potentially
involved in the oncogenic function of UPAT. (A) A549 cells were
transfected with pcDNA-UPAT (OE), UPAT siRNA (KD), or the negative
control. After 24 h, cells were transfected with pGL4-UHRF1
promoter (−2,000 bp) luciferase vector and pGL4.74 plasmids for 12
h. The luciferase activity was determined using a dual-luciferase
reporter assay system. Data are presented as mean ± SD. n=3.
*P<0.05 by ANOVA. (B) Western blotting analysis of UHRF1 protein
level in A549 cells treated with pcDNA-UPAT and UHRF1 siRNA, or the
negative control. (C) CCK-8 assay was used to determine the cell
viability for pcDNA-UPAT and UHRF1 siRNA co-transfected A549 cells.
Data are presented as mean ± SD. n=3. *P<0.05 by ANOVA vs. Ctrl
and OE/siUHRF1 groups. (D) Cell cycle assay was performed to
determine the cell cycle phase of pcDNA-UPAT and UHRF1 siRNA
co-transfected A549 cells. GAPDH was used as an internal control.
Data are presented as mean ± SD. n=3. *P<0.05 by ANOVA. UHRF1,
Ubiquitin-like with PHD and RING finger domains 1; UPAT, UHRF1
protein associated transcript; OE, overexpressed; KD, knockdown;
SD, standard deviation; si, small interfering; ctrl, control;
ANOVA, analysis of variance. |
Discussion
UHRF1, which is overexpressed in several types of
cancer, including breast, prostate and lung cancer, is a key
regulator in the G1/S transition during the cell cycle (22). Previous studies have suggested that
UHRF1/ICBP90 is associated with histone ubiquitination and DNA
methylation in the context of tumor angiogenesis and silencing of
tumor suppressor genes (23–26). UHRF1 binds hemi-methylated DNA through
its SET- and RING-associated domain, triggering the recruitment of
DNA (cystosine-5)-methyltransferase 1 and Histone deacetylase 1
(27,28). UHRF1 also mediates tumor suppressor
gene inactivation in NSCLC (19).
UHRF1 knockdown contributes to lower methylation levels of RASSF1
and CDH13 promoters in A549 NSCLC cells (19). In the present study, it was identified
that the expression of UPAT in NSCLC tissues was significantly
increased compared with paratumor tissues, suggesting that UPAT
serves a crucial role in NSCLC. Therefore, it was hypothesized that
UPAT may exert its effects through increasing UHRF1 and
epigenetically silencing RASSF1 and CDH13 transcription.
The data from the present study suggested that UPAT
significantly promoted the proliferation of NSCLC cells by
enhancing G1-S phase transition. Consistently, UPAT promotes colon
tumorigenesis by inhibiting the degradation of UHRF1 (17). Additionally, in the present study, the
downstream target genes of UHRF1 was analyzed by qPCR and western
blotting assays, and the data indicated that RASSF1 and CDH13 were
upregulated following UPAT knockdown. RASSF1 may form an endogenous
complex with the activated Ras oncoprotein and integrate pro-growth
pathways with pro-growth arrest/death pathways (21,29). CDH13
mediates cell growth arrest and regulates the cyclin-dependent
kinase inhibitor 1 pathway (30).
Loss of expression and aberrant methylation of the RASSF1 and CDH13
is observed in lung cancer (20,31). In
the present study, it was demonstrated that RASSF1 and CDH13 served
as tumor suppressors and were silenced by UPAT in NSCLC cells.
Concurrently, UPAT also increased the expression of UHRF1.
To additionally confirm that UPAT exerted its
effects through increasing UHRF1, recue experiments were performed
and it was identified that the knockdown of UHRF1 partially
diminished UPAT-promoted cell growth and G1-S phase transition.
These results revealed that the effects of UPAT on NSCLC cell
proliferation were partially dependent on increasing UHRF1
expression.
In conclusion, the present study provides evidence
that UPAT is upregulated in NSCLC tissues. The role of UPAT in the
promotion of NSCLC cell proliferation may function partly via
epigenetically inhibiting RASSF1 and CDH13 transcription by
stabilizing UHRF1 expression and increasing the UHRF1-mediated
methylation modification of these target genes. UPAT may serve as a
potential target for the diagnosis and treatment of human
NSCLC.
Acknowledgements
The authors would like to thank Dr Huanyu Ju
(Department of Laboratory Medicine, the First Affiliated Hospital
of Nanjing Medical University, Nanjing, China) for his
assistance.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FW conceived, organized and supervised the study. HW
and DC performed the experiments. DC analyzed the data and wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All experiments were approved by the Research Ethics
Committee of Jiaxing University. Written informed consent was
obtained from all patients.
Consent for publication
Written informed consent for publication was
obtained from all patients for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chang JY, Senan S, Paul MA, Mehran RJ,
Louie AV, Balter P, Groen HJ, McRae SE, Widder J, Feng L, et al:
Stereotactic ablative radiotherapy versus lobectomy for operable
stage I non-small-cell lung cancer: A pooled analysis of two
randomised trials. Lancet Oncol. 16:630–637. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Renyi-Vamos F, Tovari J, Fillinger J,
Timar J, Paku S, Kenessey I, Ostoros G, Agocs L, Soltesz I and Dome
B: Lymphangiogenesis correlates with lymph node metastasis,
prognosis, and angiogenic phenotype in human non-small cell lung
cancer. Clin Cancer Res. 11:7344–7353. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kayser G: Non-small cell lung cancer. New
biomarkers for diagnostics and therapy. Pathologe. 36(Suppl 2):
S189–S193. 2015.(In German). View Article : Google Scholar
|
|
4
|
Chen J, Wang R, Zhang K and Chen LB: Long
non-coding RNAs in non-small cell lung cancer as biomarkers and
therapeutic targets. J Cell Mol Med. 18:2425–2436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu T, Yin X, Zhou Y, Wang Z, Shen S, Qiu
Y, Sun R and Zhao Z: Roles of noncoding RNAs in metastasis of
nonsmall cell lung cancer: A mini review. J Cancer Res Ther.
11(Suppl 1): C7–C10. 2015.PubMed/NCBI
|
|
6
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sang H, Liu H, Xiong P and Zhu M: Long
non-coding RNA functions in lung cancer. Tumour Biol. 36:4027–4037.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Loewen G, Jayawickramarajah J, Zhuo Y and
Shan B: Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol.
7:902014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo F, Guo L, Li Y, Zhou Q and Li Z:
MALAT1 is an oncogenic long non-coding RNA associated with tumor
invasion in non-small cell lung cancer regulated by DNA
methylation. Int J Clin Exp Pathol. 8:15903–15910. 2015.PubMed/NCBI
|
|
11
|
Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia
R, Liu YW, Liu XH, Zhang EB, Lu KH and Shu YQ: Long noncoding RNA
ANRIL promotes non-small cell lung cancer cell proliferation and
inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer
Ther. 14:268–277. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Unoki M, Daigo Y, Koinuma J, Tsuchiya E,
Hamamoto R and Nakamura Y: UHRF1 is a novel diagnostic marker of
lung cancer. Br J Cancer. 103:217–222. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jenkins Y, Markovtsov V, Lang W, Sharma P,
Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, et al:
Critical role of the ubiquitin ligase activity of UHRF1, a nuclear
RING finger protein, in tumor cell growth. Mol Biol Cell.
16:5621–5629. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bostick M, Kim JK, Estève PO, Clark A,
Pradhan S and Jacobsen SE: UHRF1 plays a role in maintaining DNA
methylation in mammalian cells. Science. 317:1760–1764. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sadler KC, Krahn KN, Gaur NA and Ukomadu
C: Liver growth in the embryo and during liver regeneration in
zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad
Sci USA. 104:1570–1575. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Obata Y, Furusawa Y, Endo TA, Sharif J,
Takahashi D, Atarashi K, Nakayama M, Onawa S, Fujimura Y, Takahashi
M, et al: The epigenetic regulator Uhrf1 facilitates the
proliferation and maturation of colonic regulatory T cells. Nat
Immunol. 15:571–579. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Taniue K, Kurimoto A, Sugimasa H, Nasu E,
Takeda Y, Iwasaki K, Nagashima T, Okada-Hatakeyama M, Oyama M,
Kozuka-Hata H, et al: Long noncoding RNA UPAT promotes colon
tumorigenesis by inhibiting degradation of UHRF1. Proc Natl Acad
Sci USA. 113:1273–1278. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Daskalos A, Oleksiewicz U, Filia A,
Nikolaidis G, Xinarianos G, Gosney JR, Malliri A, Field JK and
Liloglou T: UHRF1-mediated tumor suppressor gene inactivation in
nonsmall cell lung cancer. Cancer. 117:1027–1037. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kontic M, Stojsic J, Jovanovic D,
Bunjevacki V, Ognjanovic S, Kuriger J, Puumala S and Nelson HH:
Aberrant promoter methylation of CDH13 and MGMT genes is associated
with clinicopathologic characteristics of primary non-small-cell
lung carcinoma. Clin Lung Cancer. 13:297–303. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee MG, Jeong SI, Ko KP, Park SK, Ryu BK,
Kim IY, Kim JK and Chi SG: RASSF1A directly antagonizes RhoA
activity through the assembly of a Smurf1-mediated destruction
complex to suppress tumorigenesis. Cancer Res. 76:1847–1859. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jeanblanc M, Mousli M, Hopfner R, Bathami
K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD and
Bronner C: The retinoblastoma gene and its product are targeted by
ICBP90: A key mechanism in the G1/S transition during the cell
cycle. Oncogene. 24:7337–7345. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Unoki M, Nishidate T and Nakamura Y:
ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG
through its SRA domain. Oncogene. 23:7601–7610. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Achour M, Jacq X, Rondé P, Alhosin M,
Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A,
Hughes AD, et al: The interaction of the SRA domain of ICBP90 with
a novel domain of DNMT1 is involved in the regulation of VEGF gene
expression. Oncogene. 27:2187–2197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Karagianni P, Amazit L, Qin J and Wong J:
ICBP90, a novel methyl K9 H3 binding protein linking protein
ubiquitination with heterochromatin formation. Mol Cell Biol.
28:705–717. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alhosin M, Sharif T, Mousli M,
Etienne-Selloum N, Fuhrmann G, Schini-Kerth VB and Bronner C:
Down-regulation of UHRF1, associated with re-expression of tumor
suppressor genes, is a common feature of natural compounds
exhibiting anti-cancer properties. J Exp Clin Cancer Res.
30:412011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Avvakumov GV, Walker JR, Xue S, Li Y, Duan
S, Bronner C, Arrowsmith CH and Dhe-Paganon S: Structural basis for
recognition of hemi-methylated DNA by the SRA domain of human
UHRF1. Nature. 455:822–825. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sharif J, Muto M, Takebayashi S, Suetake
I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T,
Okamura K, et al: The SRA protein Np95 mediates epigenetic
inheritance by recruiting Dnmt1 to methylated DNA. Nature.
450:908–912. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Donninger H, Clark JA, Monaghan MK,
Schmidt ML, Vos M and Clark GJ: Cell cycle restriction is more
important than apoptosis induction for RASSF1A protein tumor
suppression. J Biol Chem. 289:31287–31295. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang ZY, Wu Y, Hedrick N and Gutmann DH:
T-cadherin-mediated cell growth regulation involves G2 phase arrest
and requires p21(CIP1/WAF1) expression. Mol Cell Biol. 23:566–578.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yanagawa N, Tamura G, Oizumi H, Kanauchi
N, Endoh M, Sadahiro M and Motoyama T: Promoter hypermethylation of
RASSF1A and RUNX3 genes as an independent prognostic prediction
marker in surgically resected non-small cell lung cancers. Lung
Cancer. 58:131–138. 2007. View Article : Google Scholar : PubMed/NCBI
|