Introduction
Colorectal cancer is one of the most serious
malignancies threatening human health; it is ranked as the second
leading cause of gastroenteric tumor-associated mortality
worldwide, and the fourth leading cause of global tumor incidence
and mortality (1). Due to increases
in average population age, increasing CRC incidence has become a
serious global public health issue (2,3). As a
result of a lack of effective diagnostic strategies for early-stage
CRC, the majority of cases of CRC are diagnosed at advanced stages,
and consequently are not able to be completely cured by surgical
intervention. The 5-year survival rate of advanced-stage CRC is
low, at only 10% (4,5). Tumor metastasis is one of the most
common causes of cancer-associated mortality and involves cancer
cells migrating from the primary tumor site to other organs via the
lymphatic or circulatory systems. Although an increasing volume of
evidence has demonstrated that the tumorigenesis rates and
progression of CRC depends on various genetic variations (6–8),
additional studies examining the in-depth molecular mechanisms are
required. At present, novel molecular markers and targets for the
early diagnosis of CRC are become an important area of study.
MicroRNAs (miRNAs/miR), as a type of non-coding,
single-stranded RNAs ranging from 21–24 nucleotides in length, have
been previously demonstrated to be novel effective molecular
targets for cancer therapy (9–12). The
majority of miRNAs combine with the 3′ untranslated region (UTR) of
target mRNAs through incomplete complementary combination, in the
form of RNA-induced silencing complexes (13,14).
miRNAs inhibit the expression of target genes by suppressing mRNA
translation or reducing the stability of mRNAs (15). An increasing number of studies have
indicated that the aberrant expression of miRNAs may affect a
number of physiological cell processes, including cell
proliferation, differentiation, migration and invasion (16). In addition, miRNAs have also been
demonstrated to be associated with clinical CRC development and
metastasis by regulating the expression of target genes and
relevant proteins (17).
Based on previous studies, miR-128 exists in a
number of tissues with markedly different expression levels
(18,19). The aberrant expression of miR-128 is
associated with numerous types of tumors, including glioma,
prostate and breast cancer, and CRC (20). The overexpression of miR-128 may
inhibit the developmental, proliferative and invasive abilities of
prostate cancer cells (16).
Concurrently, it may also decrease the proliferation rate of acute
myeloid leukemia cells by increasing the extent of DNA damage
(21). miR-128 was suggested to
inhibit glioma cancer cell proliferation by suppressing the protein
kinase B (Akt) pathway and cyclin-dependent kinase inhibitor 1
expression (22). Ai et al
(17) identified that miR-128
inhibited murine CRC cell development in vitro and in
vivo by targeting and inhibiting the expression of matrix
metalloproteinase (MMP)3, MMP10 and MMP13 in CRC cells. Although
the mechanisms of miR-128 in tumors have been studied extensively,
its role in human CRC remains unclear.
Ribophorin-II (RPN2), encoding the essential subunit
of the oligosaccharide transferase complex, is important for cell
structure, cell signal recognition and transduction (23). The expression of RPN2 was identified
to be increased in patients with gastric cancer, CRC,
hepatocellular carcinoma, lung and breast cancer, and head and neck
neoplasms, with a potential association with clinical phenotype
(24). It is also associated with
tumor metastasis, prognosis and drug resistance in tumor cells
(25). RPN2 knockdown was
demonstrated to promote cell apoptosis, inhibit cell proliferation
and increase cell sensitivity to chemotherapeutics in lung and
breast cancer, and osteosarcoma (26). RPN2 overexpression was also verified
to inactivate glycogen synthase kinase 3β and lead to a steady
expression of mutational tumor protein (p53), and to regulate
tumorigenesis and metastasis in breast cancer stem cells (27). Whether there is any association
between miR-128 and RPN2 in regulating CRC is currently unknown,
and merits additional study.
Collectively, the present study focused on the role
of miR-128 in regulating RPN2 expression in CRC cells, and provides
novel evidence for the therapeutic potential of miRNA regulating
colorectal cancer.
Materials and methods
Patients and tumor samples
A total of 53 patients with CRC in different
Tumor-Node-Metastasis stages, as proposed by American Joint
Committee on Cancer (28–31), aged between 32–74 years old (median,
57 years old), and admitted to The Sixth Affiliated Hospital of Sun
Yat-sen University (Guangzhou, China) were enrolled from November
2015 to November 2016. No patient had received radiotherapy or
chemotherapy prior to surgery. Matched adjacent normal colorectal
tissues (taken >2 cm away from the cancer tissue) were collected
as negative controls. Basic clinical and pathological data were
collected, and all patients provided informed consent. All tissue
samples from patients were collected and protocols were performed
according to the procedures approved by the Institutional Review
Board of the Independent Ethics Committee of The Sixth Affiliated
Hospital of Sun Yat-sen University (approval no. BZ20153587).
Cell culture and grouping
Human CRCHT29, SW480, SW620 and HCT116 cell lines
and the normal human colon mucosa epithelia NCM460 cell line were
all obtained from American Type Culture Collection (Manassas, VA,
USA). Cells were cultured in RPMI-1640 medium (Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) with 1% penicillin/streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C in a 5% CO2 incubator. The
medium was changed once every 2 days. Cell passage was performed
when cells reached 80% confluence. Cells in the logarithmic phase
were used.
miR-128 mimic transfection
Human CRC HT29 cells were seeded onto 12-well plates
at a concentration of 5×104 cells/well and cultured for
24 h prior to transfection. Then, confluent cells were transfected
with miR-128 mimics (5′-UUUCUCUGGCCAAGUGACACU-3′) and control
scrambled sequences (5′-GTGACCCACGATGTGTATTCGC-3′) (Shanghai
GenePharma, Co., Ltd., Shanghai, China), at a final concentration
of 50 nmol/l with Lipofectamine®2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to manufacturer's protocol.
Human CRC HT29 cells were randomly allocated into
three groups: Control; mock (cells transfected with control
scrambled sequences); and miR-128 (cells transfected with miR-128
mimics) groups. Reverse transcription quantitative polymerase chain
reaction (RT-qPCR) was conducted to determine miR-128 expression
levels, and RT-qPCR and western blot analyses were conducted to
determine RPN2 expression levels in the control, mock and miR-128
groups, as described subsequently.
Cell Counting Kit (CCK)-8 assay
Cell viabilities in the control, mock andmiR-128
groups were measured using a CCK-8 assay (Beyotime Institute of
Biotechnology, Haimen, China). Cells were seeded onto 96-well
plates (100 µl/well) at 5×103 cells/well and cultured in
RPMI-1640 medium for 4 h at 37°C in a 5% CO2 incubator.
Subsequently, 20 µl CCK-8 reagent was added and cells were
incubated at 37°C again for 1 h. Optical density values were
measured at a wavelength of 450 nm by the iMark microplate
absorbance reader (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Transwell assay
HT29 cells (6×104 cells/well) were seeded
in the upper wells of a Transwell migration system attached with
polycarbonate filters (Corning Incorporated, Corning, NY, USA) in
RPMI-1640 supplemented with 0.1% FBS. The lower wells were filled
with RPMI-1640 with 10% FBS. Following incubation for 24 h at 37°C,
the non-migrating cells from the upper well were removed with
cotton swabs. The cells that had migrated through the membranes
were fixed with 70% cold ethanol at 4°C for 30 min, and stained by
0.1% crystal violet at 37°C for 30 min. Cells were counted in 5
separate fields of view and images were captured using light
microscopy with ×200 magnification (Olympus Corporation, Tokyo,
Japan) and cell migration rate was calculated.
The cell invasion assay was performed as
aforementioned, with the exception of the application of chambers
with Matrigel® (BD Biosciences, Franklin Lakes, NJ,
USA), which was melted at 37°C for 30 min prior to usage.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Quantification of miR-128 was performed using a
TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.). The expression of miR-128 was normalized using
U6. Analysis of relative gene expression data was conducted using
2−ΔΔCq method (32).
To determine mRNA levels of RPN2, p53, Cyclin D1,
MMP-2, MMP-9, epithelial-cadherin (E-cadherin),
metastasis-associated protein 1 (MTA1) and tissue inhibitor of
metalloproteinases 2 (TIMP2), total RNA extracted from CRC tissues,
different groups of CRC cells, or cells transfected with miR-128
mimics was firstly reverse transcribed using the Takara PrimeScript
RT reagent kit (Takara Bio, Inc., Otsu, Japan) at 25°C for 5 min,
42°C for 60 min and 72°C for 10 min. Quantification of mRNA was
determined using a TaqMan Gene Expression Assay(Thermo Fisher
Scientific, Inc.). The thermocycling conditions of the reactions
were as follows: 15 sec at 95°C, followed by 40 cycles of
denaturation at 95°C for 15 sec and annealing/extension at 60°C for
25 sec. Target gene expression was normalized to GAPDH. The primer
sequences are summarized in Table
I.
 | Table I.Primer sequences used in the present
study. |
Table I.
Primer sequences used in the present
study.
| Name | Direction | Sequences
(5′-3′) |
|---|
| hsa-miR-128 | Forward |
UCACAGUGAACCGGUCUCUUU |
| U6 | Forward |
TGCGGGTGCTCGCTTCGCAGC |
|
| Reverse |
TGCGGGTGCTCGCTTCGCAGC |
| RPN2 | Forward |
AGGAAGTGGTGTTTGTTGCC |
|
| Reverse |
ACAGTCGAGGGAGCTTCTTC |
| p53 | Forward |
TCAGTCTACCTCCCGCCATA |
|
| Reverse |
TTACATCTCCCAAACATCCCT |
| CyclinD1 | Forward |
CAATGACCCCGCACGATTTC |
|
| Reverse |
AAGTTGTTGGGGCTCCTCAG |
| MMP-2 | Forward |
TGTGTTGTCCAGAGGCAATG |
|
| Reverse |
ATCACTAGGCCAGCTGGTTG |
| MMP-9 | Forward |
TTTGAGTCCGGTGGACGATG |
|
| Reverse |
GCTCCTCAAAGACCGAGTCC |
| MTA1 | Forward |
AAACTGCCCTGAGTGTGGT |
|
| Reverse |
AAATATGTTGACCCAGCTCATCT |
| E-cadherin | Forward |
CTGAAGTGACTCGTAACGAC |
|
| Reverse |
CATGTCAGCCAGCTTCTTGAAG |
| TIMP2 | Forward |
GCCTGACGGTCATATGGTAGA |
|
| Reverse |
GAATGCGCCAAAAACCCCAT |
| GAPDH | Forward |
GAATGGGCAGCCGTTAGGAA |
|
| Reverse |
AAAAGCATCACCCGGAGGAG |
Western blot analysis
Total protein of cells in each group was extracted
using ProteoPrep® Total Extraction Sample kit
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). GAPDH was used as
a control. Concentrations of proteins were determined using the
Bradford assay (Bio-Rad Laboratories, Inc.), and protein samples
(20 µg/lane) were separated on 10–15% SDS-PAGE and electroblotted
onto polyvinylidene fluoride membranes (EMD Millipore, Billerica,
MA, USA). Following blocking with 5% non-fat dry milk dissolved in
0.01 mol/l TBST solution (Wuhan Boster Biological Technology, Ltd.,
Wuhan, China), for 1 h at 37°C, membranes were incubated with
specific primary antibodies overnight at 4°C. Then, the membranes
were treated with a goat anti-rabbit immunoglobulin G H&L
horseradish peroxidase-conjugated secondary antibody (Abcam; cat.
no., ab6721, 1:5,000 dilution) at 37°C for 1 h and exposed to X-ray
film. Finally, immunoreactive bands were detected using enhanced
chemiluminescence detection reagents (Amersham; GE Healthcare,
Chicago, IL, USA). Band densities were quantified by densitometry
Image Lab™ Software version 4.1 (Bio-Rad Laboratories, Inc.).
The antibodies used were as follows: Rabbit
anti-RPN2 (Abcam, Cambridge, UK; cat. no., ab64467; 1:1,000
dilution); anti-p53 (Abcam; cat. no., ab131442; 1:1,000 dilution);
anti-Cyclin D1 (Abcam; cat. no., ab226977; 1:2,000 dilution);
anti-MMP-2 (Abcam; cat. no., ab37150; 1:1,000 dilution); anti-MMP-9
(Abcam; cat. no., ab73734; 1:1,000 dilution); anti-MTA1 (Abcam;
cat. no., ab71153; 1:2,000 dilution); anti-E-cadherin (Abcam; cat.
no., ab15148; 1:500 dilution); anti-TIMP2 (Abcam; cat. no.,
ab180630; 1:1,000 dilution); and anti-GAPDH (Abcam; cat. no.,
ab9485; 1:2,000 dilution).
Bioinformatics target prediction and
luciferase reporter assays
Bioinformatics target prediction tools, including
Target Scan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and miRNA targets
(http://cbio.mskcc.org/mimaviewer/)
were used to define the potential targets of miR-128 in 3′-UTR
fragment of RPN2.
The miR-128-3p binding sequence of the RPN2 3′-UTR
fragment was intentionally mutated using the Gene Tailor
Site-Directed Mutagenesis System (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the protocol of the manufacturer.
Then, the human RPN2 3′-UTR or mutated RPN2 3′-UTR sequences were
ligated into the luciferase reporter vector pGL3-basic plasmid
(Promega Corporation, Madison, WI, USA), to obtain the RPN2 3′-UTR
and Mut-RPN2 3′-UTR recombinant luciferase reporter plasmids,
respectively. Then, 5×103 293 cells were seeded in
96-well plates and co-transfected with 0.2 µg luciferase reporter
plasmid (RPN2 3′-UTR, or Mut-RPN2 3′-UTR plasmid) and 50 nmol/l
miRNAs [miR-128-3p or non-specific sequence (NC)], using
Lipofectamine® 2000. Following 24 h of incubation at
37°C, cells were harvested by centrifugation (5,000 × g) at 4°C for
20 min, and luciferase activities were measured using the Dual-Glo
Luciferase Reporter Assay System (Promega Corporation) according to
the protocol of the manufacturer. Data were normalized to the
Renilla luciferase activity.
Statistical analysis
Statistical analyses were performed using SPSS 22.0
software (IBM Corp., Armonk, USA). Each experiment was repeated in
triplicate, with all data presented as mean ± standard deviation. A
one-way analysis of variance followed by a Tukey's post-hoc test
was used to compare either two or multiple groups. The
χ2 test was used to compare categorical variables. A
Spearman rank correlation coefficient was used to analyze the
correlation between variables. Kaplan-Meier and log-rank test were
used for survival analysis. P<0.05 was considered to indicate a
statistically significant difference. P<0.01 was considered to
indicate a particularly significant difference.
Results
miR-128 and RPN2 levels in CRC
tissues
The present study included a total of 53 patients,
with a median age of 57 years. The expression of miR-128 and RPN2
in CRC tissues and adjacent normal tissues was assessed using
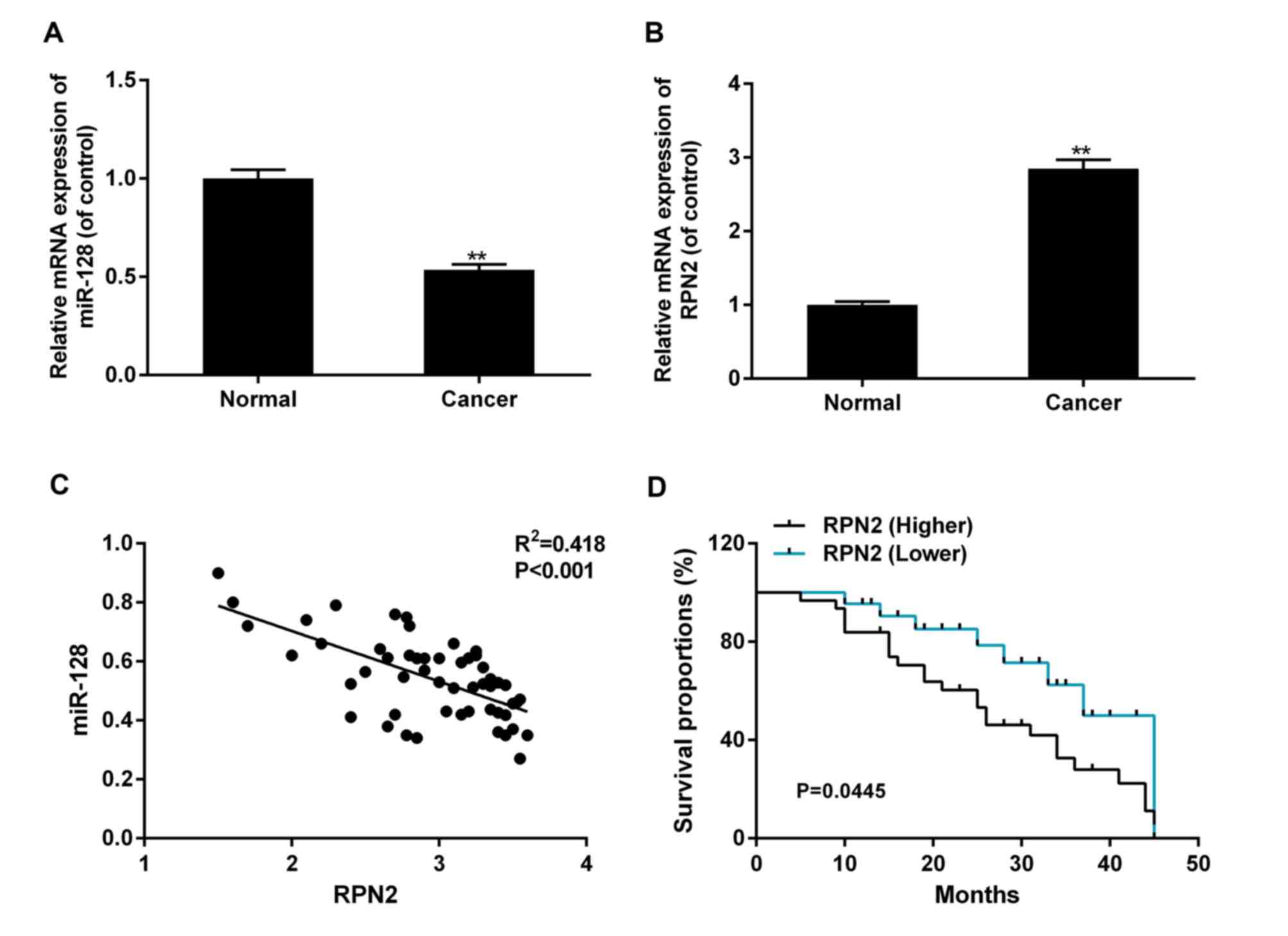
RT-qPCR. Compared with normal tissues, the levels of miR-128 in CRC
tissues were significantly decreased, while the expression of RPN2
was increased in CRC tissues (P<0.01; Fig. 1A and B). The correlation analysis
indicated a marked negative association between the expression of
miR-128 and RPN2, which suggested that miR-128 was likely to be
significant in the control of RPN2 regulation (Fig. 1C). A total of 70% (37/53) patient
tissues exhibited increased RPN2 expression in tumor tissues
compared with adjacent normal tissues, were placed in the high RPN2
expression group. Patients with decreased RPN2 expression in tumor
tissues, compared with adjacent normal tissues, were placed in the
low RPN2 expression group. As demonstrated in Table II, compared with adjacent tissue, the
increased RPN2 and corresponding reduced miR-128 were significantly
associated with poorly-differentiated histology (P=0.042), advanced
disease stages (P=0.004) and lymph node metastasis (P=0.013). It
was also detected that the survival rate was decreased in patients
with CRC with an increased expression of RPN2, and corresponding
decreased miR-128 levels, compared with patients with decreased
levels of RPN2 and corresponding increased miR-128 levels (P=0.445,
Fig. 1D).
 | Table II.Association between RPN2 and clinical
data of patients with colorectal cancer. |
Table II.
Association between RPN2 and clinical
data of patients with colorectal cancer.
| Factors | RPN2 expression
(low group/high group) | P-value |
|---|
| Age, years |
| 0.981 |
|
<50 | 14/6 |
|
|
≥50 | 23/10 |
|
| Sex |
| 0.726 |
|
Male | 26/12 |
|
|
Female | 11/4 |
|
| Histological
grade |
| 0.042a |
|
Well-moderate | 10/9 |
|
|
Poor | 27/7 |
|
| TNM |
| 0.004b |
|
I–II | 10/11 |
|
|
III–IV | 27/5 |
|
| M stage |
| 0.013a |
| M0 | 8/9 |
|
| M1 | 29/7 |
|
miR-128 and RPN2 levels in CRC cell
lines
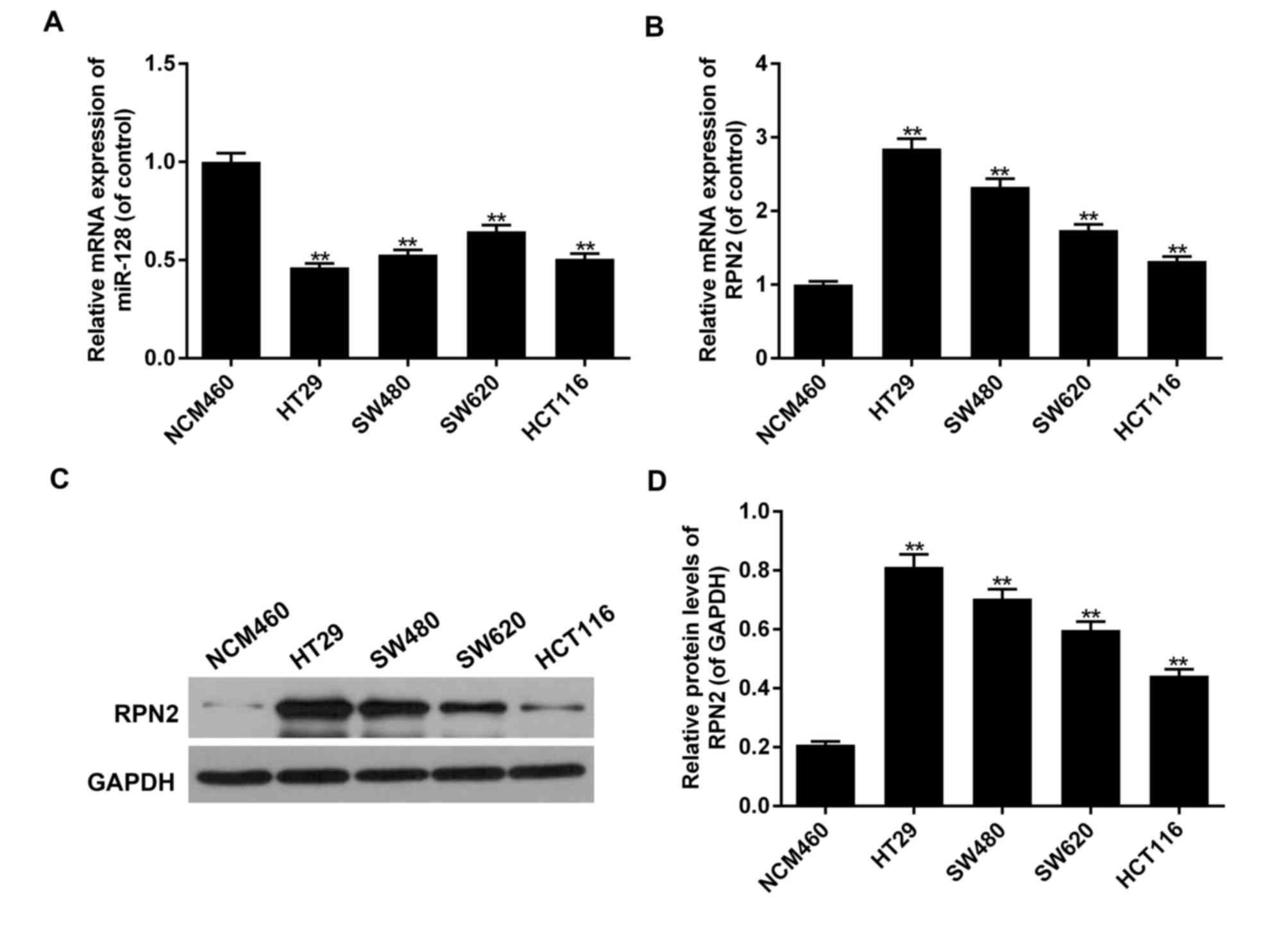
Decreased miR-128mRNA levels, as determined by
RT-qPCR, along with increased RPN2 mRNA and protein levels, as
determined by RT-qPCR and western blot analysis, were also
evaluated in 4 human CRC HT29, SW480, SW620, and HCT116 cell lines
in comparison with the normal CRC NCM460 cell line. The most
significant variations among the CRC cell lines were detected in
the HT29 cells, which was then selected to conduct subsequent
experiments (P<0.05; Fig. 2).
miR-128 downregulates mRNA and protein
expression levels of RPN2 in HT29CRC cells
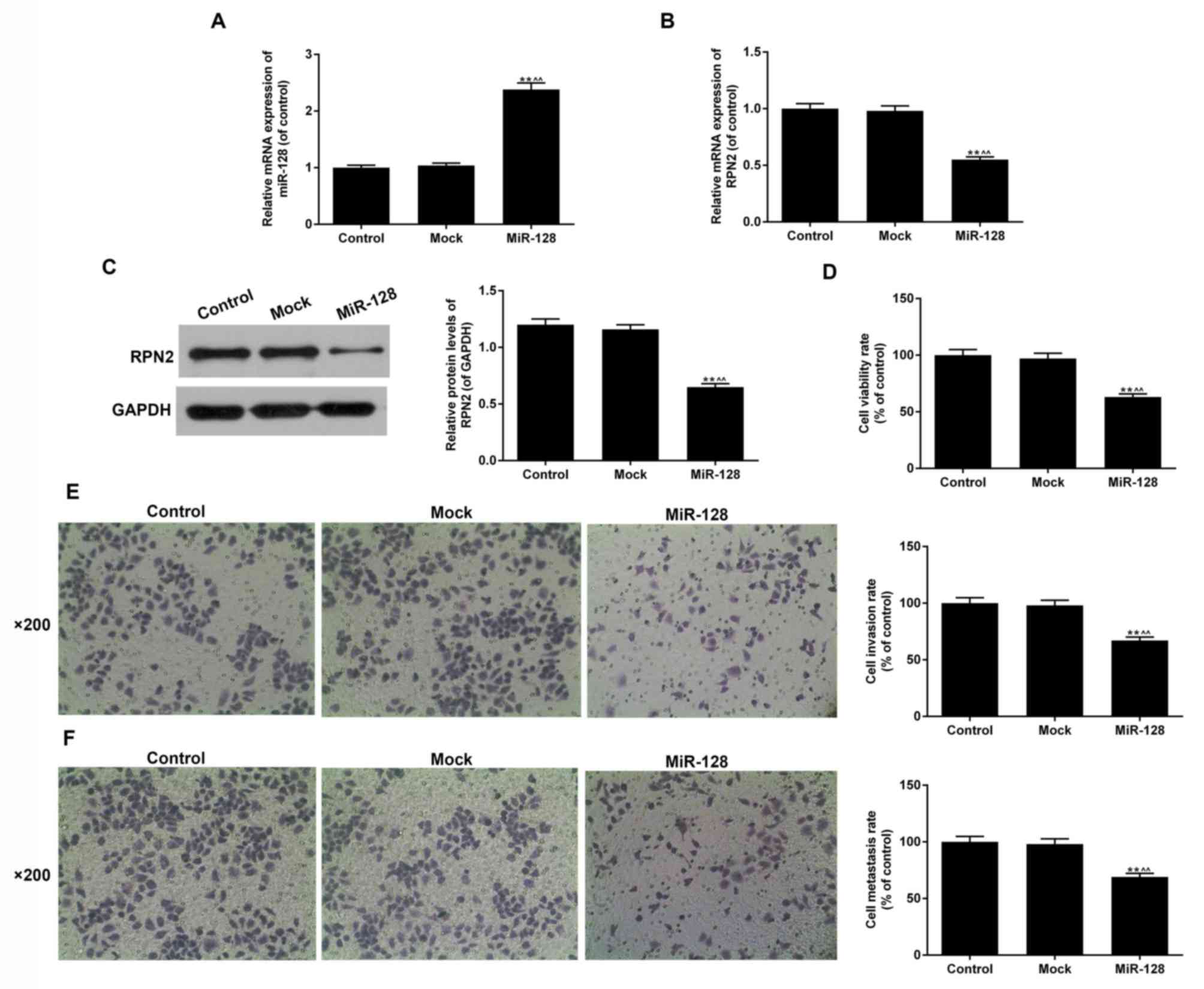
The miR-128 levels in the miR-128 group, determined
by RT-qPCR, were increased 2-fold in comparison with the control
and mock groups (P<0.05; Fig. 3A).
The effect of miR-128 mimics on RPN2 levels was subsequently
evaluated using RT-qPCR and western blot analysis (Fig. 3B and C). Aberrant increased miR-128
levels downregulated the mRNA and protein expression levels of
RPN2. As indicated in Fig. 3C, the
overexpression of miR-128 resulted in a ~44% decrease in RPN2
protein expression compared with the mock and control groups
(P<0.05).
miR-128 inhibits cell proliferation,
migration and invasion of HT29CRC cells
Then, the effect of miR-128 on the biological
processes of HT29 cells including cell proliferation, migration and
invasion was detected (Fig. 3D-F). In
the miR-128 group, cell proliferation was significantly inhibited
by miR-128 overexpression compared with the mock and control groups
(37%; P<0.05; Fig. 3D).
Additionally, increased levels of miR-128 attenuated the cell
migration and invasion rates; the inhibition rates were ~31% and
33%, respectively (P<0.01; Fig. 3E and
F). The results suggested that miR-128 serves important roles
in CRC.
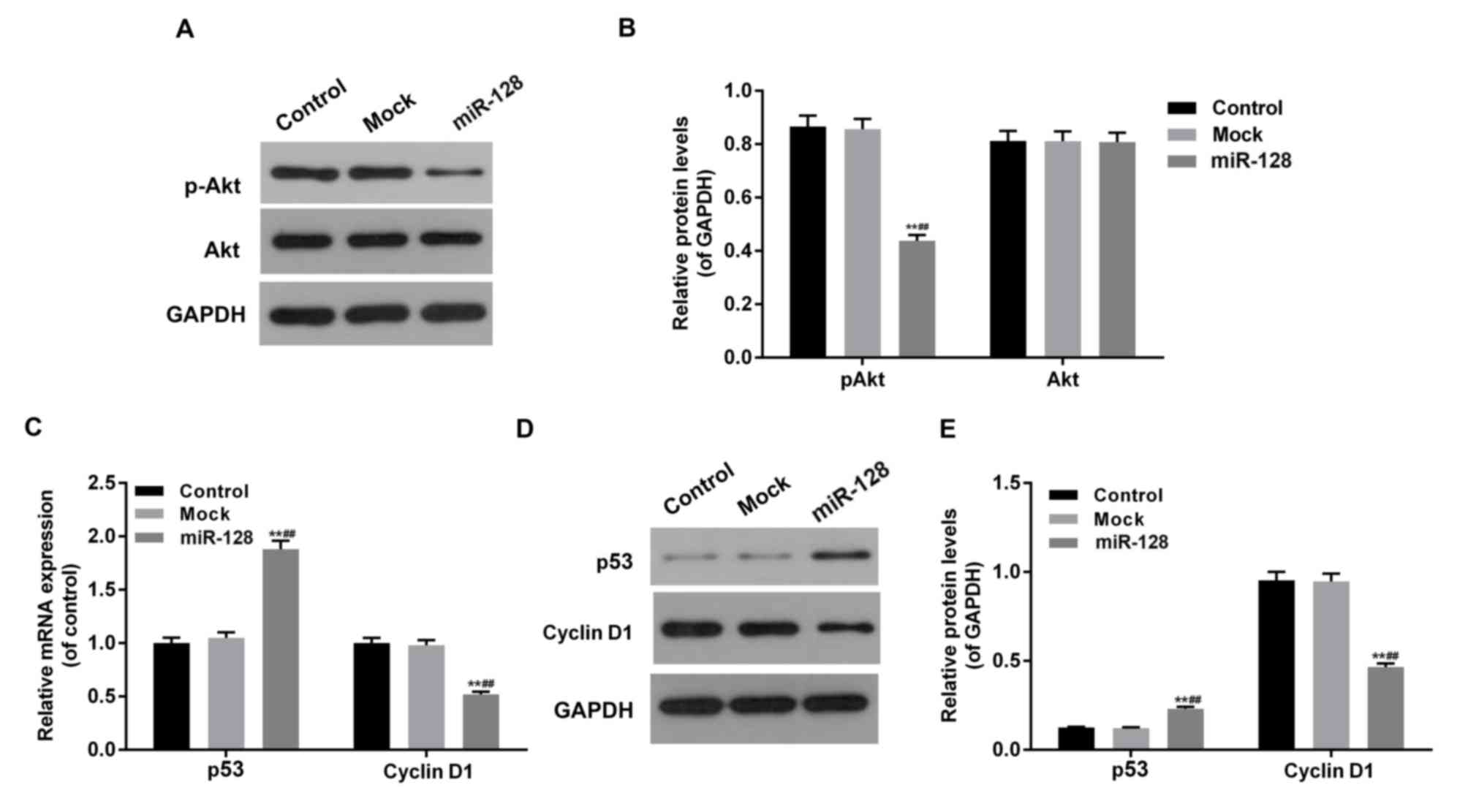
miR-128 inhibits the Akt-p53-cyclin
signal pathway in HT29 cells
To investigate the effect of miR-128 and RPN2 on the
Akt-p53-Cyclin pathway in CRC cells, the phosphorylation levels of
Akt and mRNA and protein levels of p53 and cyclin D1 were assessed
in miR-128, mock and control groups (Fig.
4). The overexpression of miR-128, combined with low levels of
RPN2, inhibited Akt phosphorylation, downregulated cyclin D1 and
upregulated p53 expression at mRNA and protein levels
(P<0.05).
miR-128 affects the expression of
invasion-associated factors including MMP-2, MMP-9, MTA1,
E-cadherin and TIMP2 in HT29 cells
The expression levels of MMP-2, MMP-9, MTA1,
E-cadherin and TIMP2 was affected by miR-128 overexpression and
RPN2 downregulation (Fig. 5). miR-128
mimic transfection inhibited the expression of MMP-2, MMP-9 and
MTA1, and promoted the expression of E-cadherin and TIMP2 at mRNA
and protein levels (P<0.05).
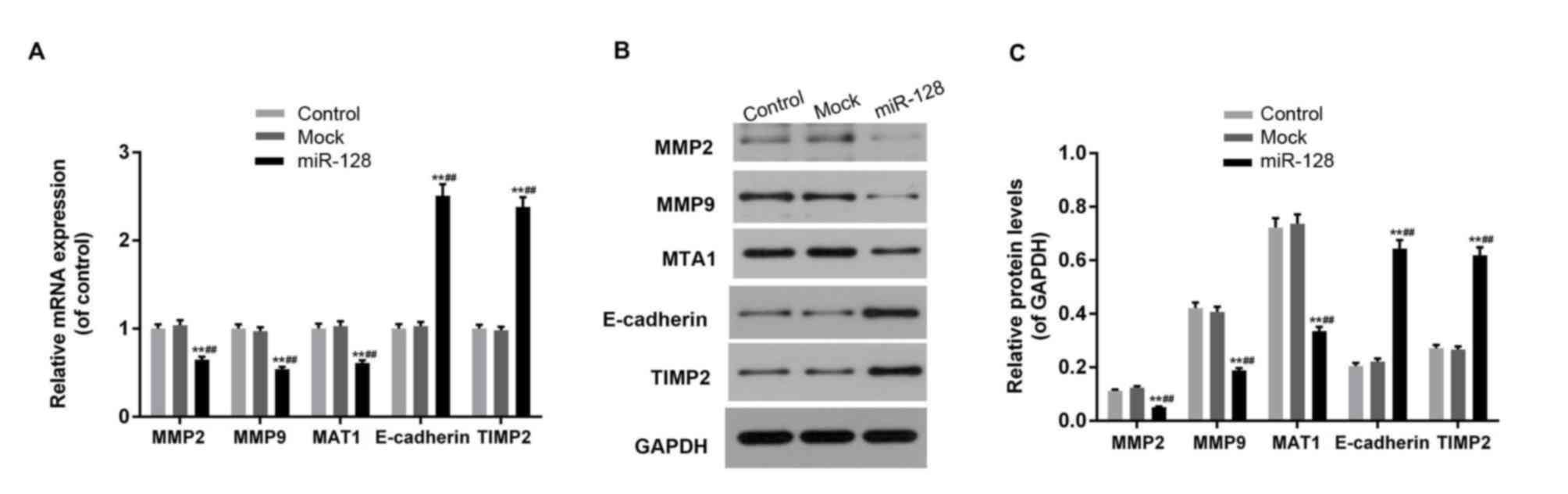 | Figure 5.miR-128 overexpression affects the
expression of cell invasion-associated factors. (A) Aberrant
increased miR-128 levels and inhibited Ribophorin-II expression
downregulated the mRNA expression of MMP-2, MMP-9 and MTA1, and
upregulated E-cadherin and TIMP2 mRNA levels. (B and C) MMP-2,
MMP-9 and MTA1 proteins were downregulated, while E-cadherin and
TIMP2 proteins were increased in the miR-128 mimic-transfected
cells. Data are expressed as the mean ± standard deviation from
three independent experiments. **P<0.01 vs. control,
^^P<0.01 vs. mock. miR, microRNA; MMP, matrix
metalloproteinase; MTA1, metastasis-associated protein 1;
E-cadherin, epithelial cadherin; TIMP1, Tissue inhibitor of
metalloproteinases 2. |
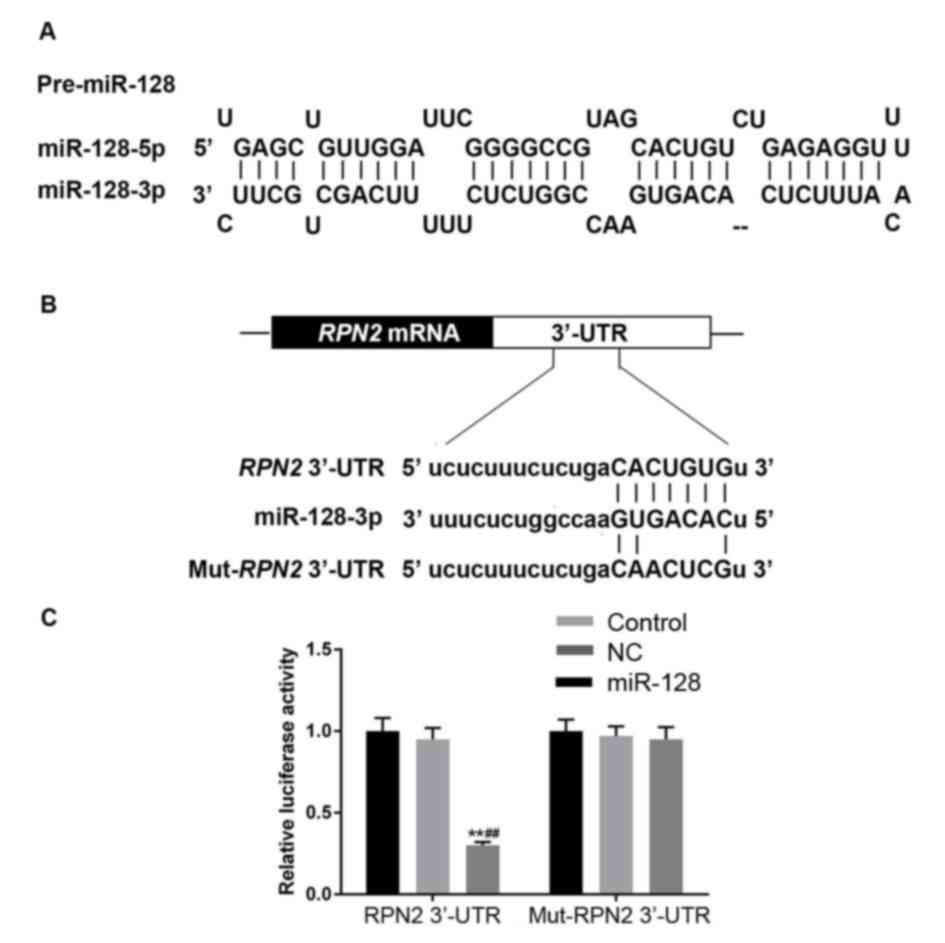
Bioinformatics target prediction and
luciferase reporter assays
The target prediction databases indicated that one
highly conserved miR-128 binding site was present in the RPN2
3′-UTR (Fig. 6A and B). Direct
interaction between miR-128 and RPN2 was verified by
dual-luciferase activity assay. miR-128 mimic transfection
significantly inhibited the relative luciferase activity of RPN2
3′-UTR in 293 cells (P<0.05), while there was no significant
difference among the control, NC and miR-128 + Mut-RPN2 3′-UTR
groups (P>0.05; Fig. 6C).
Discussion
Colorectal cancer (CRC) is a malignancy with high
mortality rates worldwide, which may be due to high metastasis
rates (33,34). Previous studies have suggested that
the aberrant expression of miRNAs, including miR-128, was
identified to be associated with clinical CRC development and
metastasis by regulating target genes and mediating protein
expression in cancer-associated pathways (35–37).
Concomitantly, RPN2 has been suggested to be closely associated
with the tumorigenesis of CRC (38,39).
However, whether miR-128 exhibits a direct regulatory effect on
RPN2 in CRC cell proliferation and metastasis remains unknown.
In the present study, it was identified that the
aberrant expression of miR-128 was negatively associated with RPN2
in CRC tissues and cell lines including HT29, SW480, SW620, and
HCT116 cells. The increased RPN2 levels and corresponding decreased
miR-128 levels were significantly associated with
poorly-differentiated histology, advanced disease stages and lymph
node metastasis in patients with CRC. In addition, the survival
rate of patients with CRC was associated with RPN2 levels. These
data suggest that miR-128 and RPN2 serve critical roles in CRC
development and tumor metastasis.
In order to detect the molecular mechanisms of
miR-128 and RPN2 regulation during CRC development, the miR-128
overexpression model was constructed by miR-128 mimic transfection
in HT29 cells, in order to induce the highest RPN2 expression and
lowest miR-128 expression levels. The results of this assay
verified that miR-128 overexpression downregulated the mRNA and
protein expression levels of RPN2, and significantly inhibited CRC
cell proliferative, migratory and invasive abilities. It suggested
that the negative regulatory role of miR-128 on RPN2 may serve as a
suppressor of CRC development and metastasis.
As an essential subunit of the oligosaccharide
transferase complex, RPN2 induces cell signal recognition and
transduction and cooperates with numerous signal pathways,
including the phosphatidylinositol 3-kinase/Akt signaling pathway,
which is suggested to be closely associated with tumorigenesis
(40,41). The phosphorylation of Akt will
activate downstream target genes to regulate the cell cycle,
proliferation and migration (42).
This is achieved primarily through activation of cyclin D1 by
phosphorylated Akt to promote cell cycle G1/S transition (43). Cyclin D1, a downstream target of the
Akt-p53 signaling pathway, is a key in nuclear transcription factor
in cell cycle regulation. The activation of Akt and Cyclin D1 has
been demonstrated to be associated with CRC occurrence and
development (44). The present study
identified that aberrant high miR-128 expression, along with low
RPN2 expression, decreased the phosphorylation level of Akt,
downregulated the mRNA and protein levels of cyclin D1 and promoted
p53 expression levels markedly, which indicated an inhibition of
the Akt-p53-cyclin signal pathway by miR-128 mimics. It suggested
that the suppressive function of miR-128 mimics on the migratory
ability of HT29 cells may depend on the inhibition of
Akt-p53-cyclin signal pathway.
To additionally verify this hypothesis, the effect
of the ectopic expression of miR-128 on epithelial-mesenchymal
transition (EMT) and metastasis-associated genes including
E-cadherin, MMP-2, MMP-9, MTA1 andTIMP2 were evaluated. EMT is an
essential process for cell metastasis (45), in which cancer cells obtain migratory
capabilities to enter into circulatory system via the extracellular
matrix and basement membrane of blood vessels (46). E-cadherin is the marker of epithelial
cells. As a cell adhesion molecule, the protein complexes of
E-cadherin combine with actin cytoskeletons to weaken cell-cell
adhesion and induce migration and invasion of tumor cells (47). MMPs are structurally analogous,
zinc-dependent endopeptidases (48).
MMPs and their inhibitors (TIMPs) serve important roles in
extracellular matrix degradation (49,50), which
may induce tumor invasion and metastasis (51). MMP2 and MMP9 are able to degrade the
primary component of type IV collagen to induce basement membranes
degradation (52), which results in
cell migration and eventual tumor metastasis. MMPs serve important
roles in EMT-associated regulation of cell migration (53). The mRNA expression of MMP9 was
verified to be notably improved during murine colitis-associated
cancer progression following administering azoxymethane and dextran
sulfate sodium ingestion (54). MTA1
was demonstrated to be persistently highly expressed in lymph nodes
metastasis in non-small cell lung and breast cancer, CRC and
pancreatic cancer. It may inhibit the expression of tumor
suppressors and contribute to cell migration and invasion (55). In the present study, it was confirmed
that the expression levels of MMP-2, MMP-9, MTA1, E-cadherin and
TIMP2 were significantly affected by miR-128 overexpression not
only at mRNA levels but also at protein levels. Cells transfected
with miR-128 mimics expressed decreased levels of MMP-2, MMP-9 and
MTA1, but increased levels of E-cadherin and TIMP2, which inhibited
cell migration. It was also revealed that the regulation of
metastasis-associated proteins was necessary in the molecular
mechanisms underlying the miR-128-associated modification of HT29
cell metastasis.
To comprehensively investigate the association
between miR-128 and RPN2 in regulating CRC cell proliferation and
migration, additional experiments were performed. According to the
bioinformatics analysis, it was identified that the RPN2 3′-UTR
contained the binding site of miR-128. Luciferase reporter gene
analysis confirmed that RPN2 was a direct target of miR-128: The
RPN2 3′-UTR or Mut-RPN2 3′-UTR recombinant plasmids were
co-transfected with miR-128 mimics into 293 cells. The results
demonstrated that the miR-128 mimics significantly inhibited the
relative luciferase activity of RPN2 3′-UTR cells, but exhibited no
effect on the Mut-RPN2 3′-UTR plasmid. This confirmed that miR-128
directly targeted the 3′-UTR of RPN2. It also suggested that
miR-128 targeted RPN2 during the regulatory process of CRC cell
proliferation and migration.
In conclusion, the results of the present study
suggested that miR-128 was a specific negative regulator of RPN2,
which attenuated colorectal cancer cell proliferation and
migration. This regulation may be associated with the Akt-p53
signal pathway and metastasis-associated factors. These data may
provide novel evidence for potential target biomarkers in the
treatment of colorectal cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by Guangdong
Provincial Science and Technology Plan (grant no.
2017A020215036).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TZ constructed miR-128 overexpressed cells and
detected the invasion and migration abilities. LW studied the
expression levels of RPN2 in CRC cells. QW investigated the effect
on P53/Cyclin D1. ZJ, NM and YL collected the tissue samples and
conducted associated RT-qPCR and Western blot analysis. WC, ZH, WG
investigated the levels of metastasis associated factors. SC
conceived the study and drafted the manuscript.
Ethics approval and consent to
participate
All tissue samples from patients were collected and
protocols were performed according to the procedures approved by
the Institutional Review Board of the Independent Ethics Committee
of the Sixth Affiliated Hospital of Sun Yat-sen University
(approval no. BZ20153587). All patients provided informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shroff J, Thosani N, Batra S, Singh H and
Guha S: Reduced incidence and mortality from colorectal cancer with
flexible-sigmoidoscopy screening: A meta-analysis. World J
Gastroenterol. 20:18466–18476. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Zeng H and Zhang S: The
updated incidences and mortalities of major cancers in China, 2011.
Chin J Cancer. 34:502–507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Labianca R and Merelli B: Screening and
diagnosis for colorectal cancer: Present and future. Tumori.
96:889–901. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Levi F, Lucchini F, Negri E, Zatonski W,
Boyle P and La Vecchia C: Trends in cancer mortality in the
European Union and accession countries, 1980–2000. Ann Oncol.
15:1425–1431. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chouhan V, Mansoor E, Parasa S and Cooper
GS: Rates of prevalent colorectal cancer occurrence in persons 75
years of age and older: A population-based national study. Dig Dis
Sci. 63:1929–1936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kumar A, Cherukumilli M, Mahmoudpour SH,
Brand K and Bandapalli OR: ShRNA-mediated knock-down of CXCL8
inhibits tumor growth in colorectal liver metastasis. Biochem
Biophys Res Commun. 500:731–737. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peng Y, Zhao BC, Kang Q, Liu J, Chen C, Li
BS, Xie YP and Wu Q: Colorectal cancer preventive effect of
combined administration of phenolic acids and supercritical
extracts from Angelica sinensis. Zhongguo Zhong Yao Za Zhi.
43:1235–1240. 2018.(In Chinese). PubMed/NCBI
|
|
9
|
Costa FF, Bischof JM, Vanin EF, Lulla RR,
Wang M, Sredni ST, Rajaram V, Bonaldo Mde F, Wang D, Goldman S, et
al: Identification of microRNAs as potential prognostic markers in
ependymoma. PLoS One. 6:e251142011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gattolliat CH, Thomas L, Ciafrè SA,
Meurice G, Le Teuff G, Job B, Richon C, Combaret V, Dessen P,
Valteau-Couanet D, et al: Expression of miR-487b and miR-410
encoded by 14q32.31 locus is a prognostic marker in neuroblastoma.
Br J Cancer. 105:1352–1361. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Haller F, von Heydebreck A, Zhang JD,
Gunawan B, Langer C, Ramadori G, Wiemann S and Sahin O:
Localization- and mutation-dependent microRNA (miRNA) expression
signatures in gastrointestinal stromal tumours (GISTs), with a
cluster of co-expressed miRNAs located at 14q32.31. J Pathol.
220:71–86. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lavon I, Zrihan D, Granit A, Einstein O,
Fainstein N, Cohen MA, Cohen MA, Zelikovitch B, Shoshan Y, Spektor
S, et al: Gliomas display a microRNA expression profile reminiscent
of neural precursor cells. Neuro Oncol. 12:422–433. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: microRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gong J, Zhang JP, Li B, Zeng C, You K,
Chen MX, Yuan Y and Zhuang SM: MicroRNA-125b promotes apoptosis by
regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene.
32:3071–3079. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Corté H, Manceau G, Blons H and
Laurent-Puig P: MicroRNA and colorectal cancer. Dig Liver Dis.
44:195–200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu N, Zhao X, Liu M, Liu H, Yao W, Zhang
Y, Cao S and Lin X: Role of microRNA-26b in glioma development and
its mediated regulation on EphA2. PLoS One. 6:e162642011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ai F, Zhang X, Li X, Qin Z, Ye Q, Tian L,
Tang A, Li N, Li G, Ma J and Shen S: Up-regulation of matrix
metalloproteinases in a mouse model of chemically induced
colitis-associated cancer: The role of microRNAs. Oncotarget.
6:5412–5425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takahashi Y, Iwaya T, Sawada G, Kurashige
J, Matsumura T, Uchi R, Ueo H, Takano Y, Eguchi H and Sudo T:
Up-regulation of NEK2 by MicroRNA-128 methylation is associated
with poor prognosis in colorectal cancer. Ann Surg Oncol.
21:205–212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao Y, Pang H and Chen C: MicroRNA-128
inhibits invasion of colon cancer cells. Zhejiang Med J. 2016.(In
Chinese).
|
|
20
|
Ciafrè SA, Galardi S, Mangiola A, Ferracin
M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM and Farace MG:
Extensive modulation of a set of microRNAs in primary glioblastoma.
Biochem Biophys Res Commun. 334:1351–1358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tutar Y: miRNA and cancer; computational
and experimental approaches. Curr Pharm Biotechnol. 15:4292014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Godlewski J, Nowicki MO, Bronisz A,
Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca
EA and Lawler S: Targeting of the Bmi-1 oncogene/stem cell renewal
factor by microRNA-128 inhibits glioma proliferation and
self-renewal. Cancer Res. 68:9125–9130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koulich E, Li X and DeMartino GN: Relative
structural and functional roles of multiple deubiquitylating
proteins associated with mammalian 26S proteasome. Mol Biol Cell.
19:1072–1082. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fujita Y, Yagishita S, Takeshita F,
Yamamoto Y, Kuwano K and Ochiya T: Prognostic and therapeutic
impact of RPN2-mediated tumor malignancy in non-small-cell lung
cancer. Oncotarget. 6:3335–3345. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Honma K, Iwao-Koizumi K, Takeshita F,
Yamamoto Y, Yoshida T, Nishio K, Nagahara S, Kato K and Ochiya T:
RPN2 gene confers docetaxel resistance in breast cancer. Nat Med.
14:939–948. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fujita Y, Takeshita F, Mizutani T, Ohgi T,
Kuwano K and Ochiya T: A novel platform to enable inhaled naked
RNAi medicine for lung cancer. Sci Rep. 3:33252013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takahashi RU, Takeshita F, Honma K, Ono M,
Kato K and Ochiya T: Ribophorin II regulates breast tumor
initiation and metastasis through the functional suppression of
GSK3β. Sci Rep. 3:24742013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao Y, Deng Y, Peng J, Sui Q, Lin J, Qiu
M and Pan Z: Does the preoperative prognostic nutritional index
predict survival in patients with liver metastases from colorectal
cancer who underwent curative resection? J Cancer. 9:2167–2174.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hendricks A, Eggebrecht GL, Bernsmeier A,
Geisen R, Dall K, Trauzold A, Becker T, Kalthoff H, Schafmayer C,
Röder C and Hinz S: Identifying patients with an unfavorable
prognosis in early stages of colorectal carcinoma. Oncotarget.
9:27423–27434. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han HQ, Liu T, Zhao LZ, Qi F and Wang PZ:
Clinical value of the sixth edition TNM stages analysing prognosis
of colorectal cancer. Zhonghua Yi Xue Za Zhi. 86:819–821. 2006.(In
Chinese). PubMed/NCBI
|
|
31
|
Chen JX, Tang XD, Xiang DB, Dong XL, Peng
FY and Sun GY: TNM stages and prognostic features of colorectal
mucinous adenocarcinomas: A meta analysis. Asian Pac J Cancer Prev.
13:3427–3430. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
D'Haene N, Fontanges Q, De Nève N,
Blanchard O, Melendez B, Delos M, Dehou MF, Maris C, Nagy N,
Rousseau E, et al: Clinical application of targeted next-generation
sequencing for colorectal cancer patients: A multicentric Belgian
experience. Oncotarget. 9:20761–20768. 2018.PubMed/NCBI
|
|
34
|
Sun J, Hu J, Wang G, Yang Z, Zhao C, Zhang
X and Wang J: LncRNA TUG1 promoted KIAA1199 expression via miR-600
to accelerate cell metastasis and epithelial-mesenchymal transition
in colorectal cancer. J Exp Clin Cancer Res. 37:1062018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Feng H, Xu M, Zhang Y, Han B, Wang J and
Sun P: Identification of differentially expressed MicroRNAs
involved in the pathogenesis of colorectal cancer. Clin Lab.
64:797–804. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Xu J, Yan X, Jin K, Li W and Zhang
R: MicroRNA-485 plays tumour-suppressive roles in colorectal cancer
by directly targeting GAB2. Oncol Rep. 40:554–564. 2018.PubMed/NCBI
|
|
37
|
Sun W, Wang X, Li J, You C, Lu P, Feng H,
Kong Y, Zhang H, Liu Y, Jiao R, et al: MicroRNA-181a promotes
angiogenesis in colorectal cancer by targeting SRCIN1 to promote
the SRC/VEGF signaling pathway. Cell Death Dis. 9:4382018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang J, Yan B, Späth SS, Qun H, Cornelius
S, Guan D, Shao J, Hagiwara K, Van Waes C, Chen Z, et al:
Integrated transcriptional profiling and genomic analyses reveal
RPN2 and HMGB1 as promising biomarkers in colorectal cancer. Cell
Biosci. 5:532015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li H, Al-Japairai K, Tao Y and Xiang Z:
RPN2 promotes colorectal cancer cell proliferation through
modulating the glycosylation status of EGFR. Oncotarget.
8:72633–72651. 2017.PubMed/NCBI
|
|
40
|
Liu GL, Yang HJ, Liu B and Liu T: Effects
of microrna-19b on the proliferation, apoptosis, and migration of
Wilms' Tumor cells via the PTEN/PI3K/AKT signaling pathway. J Cell
Biochem. 118:3424–3434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lu X, Lv S, Mi Y, Wang L and Wang G:
Neuroprotective effect of miR-665 against sevoflurane
anesthesia-induced cognitive dysfunction in rats through PI3K/Akt
signaling pathway by targeting insulin-like growth factor 2. Am J
Transl Res. 9:1344–1356. 2017.PubMed/NCBI
|
|
42
|
Manning BD and Cantley LC: AKT/PKB
signaling: Navigating downstream. Cell. 129:1261–1274. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liang J and Slingerland JM: Multiple roles
of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell
Cycle. 2:339–345. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu Y, Bi T, Wang Z, Wu G, Qian L, Gao Q
and Shen G: Oxymatrine synergistically enhances antitumor activity
of oxaliplatin in colon carcinoma through PI3K/AKT/mTOR pathway.
Apoptosis. 21:1398–1407. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sabe H: Cancer early dissemination:
Cancerous epithelial-mesenchymal transdifferentiation and
transforming growth factor beta signalling. J Biochem. 149:633–639.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Acloque H, Adams MS, Fishwick K,
Bronner-Fraser M and Nieto MA: Epithelial-mesenchymal transitions:
The importance of changing cell state in development and disease. J
Clin Invest. 119:1438–1449. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gao H, Lan X, Li S and Xue Y:
Relationships of MMP-9, E-cadherin, and VEGF expression with
clinicopathological features and response to chemosensitivity in
gastric cancer. Tumour Biol. 39:10104283176983682017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Visse R and Nagase H: Matrix
metalloproteinases and tissue inhibitors of metalloproteinases:
Structure, function, and biochemistry. Circ Res. 92:827–839. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Knapinska AM, Estrada CA and Fields GB:
The roles of matrix metalloproteinases in pancreatic cancer. Prog
Mol Biol Transl Sci. 148:339–354. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Parrish AR: Matrix metalloproteinases in
kidney disease: Role in pathogenesis and potential as a therapeutic
target. Prog Mol Biol Transl Sci. 148:31–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Curran S and Murray GI: Matrix
metalloproteinases: Molecular aspects of their roles in tumour
invasion and metastasis. Eur J Cancer. 36:1621–1630. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hendrix AY and Kheradmand F: The role of
matrix metalloproteinases in development, repair, and destruction
of the lungs. Prog Mol Biol Transl Sci. 148:1–29. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tang G, Du R, Tang Z and Kuang Y:
MiRNALet-7a mediates prostate cancer PC-3 cell invasion, migration
by inducing epithelial-mesenchymal transition through CCR7/MAPK
pathway. J Cell Biochem. 119:3725–3731. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shang K, Bai YP, Wang C, Wang Z, Gu HY, Du
X, Zhou XY, Zheng CL, Chi YY, Mukaida N and Li YY: Crucial
involvement of tumor-associated neutrophils in the regulation of
chronic colitis-associated carcinogenesis in mice. PLoS One.
7:e518482012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cagatay Tuncay S, Cimen I, Savas B and
Banerjee S: MTA-1 expression is associated with metastasis and
epithelial to mesenchymal transition in colorectal cancer cells.
Tumor Biol. 34:1189–1204. 2013. View Article : Google Scholar
|