Introduction
Brain metastasis occurs in ~30% of patients with
breast cancer (1). Owing to the lack
of effective therapeutic treatments, breast cancer brain metastasis
can cause significant morbidity, resulting in a poor prognosis. A
number of characteristics are associated with the development of
primary breast cancer into brain metastases, including being
diagnosed at a young age, hormone receptor-negative status, tumors
>20 mm in diameter, lymph node invasion, grade 3 and human
epidermal growth factor receptor 2 (HER2)+ status
(2–4).
In another study it has been reported that patients with
HER2+ breast cancer have a 2–4-fold increased frequency
of brain metastasis (25–37%) compared with patients with
HER2− breast cancer (2).
The reason HER2 is associated with breast cancer brain metastasis
has yet to be determined; however, previous studies identified that
the improved extracranial control of disease, inability of
treatments to access or be active in the central nervous system
(CNS) and the natural predilection for tumor cells to deposit in
the CNS may contribute (5,6). Despite the availability of HER2-targeted
therapies, e.g., trastuzumab, it is expected that brain metastasis
from breast cancer will continue to be a significant clinical
problem in the long term.
To address this urgent situation, more biomarkers
for the early detection of breast cancer-associated brain
metastasis are required. Owing to the development of microarray
assays, there has been a significant increase in the data
available, broadening and deepening the research concerning
biomarkers for breast cancer-associated brain metastasis. Weighted
gene co-expression network analysis (WGCNA) is a powerful
instrument to identify hub genes in the data from microarray
assays. The concept of WGCNA is straightforward: Nodes represent
genes and nodes are connected when the corresponding genes are
significantly co-expressed across appropriately selected tissue
samples (7,8).
In the present study, WGCNA was performed to detect
the significant hub genes in a set of samples collected from
patients with HER2+ breast cancer, including patients
with brain metastasis. Additionally, the potential candidate genes
were verified in a set of patient samples using the reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Materials and methods
Ethics statement
Written informed consent for research was obtained
from all patients included in the present study. All experiments
were conducted according to the relevant guidelines and regulations
of the Institutional Ethical Review Committee of Hubei Cancer
Hospital (Wuhan, China). The present study was approved by the
Institutional Ethical Review Committee of Hubei Cancer
Hospital.
Data collection
Normalized gene expression data were downloaded from
the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo). The dataset GSE43837 was
used as a training set to construct expression networks and
identify hub genes in the present study. This dataset was based on
the microarray platform of Affymetrix Human X3P Array (U133_X3P),
which consisted of the expression data of 38 human samples,
containing 19 HER2+ human breast cancer-associated brain
metastases and 19 HER2+ primary human breast tumors. The
gene expression data were based on the RNA microdissection and
hybridization technology, as reported previously (9).
Study population
A total of 28 patients between 28 and 65 years old
with HER2+ breast cancer treated at the Hubei Cancer
Hospital from 1 January 2012 to 31 December 2016 were included in
the present study and utilized as the testing set for the
confirmation of hub genes determined from the training set. The
inclusion criteria for the patients participating in the present
study were the following: Primary diagnosis for HER2+ breast cancer
with tumours harvested during operation and all patients were
diagnosed with brain metastasis at different time points following
operation, with the brain being the first site of relapse.
Harvested tumors were subsequently stored in liquid nitrogen (25
kpa) at −195.79°C for long-term storage. In addition, the brain
metastasis samples were harvested following diagnosis and kept in
liquid nitrogen. The clinical characteristics of the patients
included in the present study are indicated in Table I.
 | Table I.General characteristics of the
patients included in the present study for testing the hub
genes. |
Table I.
General characteristics of the
patients included in the present study for testing the hub
genes.
| Characteristic | n |
|---|
| Patients in
total | 28 |
| Sex |
|
|
Male | 0 |
|
Female | 28 |
| HER2 |
|
|
Positive | 28 |
|
Negative | 0 |
| ER |
|
|
Positive | 16 |
|
Negative | 12 |
| PR |
|
|
Positive | 19 |
|
Negative | 9 |
| AJCC grade |
|
| I | 4 |
| II | 13 |
|
III | 11 |
| Lymph metastasis
prior to BM |
|
|
Positive | 17 |
|
Negative | 11 |
| Chemotherapy prior
to BM |
|
|
Presence | 28 |
|
Absence | 0 |
| Trastuzumab
treatment prior to BM |
|
|
Presence | 7 |
|
Absence | 21 |
| Endocrine treatment
prior to BM |
|
|
Presence | 13 |
|
Absence | 15 |
| Radiation therapy
prior to BM |
|
|
Presence | 26 |
|
Absence | 2 |
Data preprocessing
For each patient included in the training set, when
brain metastasis occurred, a value of 1 was assigned; otherwise, a
value of 0 was assigned for patients without brain metastasis.
Co-expression network
construction
The WGCNA R package software (R version 2.15.3 for
Windows; www.r-project.org) was used to perform
the analysis. Primarily, the genomic expression information of each
patient was associated with brain metastasis. Student's t-test was
used to determine the association between gene expression and brain
metastasis. The significantly altered genes were selected as
primary hub genes. In the following steps, according to the
official tutorial for the WGCNA R 2.15.3 package software
(www.r-project.org) (10), the automatic block-wise network
construction and module detection methods were used to construct
co-expression networks. Analysis of network topology was also
performed to select the feasible soft-thresholding power. Following
detection of the gene modules, the corresponding association with
brain metastasis was determined. The relative association of the
individual gene with brain metastasis was termed ‘gene
significance’. This was defined by the absolute value of the
association between the gene and brain metastasis. Module
membership was also defined as the association between the module
eigengene and the gene expression profile to quantify the
significance of the association between the gene profile and the
module. The module with the strongest association was further
analyzed to locate the genes associated with brain metastasis.
Visualization of the network
Following confirmation of the significantly
associated genes of the training set, the network of genes was
visualized by integrating the information from the process of
network construction into the software of Cytoscape (version 3.4;
www.cytoscape.org). Cytoscape is open source
software for the visualization of molecular interaction networks
and biological pathways.
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) analyses
Subsequently, the relatively significant genes of
the networks were added into GO and KEGG (https://david.ncifcrf.gov/) analyses to determine the
significant biological processes of the cancer cells. The online
analysis tool Database for Annotation, Visualization and Integrated
Discovery (david.ncifcrf.gov/tools.jsp) was used to transform the
network data in order to conduct GO and KEGG analyses.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA of the patients in the testing set was
extracted from the frozen tissues as previously described (9). For RT-qPCR, a PrimeScript RT Master Mix
(Takara Biotechnology Co., Ltd., Dalian, China) was used to
synthesize cDNA. SYBR® Real-time PCR Master mix (Toyobo
Co., Ltd., Shanghai, China) was used to perform qPCR analyses. The
results were normalized with the expression of GAPDH as a reference
and quantified using the 2−ΔΔCq method (11). The GAPDH sequence was as follows:
Forward, 5′-ACCATCTTCCAGGAGCGAGA-3′ and reverse,
5′-GCAAATGAGCCCCAGCCTTC-3′. The primers are detailed in Table II. RT-qPCR and data collection were
performed using a Bio-Rad CFX Manager 3.0 instrument (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). All RT-qPCRs were performed
in triplicate. The following thermocycling conditions were used for
qPCR: 94°C for 30 sec; 40 cycles of 60°C for 30 sec and 72°C for 30
sec. The data were processed with GraphPad Prism (version 7;
GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was
considered to indicate a statistically significant difference.
 | Table II.Primers for the reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Primers for the reverse
transcription-quantitative polymerase chain reaction.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| TP63 |
TACAGTACTGCCCTGACCCT |
CTCTGGGACATGGTGGATCG |
| CypA |
CCGTGTTCTTCGACATTGCC |
TTGTCTGCAAACAGCTCAAAGG |
| VWA8 |
GGCTACAACATTGGTCTGGT |
ATGCAGAACTGAGAGTGGGC |
| RPL17 |
TGGCCCAAAAAGAGTGCTGA |
GGCGCATCTTAGGTGCTTTG |
Results
Construction of the network
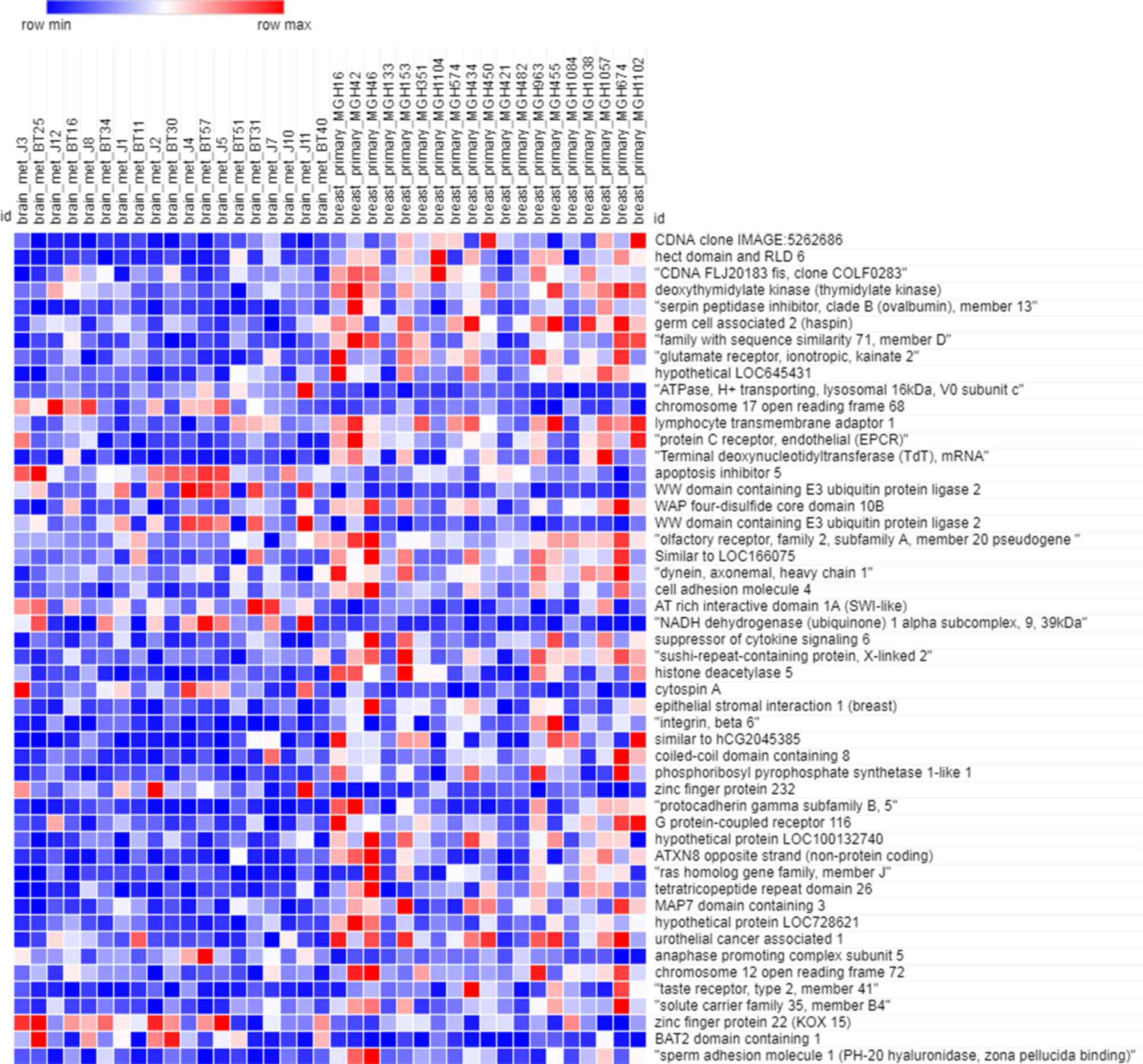
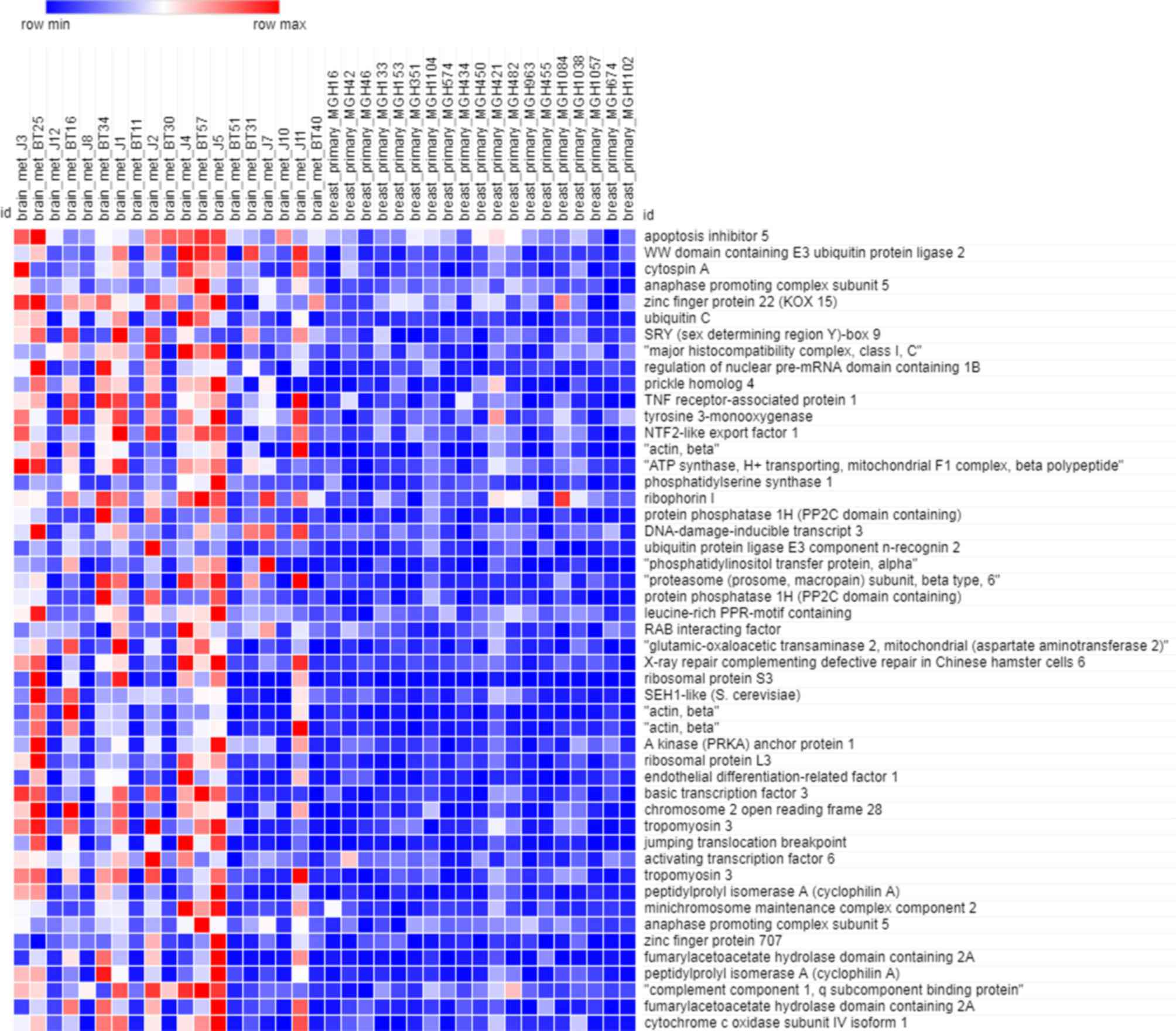
As depicted in Fig. 1,
the heatmap of the training dataset indicated that the expression
profiles in primary breast cancer and brain metastasis were
significantly different. There were 4,168 genes included in the
training dataset. Owing to the large amount of data, only the
expression of the 50 most significant (P<0.05) genes was
depicted in the heatmap.
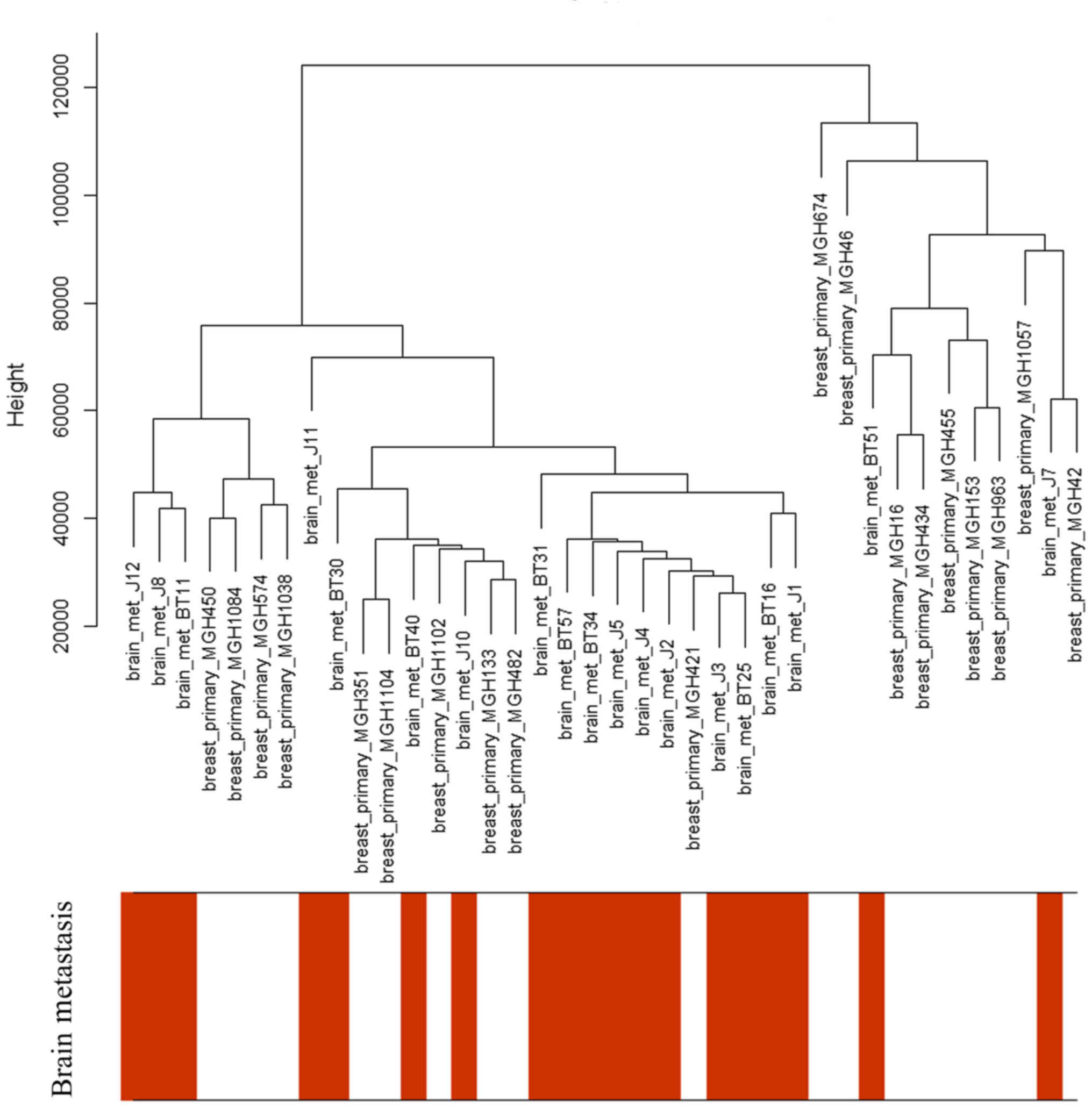
Only the clinical characteristics of brain
metastasis were associated with the sample dendrogram, as depicted
in Fig. 2. The samples were clustered
according to the expression microarray, and there was no notable
outlier; therefore, all the patients were included in this
analysis.
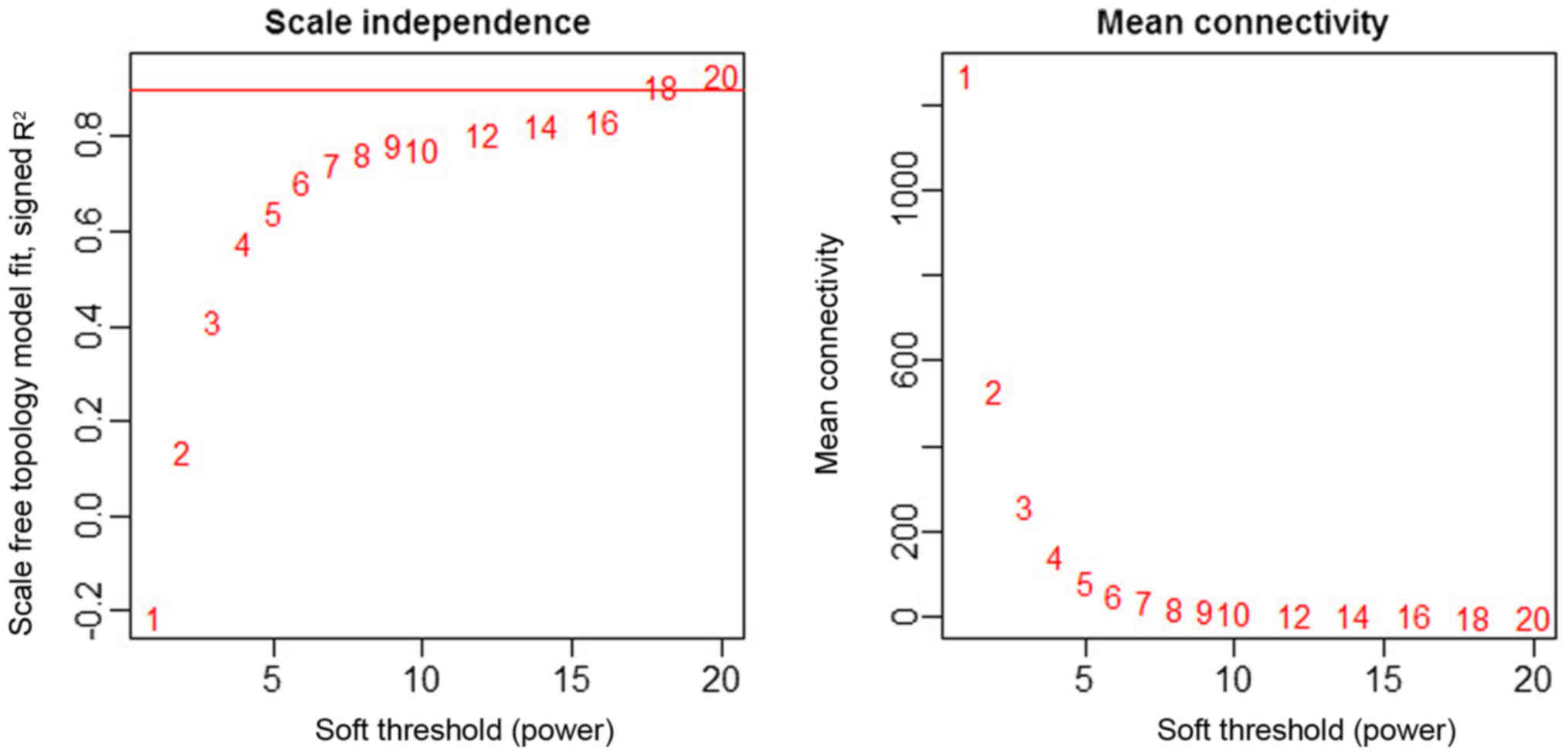
As depicted in Fig. 3,
the power 18 was selected as the lowest power for the scale-free
topology for which the fitting index reached 0.90. Different genes
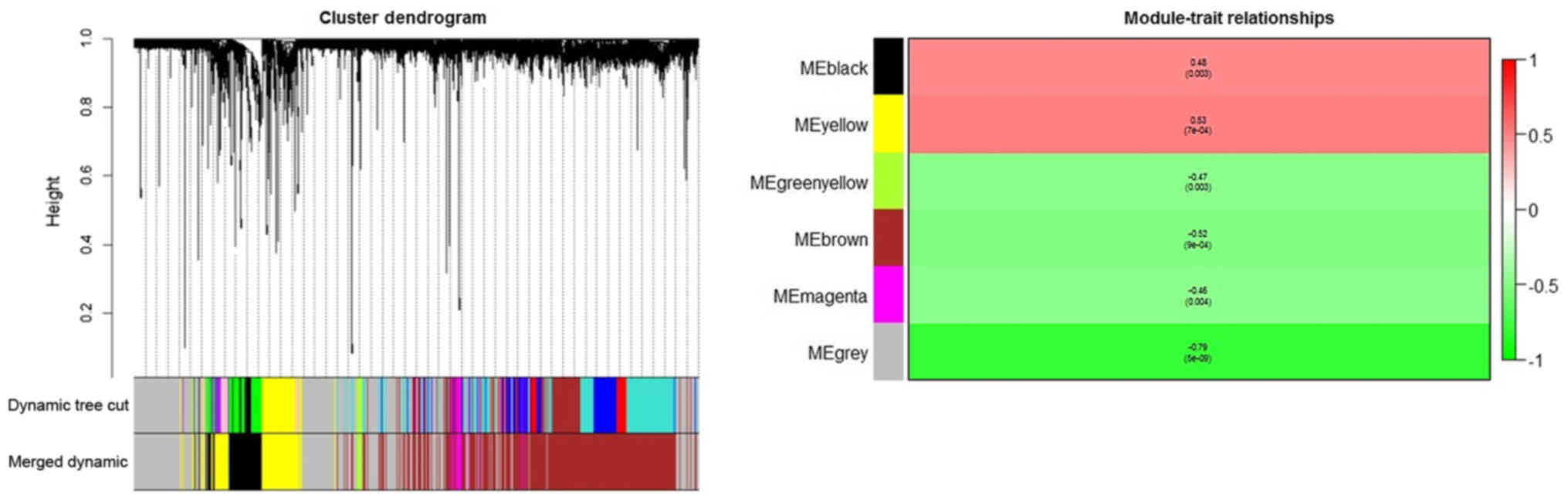
were subsequently grouped into modules according to the association
of expression (Fig. 4). Each color
represented a different module, comprised of gene sets of multiple
different genes.
In the clustering tree (dendrogram), each leaf,
represented by a short vertical line, corresponded to a gene.
Branches of the dendrogram group together to form densely
interconnected, highly co-expressed genes. Module identification
amounted to the identification of individual branches. Owing to the
marked association among the genes inside each module, the modules
with similar expression profiles were merged. To quantify the
co-expression similarity of entire modules, their eigengenes were
calculated and were clustered according to their association. The
modules were associated with brain metastasis according to the
overall gene significance inside each module, and the most
significant associations were determined. The further the value was
from 0, the more significant the association was between brain
metastasis and the module. Red was used to depict the positive
associations, and green depicts negative associations. It was
indicated that the yellow module had the greatest positive
association with breast cancer-associated brain metastasis, whereas
the grey module had the lowest negative association.
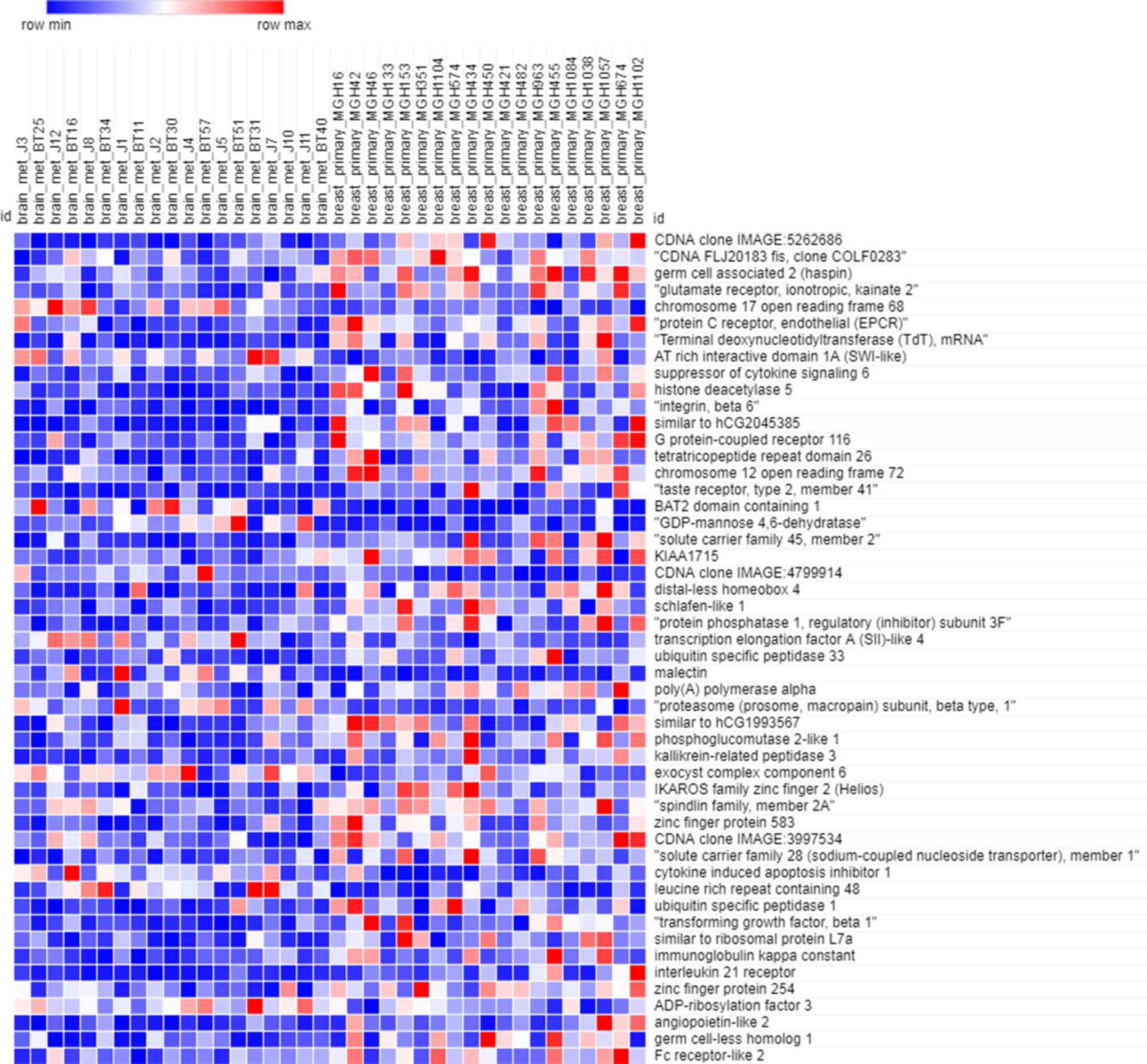
Figs. 5 and 6 depict the heatmaps of the expression
profiles of the yellow module and grey module, respectively. There
were 449 genes included in the yellow module and 1,550 genes
included in the grey module. Owing to the large amount of data,
only the expression of top 50 genes were depicted in the heatmap.
According to these heatmaps, the expression profiles in primary
breast cancer and brain metastasis were significantly different in
each module (P<0.05).
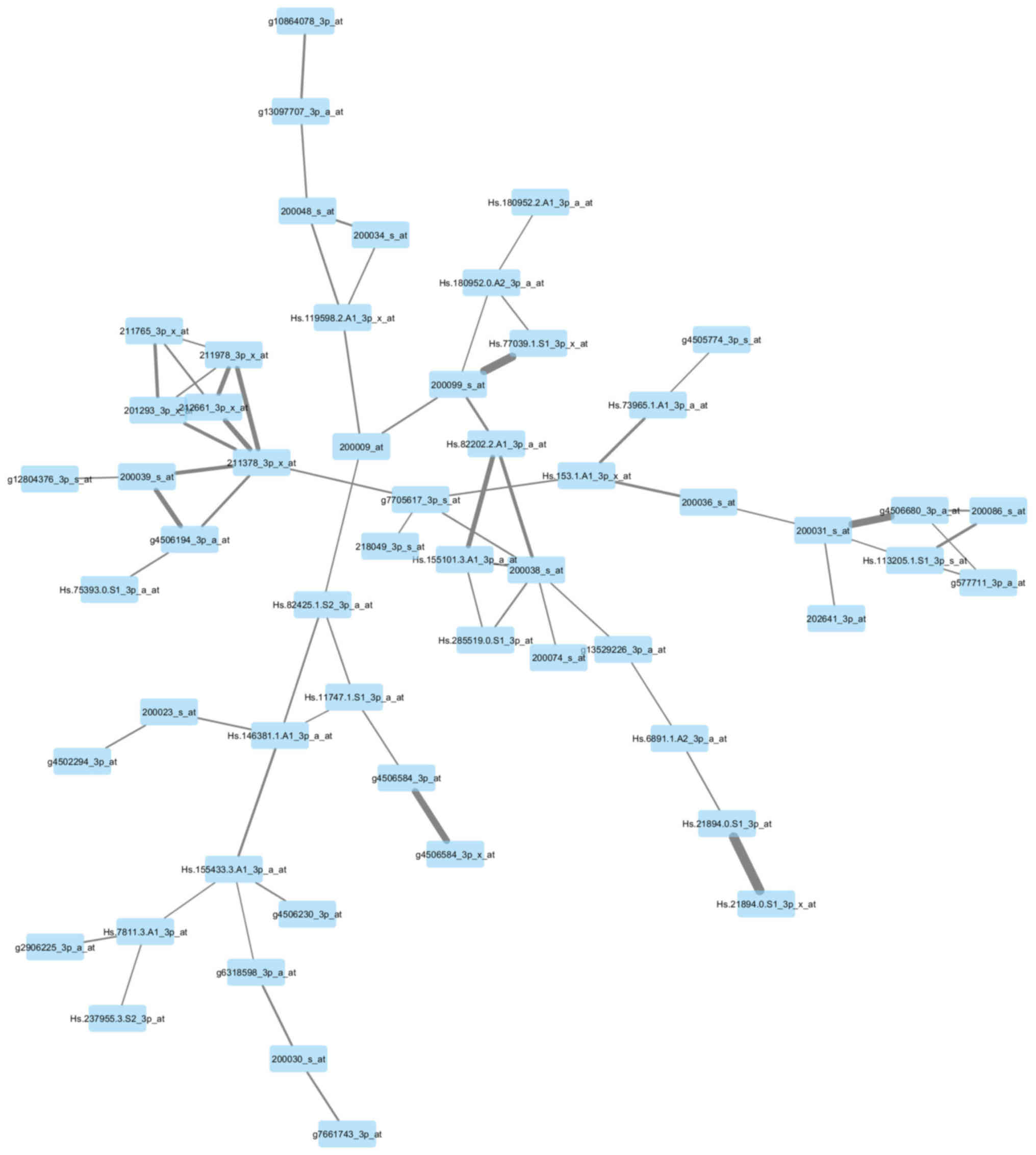
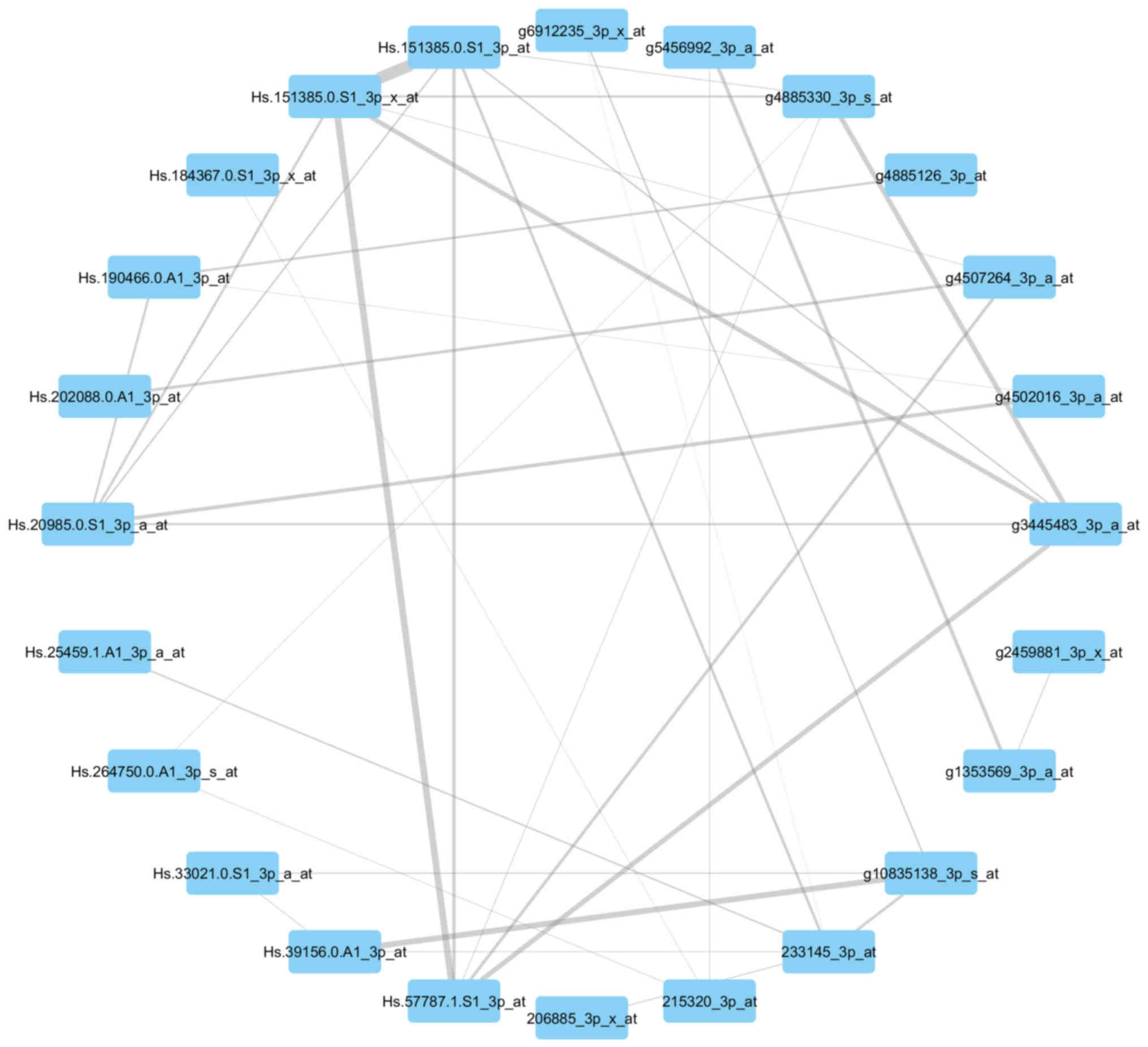
Visualization of the networks
In Figs. 7 and
8, the genes of the yellow module and
grey module that had the greatest association with each other were
correspondingly integrated into the network. According to the
networks, there were two hub genes situated in the center:
211378_39_x_at, representing cyclophilin A (CypA) (also known as
peptidylprolyl isomerase A); Hs.82202.2.A1_3p_a_at, representing
ribosomal protein L17 (RPL17); g3445483_3p_a_at, representing tumor
protein p63 (TP63); and Hs.57787.1.S1_3p_at, representing von
Willebrand factor A domain-containing 8 (VWA8).
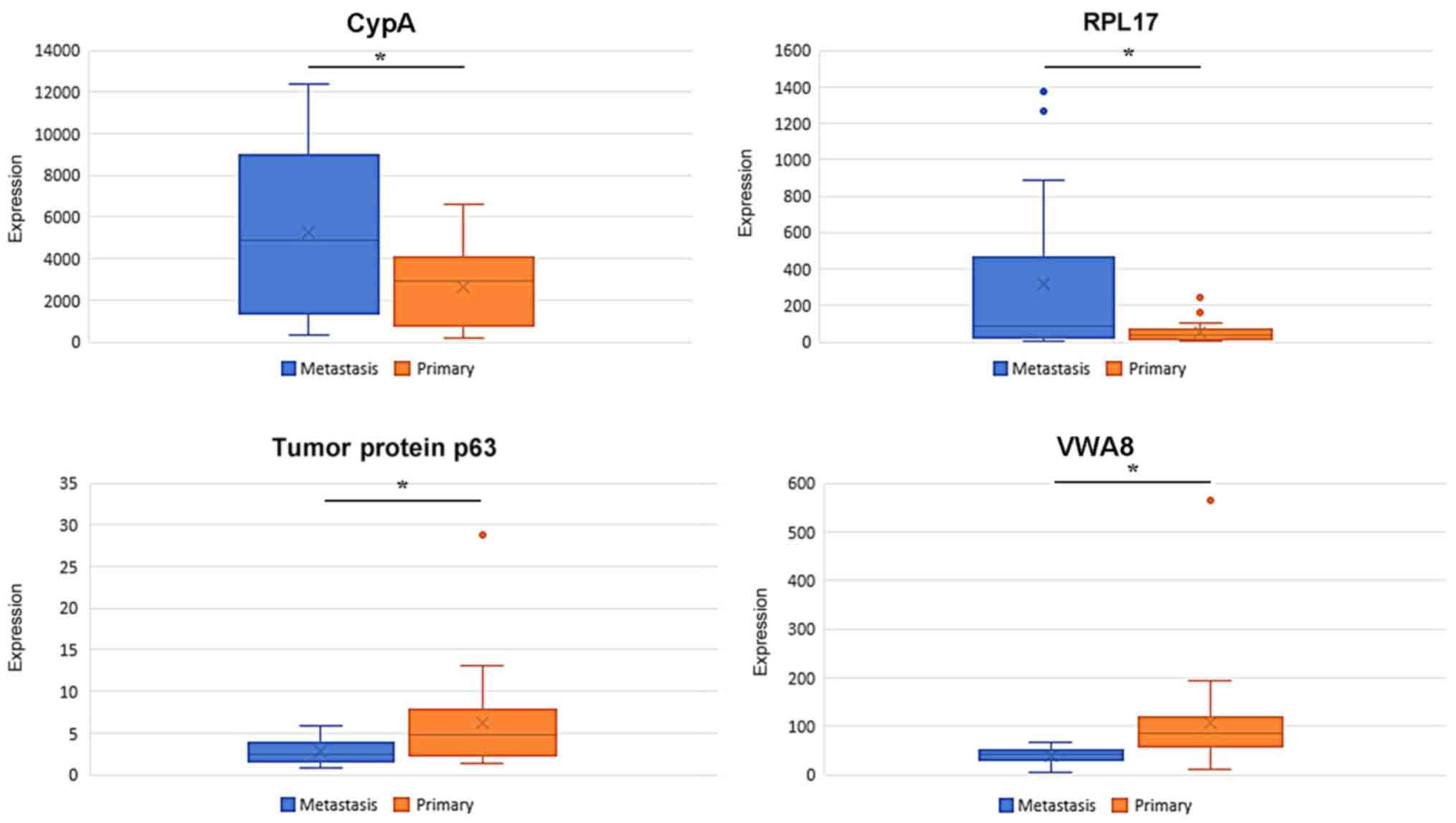
The expression data of CypA, RPL17, TP63 and VWA8
were extracted from the respective gene modules. As depicted in
Fig. 9, in the training set, CypA and
RPL17 were overexpressed in breast cancer-associated brain
metastasis, whereas the expression of TP63 and VWA8 was
significantly downregulated in breast cancer-associated brain
metastasis.
GO and KEGG pathway analysis
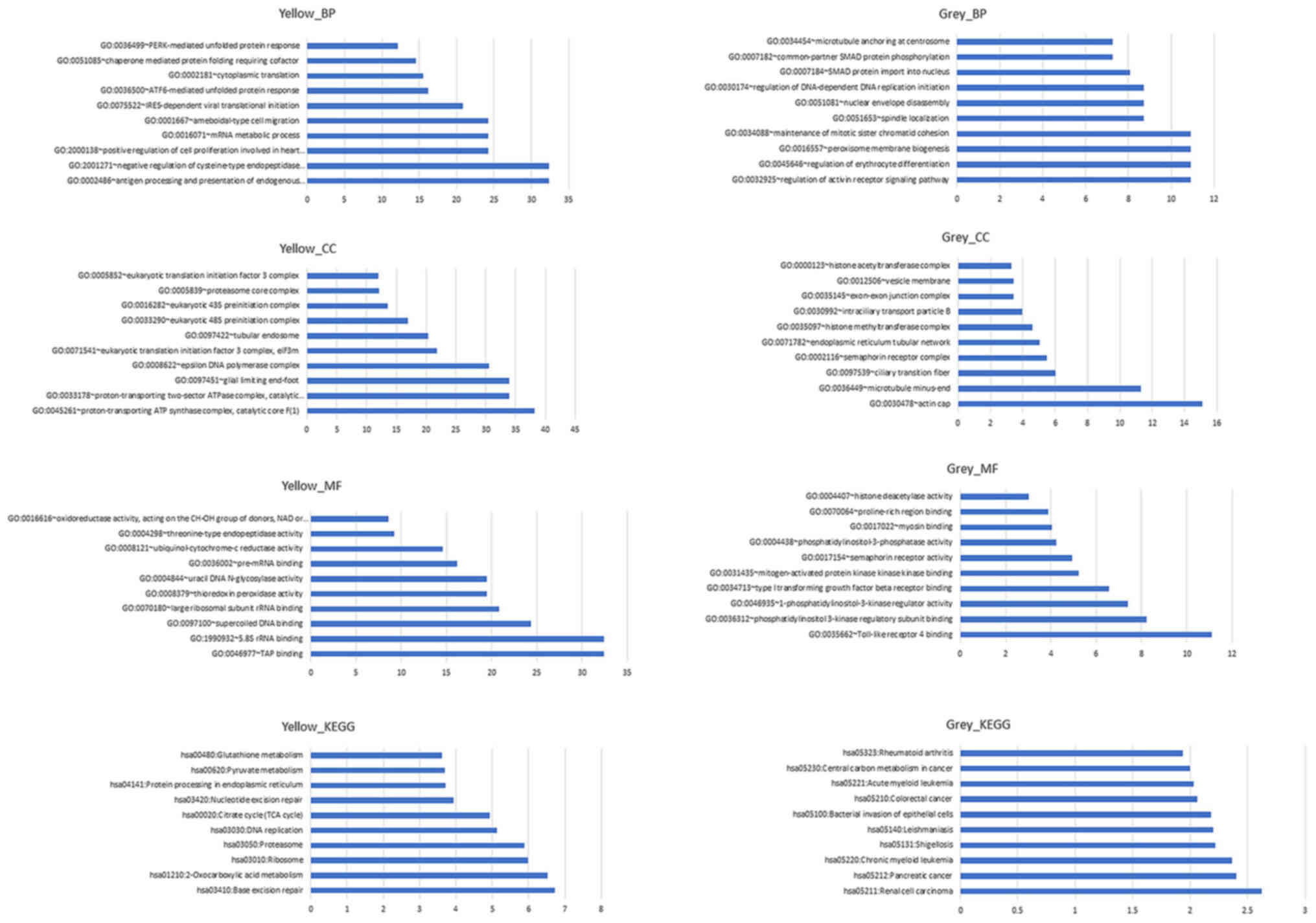
Fig. 10 depicts the
results of GO and KEGG pathway analysis of the yellow and grey
modules, respectively. According to the results, a number of
pathways changed significantly when breast cancer metastasized to
the brain. For the yellow module, the process of antigen processing
and presentation of endogenous peptide antigen was the most
significant biological process (BP), the component of the
proton-transporting ATP synthase complex was the most significant
cell component (CC), the function of transporter associated with
antigen processing binding was the most significant molecular
function (MF) and the pathway of base excision repair was the most
significant KEGG pathway. For the grey module, the process of
regulation of activin receptor signaling pathway was the most
significant BP, the component of actin cap was the most significant
CC, the function of Toll-like receptor 4 binding was the most
significant MF and the pathway of renal cell carcinoma was the most
significant KEGG pathway. However, none of the aforementioned genes
were included in the significantly influenced pathways.
Hub gene confirmation in patient
samples
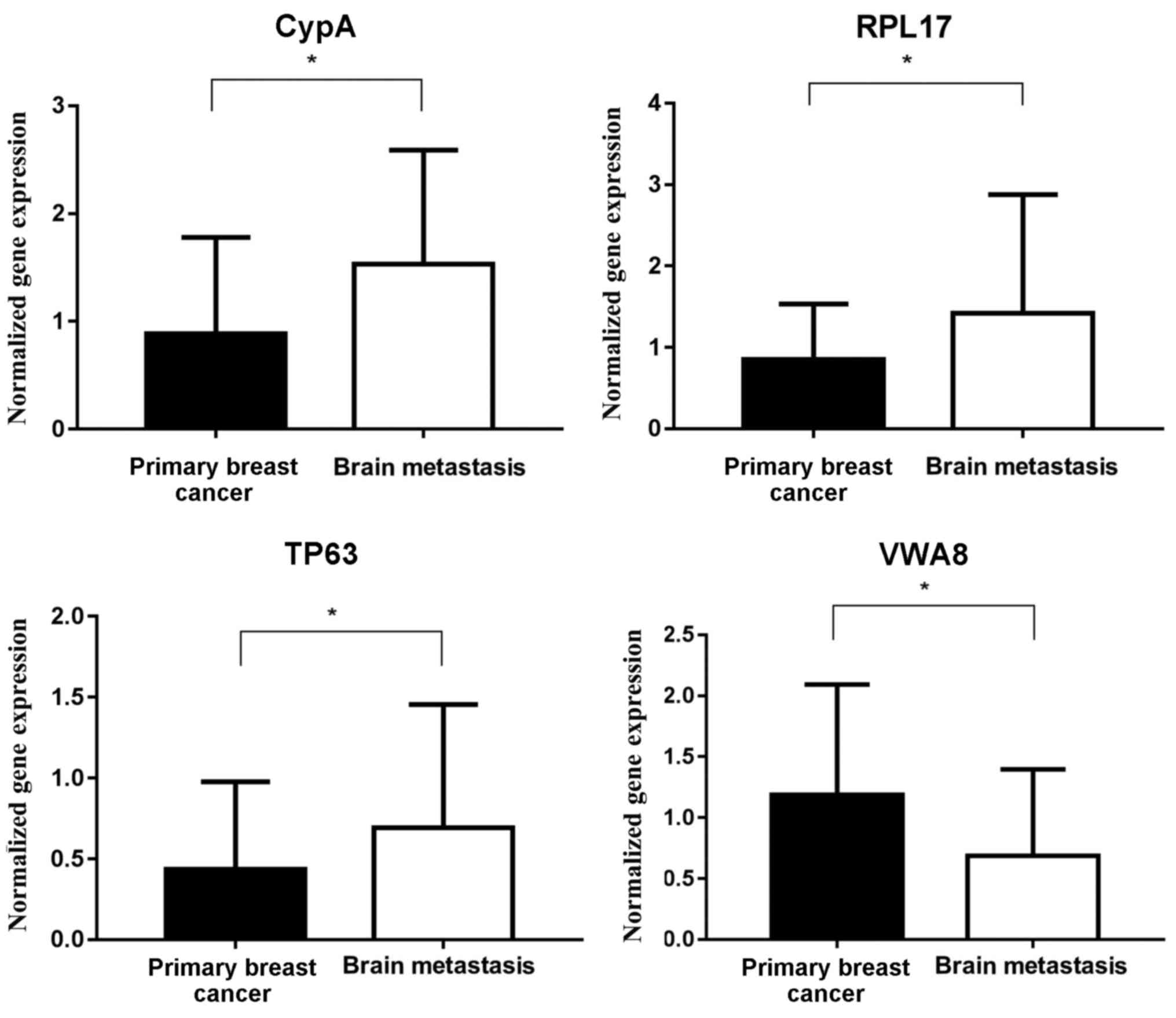
As depicted in Fig.
11, in the patient samples from the testing set, which included
all 28 patients, the expression of CypA and RPL17 in brain
metastases was significantly increased, compared with in primary
breast tumors, and the expression of TP63 and VWA8 in brain
metastases was significantly decreased, compared with in primary
breast tumors.
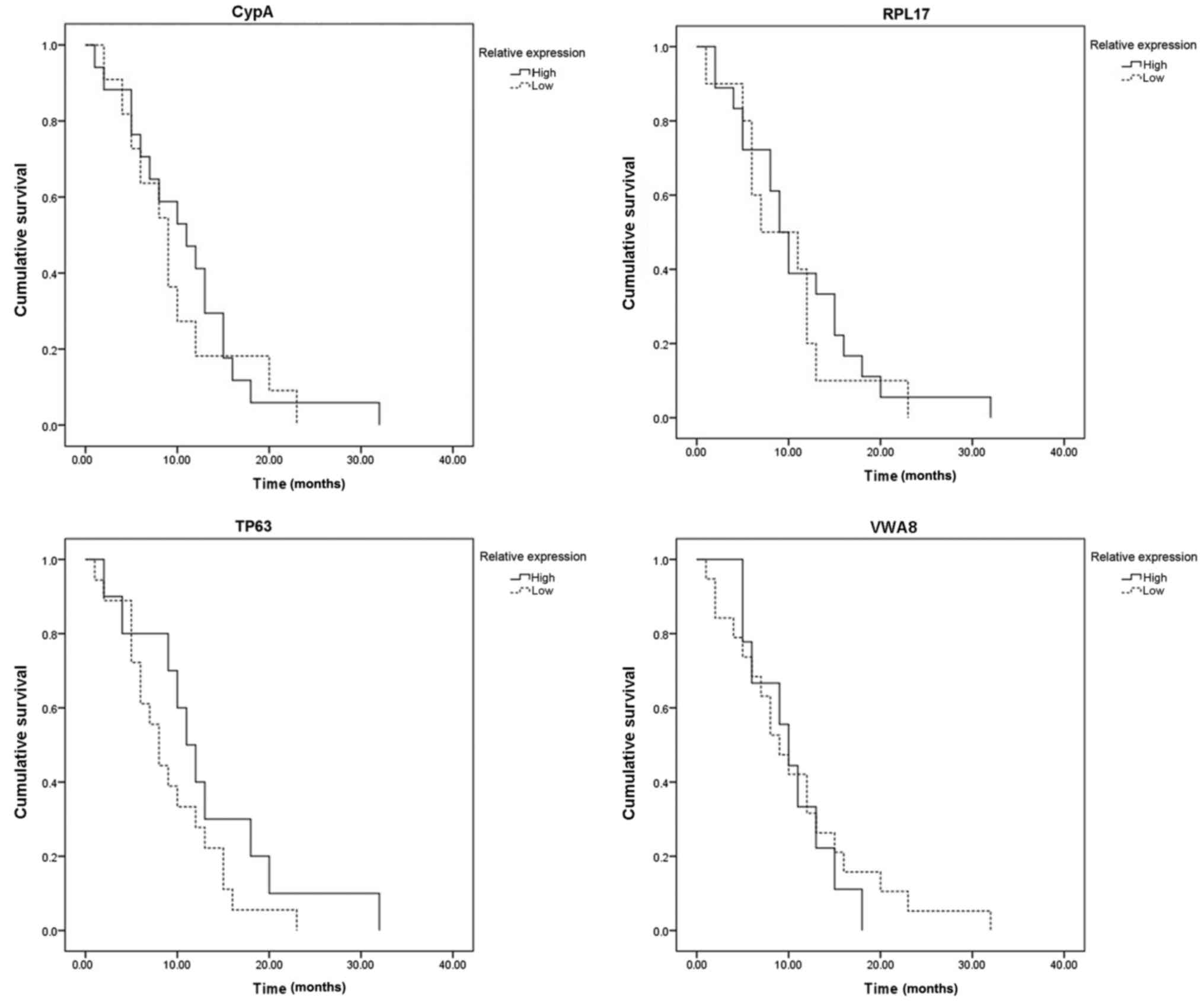
Fig. 12 indicated
that the expression of the four hub genes was associated with
progression to brain metastasis following surgery for primary
breast cancer; however, a positive marker associated with brain
metastasis-free survival time was not determined.
Discussion
Cyclophilins (Cyps) are proteins that bind to
cyclosporin and serve an immunosuppressive role following organ
transplantation (12). CypA is one of
the 16 family members constituting human Cyps (13). The expression of CypA was reported to
be upregulated in a number of malignant tumor types, including
small-cell lung cancer (14),
pancreatic cancer (15),
hepatocellular carcinoma (16) and
breast cancer (17). The presence of
CypA was considered to promote cellular proliferation and protect
those cancer cells against apoptosis or oxidative stress (13). In the present study, the results of
the network analysis demonstrated that the upregulated CypA
expression was significantly associated with breast
cancer-associated brain metastasis in patients with
HER2+ breast cancer. The identification testing in
patient samples also determined that CypA may promote breast cancer
cells to metastasize to brain tissue.
RPL17, a member of the L22P family of ribosomal
proteins, has been indicated to be located in the cytoplasm
(18). Due to the encoded protein
sharing an amino acid identity with RPL23 from Halobacterium
marismortui, the official name of this gene is RPL17 (19). RPL17 could promote multidrug
resistance in gastric cancer cells by suppressing drug-induced
apoptosis (20,21). In human osteosarcoma, the
overexpression of RPL17 may result in stabilization and activation
of p53, which serves a pivotal role in suppressing cancer cell
proliferation (22). In the network
analysis of the present study, the results indicated that the
expression of RPL17 was significantly downregulated in primary
breast tumors, compared with in metastatic brain tumors.
Furthermore, in the patient samples, the expression of RPL17 was
more suppressed in primary tumors, compared with brain metastatic
neoplasms. The results may indicate that in breast cancer, RPL17
could promote tumor cell invasion and migration.
TP63 is a member of the p53 family, which consists
of numerous transcription factors including, p53, p63 and p73
(21), and is reported to be involved
in the process of epithelial development and carcinogenesis
(23). In normal breast tissue, TP63
expression is restricted to basal/myoepithelial cells, and p63 is
essential for mammary gland morphogenesis during embryonic
development (24–26). TP63 is highly expressed in a subset of
tumors lacking the expression of the estrogen receptor,
progesterone receptor and HER2, which is denoted as triple-negative
breast cancer (27–29). As aforementioned, patients with
HER2+ breast cancer have an increased probability of
developing brain metastasis, compared with patients without HER2
expression. Orzol et al (30)
introduced in their research that knocking out TP63 caused breast
cancer cells to proliferate at a decreased rate and adhere less
tightly, which resulted in an increased rate of migration. In the
present study, the network analysis indicated that the expression
of TP63 was negatively associated with HER2+ breast
cancer-associated brain metastasis, which indicated that an
increased expression of TP63 was associated with decreased
frequency of brain metastasis occurrence. Furthermore, in the
patient samples, compared with primary breast tumors, the
expression of TP63 in brain metastasis tumors was notably
decreased.
The VWA8 gene was originally identified by the
Kazusa cDNA project, where it was termed VWA8 (31). Sequence analysis demonstrated that the
protein encoded by the VWA8 gene may contain a VWA domain in its
C-terminus (31). Data from curated
expression analysis of VWA8 in human tissues indicated that VWA8
expression was marked in organs that had high-energy requirements,
including organs with a high density of mitochondria (31). The presence of VWA8 exclusively in
mitochondria raised the possibility that this protein had a role in
metabolic regulation or bioenergetic events (31); however, to date, there is no research
concerning its role in malignant tumor types. In the network
analysis of the present study, the results indicated that the
upregulated expression of VWA8 was negatively associated with
breast cancer-associated brain metastasis. Additionally, in the
patient samples, the expression of VWA8 in primary breast tumors
was notably increased, compared with the metastatic brain
tumors.
Furthermore, none of the four genes aforementioned
were associated with brain metastasis-free survival time. This may
be attributable to the following reasons. First, the quantity of
patients with HER2+ brain metastasis was too small. At
present, there are only 28 patients with HER2+ breast
cancer-associated brain metastasis that could be included in the
analysis. Secondly, the treatments the patients received following
surgery may influence the occurrence of brain metastasis. According
to Romond et al (32), the
application of trastuzumab for patients with HER2+ may
significantly delay the occurrence of brain metastasis, while its
effectiveness remains unclear. Thirdly, the different grades of
breast cancer prior to surgery and early lymph-node metastasis may
also influence the occurrence of brain metastasis (33). To determine how the actual mechanism
regulates the occurrence of breast cancer-associated brain
metastasis, further clinical and basic research is required.
Primarily, the results of the network analysis in
the present study were confirmed in the patient samples to a
certain extent. A number of novel target genes were identified,
which may provide a novel foundation for targeted therapeutic
treatments of HER2+ breast cancer-associated brain
metastasis. Although numerous genes that function together and
constitute networks were determined, the core hub genes were not
identified as the most significantly changed genes, and they did
not serve pivotal functions in the most significantly influenced
signaling pathways. However, as aforementioned, the significant
differences observed between primary breast tumors and brain
metastatic neoplasms were derived from significantly altered
systems, specifically, gene modules rather than single
molecules.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets (GSE43837) generated and analyzed
during the study are available from the corresponding author, on
reasonable request.
Authors' contributions
FY and WW wrote the manuscript and conducted related
experiments and data analysis. HC designed the study and discussed
the findings. All authors have read and approved the
manuscript.
Ethics approval and consent to
participate
Written informed consent for research was obtained
from all patients included in the present study. All experiments
were conducted according to the relevant guidelines and regulations
of the Institutional Ethical Review Committee of Hubei Cancer
Hospital (Wuhan, China). The present study was approved by the
Institutional Ethical Review Committee of Hubei Cancer
Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tsukada Y, Fouad A, Pickren JW and Lane
WW: Central nervous system metastasis from breast carcinoma.
Autopsy study. Cancer. 52:2349–2354. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kennecke H, Yerushalmi R, Woods R, Cheang
MC, Voduc D, Speers CH, Nielsen TO and Gelmon K: Metastatic
behavior of breast cancer subtypes. J Clin Oncol. 28:3271–3277.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aversa C, Rossi V, Geuna E, Martinello R,
Milani A, Redana S, Valabrega G, Aglietta M and Montemurro F:
Metastatic breast cancer subtypes and central nervous system
metastases. Breast. 23:623–628. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soni A, Ren Z, Hameed O, Chanda D, Morgan
CJ, Siegal GP and Wei S: Breast cancer subtypes predispose the site
of distant metastases. Am J Clin Pathol. 143:471–478. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brufsky AM, Mayer M, Rugo HS, Kaufman PA,
Tan-Chiu E, Tripathy D, Tudor IC, Wang LI, Brammer MG, Shing M, et
al: Central nervous system metastases in patients with
HER2-positive metastatic breast cancer: Incidence, treatment, and
survival in patients from registHER. Clin Cancer Res. 17:4834–4843.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen Q, Sahin AA, Hess KR, Suki D, Aldape
KD, Sawaya R and Ibrahim NK: Breast cancer with brain metastases:
Clinicopathologic features, survival, and paired biomarker
analysis. Oncologist. 20:466–473. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang B and Horvath S: A general framework
for weighted gene co-expression network analysis. Stat Appl Genet
Mol Biol. 4:Article172005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Horvath S, Zhang B, Carlson M, Lu KV, Zhu
S, Felciano RM, Laurance MF, Zhao W, Qi S, Chen Z, et al: Analysis
of oncogenic signaling networks in glioblastoma identifies ASPM as
a molecular target. Proc Natl Acad Sci USA. 103:17402–17407. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang L, Qi H, Xiao Y, Li C, Wang Y, Guo
T, Liu Z and Liu Q: Integrated analysis of noncoding RNAs and mRNAs
reveals their potential roles in the biological activities of the
growth hormone receptor. Growth Horm IGF Res. 29:11–20. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee J: Role of cyclophilin a during
oncogenesis. Arch Pharm Res. 33:181–187. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng J, Koblinski JE, Dutson LV, Feeney
YB and Clevenger CV: Prolyl isomerase cyclophilin A regulation of
Janus-activated kinase 2 and the progression of human breast
cancer. Cancer Res. 68:7769–7778. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Campa MJ, Wang MZ, Howard B, Fitzgerald MC
and Patz EF Jr: Protein expression profiling identifies macrophage
migration inhibitory factor and cyclophilin A as potential
molecular targets in non-small cell lung cancer. Cancer Res.
63:1652–1656. 2003.PubMed/NCBI
|
|
15
|
Cecconi D, Astner H, Donadelli M, Palmieri
M, Missiaglia E, Hamdan M, Scarpa A and Righetti PG: Proteomic
analysis of pancreatic ductal carcinoma cells treated with
5-aza-2′-deoxycytidine. Electrophoresis. 24:4291–4303. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Corton JC, Moreno ES, Merritt A, Bocos C
and Cattley RC: Cloning genes responsive to a hepatocarcinogenic
peroxisome proliferator chemical reveals novel targets of
regulation. Cancer Lett. 134:61–71. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chevalier F, Depagne J, Hem S, Chevillard
S, Bensimon J, Bertrand P and Lebeau J: Accumulation of cyclophilin
A isoforms in conditioned medium of irradiated breast cancer cells.
Proteomics. 12:1756–1766. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kenmochi N, Kawaguchi T, Rozen S, Davis E,
Goodman N, Hudson TJ, Tanaka T and Page DC: A map of 75 human
ribosomal protein genes. Genome Res. 8:509–523. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mager DL and Freeman JD: A human gene
related to the ribosomal protein L23 gene of Halobacterium
marismortui. Nucleic Acids Res. 18:53011990. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi Y, Zhai H, Wang X, Han Z, Liu C, Lan
M, Du J, Guo C, Zhang Y, Wu K and Fan D: Ribosomal proteins S13 and
L23 promote multidrug resistance in gastric cancer cells by
suppressing drug-induced apoptosis. Exp Cell Res. 296:337–346.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pflaum J, Schlosser S and Müller M: p53
family and cellular stress responses cancer. Front Oncol.
4:2852014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jin A, Itahana K, O'Keefe K and Zhang Y:
Inhibition of HDM2 and activation of p53 by ribosomal protein L23.
Mol Cell Biol. 24:7669–7680. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang A, Kaghad M, Wang Y, Gillett E,
Fleming MD, Dötsch V, Andrews NC, Caput D and McKeon F: p63, a p53
homolog at 3q27-29, encodes multiple products with transactivating,
death-inducing, and dominant-negative activities. Mol Cell.
2:305–316. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Batistatou A, Stefanou D, Arkoumani E and
Agnantis NJ: The usefulness of p63 as a marker of breast
myoepithelial cells. In Vivo. 17:573–576. 2003.PubMed/NCBI
|
|
25
|
Barbareschi M, Pecciarini L, Cangi MG,
Macrì E, Rizzo A, Viale G and Doglioni C: p63, a p53 homologue, is
a selective nuclear marker of myoepithelial cells of the human
breast. Am J Surg Pathol. 25:1054–1060. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nylander K, Vojtesek B, Nenutil R,
Lindgren B, Roos G, Zhanxiang W, Sjöström B, Dahlqvist A and Coates
PJ: Differential expression of p63 isoforms in normal tissues and
neoplastic cells. J Pathol. 198:417–427. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leong CO, Vidnovic N, DeYoung MP, Sgroi D
and Ellisen LW: The p63/p73 network mediates chemosensitivity to
cisplatin in a biologically defined subset of primary breast
cancers. J Clin Invest. 117:1370–1380. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Koker MM and Kleer CG: p63 expression in
breast cancer-a highly sensitive and specific marker of metoplastic
carcinoma. Am J Surg Pathol. 28:1506–1512. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du Z, Li J, Wang L, Bian C, Wang Q, Liao
L, Dou X, Bian X and Zhao RC: Overexpression of Delta Np63 alpha
induces a stem cell phenotype in MCF7 breast carcinoma cell line
through the Notch pathway. Cancer Sci. 101:2417–2424. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Orzol P, Nekulova M, Holcakova J, Muller
P, Votesek B and Coates PJ: ΔNp63 regulates cell proliferation,
differentiation, adhesion, and migration in the BL2 subtype of
basal-like breast cancer. Tumour Biol. 37:10133–10140. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luo M, Mengos AE, Ma W, Finlayson J,
Bustos RZ, Xiao Zhu Y, Shi CX, Stubblefield TM, Willis WT and
Mandarino LJ: Characterization of the novel protein KIAA0564 (Von
Willebrand Domain-containing Protein 8). Biochem Biophys Res
Commun. 487:545–551. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Romond EH, Jeong JH, Rastogi P, Swain SM,
Geyer CE Jr, Ewer MS, Rathi V, Fehrenbacher L, Brufsky A, Azar CA,
et al: Seven-year follow-up assessment of cardiac function in NSABP
B-31, a randomized trial comparing doxorubicin and cyclophosphamide
followed by paclitaxel (ACP) with ACP plus trastuzumab as adjuvant
therapy for patients with node-positive, human epidermal growth
factor receptor 2-positive breast cancer. J Clin Oncol.
30:3792–3799. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Anders CK, Deal AM, Miller CR, Khorram C,
Meng H, Burrows E, Livasy C, Fritchie K, Ewend MG, Perou CM and
Carey LA: The prognostic contribution of clinical Breast cancer
subtype, age, and race among patients with breast cancer brain
metastases. Cancer. 117:1602–1611. 2011. View Article : Google Scholar : PubMed/NCBI
|