Introduction
Liver cancer is one of the most common malignant
tumor types, exhibiting the highest rate of cancer-associated
mortality globally in 2014 due to uncontrolled cell growth in the
liver and high metastatic ability (1). Anticancer drugs are frequently applied
to liver cancer treatment combined with surgical therapy and/or
radiation therapy (2). However, the
majority of patients are diagnosed at an advanced stage, when liver
cancer is highly resistant to the existing therapeutics, which
results in poor prognosis (3,4).
5-Fluorouracil (5-FU) is an anticancer drug that is used as one of
the standard chemotherapies for liver cancer treatment (5). However, 5-FU therapy has been
demonstrated to frequently be inefficient due to the acquisition of
drug resistance and cytotoxicity at high concentrations (6). Therefore, investigating the mechanism
underlying 5-FU resistance and investigating therapeutic
strategies, to reverse drug resistance and resensitize cancer cells
to 5-FU, is vital.
Tight junctions are a type of cell-to-cell adhesion
in epithelial or endothelial cell sheets, forming continuous seals
around cells and functioning as a physical barrier to prevent
solutes and water from passing freely through the paracellular
space (7). Tight junctions maintain
cell polarity by preventing the lateral diffusion of integral
membrane proteins between the lateral/basal and apical surfaces,
allowing the specialized functions of each surface to be preserved
(8). It has also been reported that
tight junctions are associated with cellular survival,
proliferation, migration and differentiation (9). Claudin-1 (CLDN1), a protein that is
encoded by the CLDN1 gene in humans, belongs to the group of
CLDNs and serves a crucial role in tight junctions (10,11).
Abnormal expression of CLDN1 has been demonstrated to destroy the
epithelial permeability barrier and disrupt cellular polarity,
which results in decreased cell adhesion (12). Additionally, abnormal expression of
CLDN1 has been revealed to be associated with mechanisms of tumor
progression and development, including proliferation, migration,
invasion and chemotherapy resistance (13–16).
CLDN1 has been identified to be expressed in multiple tumor tissue
types and is involved in tumor growth, metastasis and prognosis
(15,16). However, the function of CLDN1 is
distinct in different types of tumor (17). To the best of our knowledge, the role
of CLDN1 in the development of 5-FU resistance in liver cancer
remains unclear (18,19). The present study developed a
5-FU-resistant liver cancer HepG2 cell line and investigated the
effect of CLDN1 and the underlying mechanism in 5-FU resistance of
HepG2 cells. Additionally, CLDN1 was investigated as a potential
therapeutic target for enhancing the sensitivity of HepG2 cells to
5-FU.
Materials and methods
Cell culture
The human liver cancer cell line HepG2 was purchased
from the Cell Bank of Type Culture Collection the Chinese Academy
of Sciences (Shanghai, China). Cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Lonza Group, Ltd., Basel, Switzerland), 5 mM L-glutamine, 5
mM non-essential amino acids, and 100 U/ml penicillin and
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.), in a
humidified 5% CO2 incubator at 37°C.
Cultivation of a 5-FU-resistant cell
line
5-FU-resistant HepG2 cells were developed by
exposing HepG2 cells to increasing concentrations of 5-FU ranging
from 10 to 50 mg/l in complete medium (Gibco; Thermo Fisher
Scientific, Inc.), as described previously (20). Briefly, HepG2 cells (2×106
cells/plate) were seeded in 60 mm culture plates and allowed to
grow. Following incubation for 24 h at 37°C, 10 mg/l 5-FU was added
for a further 48 h at 37°C. Subsequently, the medium was removed
and fresh drug-free medium (cat. no. C11995500BT; Gibco; Thermo
Fisher Scientific, Inc.) was added. The cells were incubated at
37°C. When 90% confluence was reached, cells were trypsinized,
replated at a density of 2×106 cells/plate and
re-exposed to 20 mg/l 5-FU as previously described. This process
was repeated with increasing doses (40 and 80 mg/l) until clones
developed resistance to 50 mg/l 5-FU. Following exposure to 5-FU
for ≥3 months, living cells were collected, termed drug-resistant
cells (Hep/5FU) and used for subsequent experiments.
Proliferation assay
Cell proliferation was evaluated with an MTT assay,
for which MTT was obtained from Sigma-Aldrich; Merck KGaA
(Darmstadt, Germany). A total of 1×104 Hep/5FU cells and
HepG2 cells with 100 µl Dulbecco's Modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc.) were plated in each well of
a 96-well plate and incubated for 24 h at 37°C. The cells were then
treated with 0, 10, 25, 50, 100, 200 or 400 mg/l 5-FU, 5 mM
3-methyladenine (3-MA; Sigma-Aldrich; Merck KGaA) or 10 nM
Rapamycin (Selleck Chemicals, Houston, TX, USA) for 48 h at 37°C in
a 5% CO2 incubator. Subsequently, cells were incubated
with 20 µl 5 mg/ml MTT for 4 h and lysed for 10 min at room
temperature by addition of 200 µl dimethyl sulfoxide (OriGene
Technologies, Inc., Rockville, MD, USA). Absorbance was measured at
490 nm using a Rainbow microplate reader (Tecan Group, Ltd.,
Mannedorf, Switzerland). Cell proliferation was expressed as a
percentage of the untreated control cells.
Migration and invasion assays
Cell migration was assessed with a Transwell assay
using 6.5 mm chambers (Corning Incorporated, Corning, NY, USA) with
8-µm pore membranes (17). A total
of 600 µl DMEM with 40 mg/l 5-FU was added to the lower chamber. A
suspension of 5×104 cells (Hep/5FU and HepG2 cells) in
100 µl DMEM with 1% FBS was plated into the upper chamber followed
by addition of 5-FU. Cells on the undersurface of the polycarbonate
membranes were stained with crystal violet (Amresco, LCC, Solon,
OH, USA) for 10 min at room temperature. Stained cells were
observed using a light microscope (magnification, ×100) and six
fields were selected at random to measure the mean cell coverage
using Image J software version 1.8.0 (National Institutes of
Health, Bethesda, MD, USA). The migration of Hep/5FU cells was
calculated relative to the migration of control HepG2 cells.
Invasion was assayed using the same protocol as the migration
assay, with the exception that 70 µl 1 mg/ml Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) was added into the upper face
of the membrane.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA in Hep/5FU cells and HepG2 cells was
extracted using the Total RNA isolation kit (A&A Biotechnology,
Gdynia, Poland). cDNA was obtained by RT using a RevertAidi cDNA
synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.) and was
amplified using a TaqMan® Gene Expression assay (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The reverse
transcription reaction parameters were 37°C for 60 min, followed by
85°C for 5 min. The PCR parameters were as follows: 95°C for 10
min, followed by 40 cycles of 95°C for 10 sec, 60°C for 20 sec and
72°C for 3 sec. The primer sequences used for RT-qPCR were as
follows: CLDN1 forward, 5′-CAGAAGATGAGGATGGCTGTCATT-3′ and reverse,
5′-AAGGGGGGCACAGCCTCTATTA-3′; GAPDH forward,
5′-TCCCTGAGCTGAACGGGAAG-3′ and reverse, 5′-GGAGGAGTGGGTGTCGCTGT-3′.
RT-qPCR was performed using an ABI PRISM 7700 Sequence Detector
(PerkinElmer, Inc., Waltham, MA, USA). mRNA expression levels were
calculated using the 2−ΔΔCq method and were normalized
to the expression level of GAPDH (21). The expression level of mRNA in cells
was made relative to the expression level of mRNA in control
cells.
Western blot analysis
Hep/5FU cells and HepG2 cells were seeded in a
culture dish (2×106 cells/dish) with DMEM, cultured
overnight and then incubated at room temperature with 50 mg/l 5-FU
for 48 h. Cells were harvested and washed twice with cold PBS, and
then lysed in cell lysis buffer (catalog no. P0013; Beyotime
Institute of Biotechnology, Haimen, China) containing a protease
inhibitor cocktail (Roche Diagnostics, Basel, Switzerland) at 4°C
for 15 min. Following centrifugation at 12,000 × g for 10 min at
room temperature, the supernatant was collected and quantified
using a bicinchoninic acid quantification kit (Beyotime Institute
of Biotechnology). The proteins (50 µg/lane) were separated by 12%
SDS-PAGE (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China) and transferred to polyvinylidene fluoride
membranes (EMD Millipore, Billerica, MA, USA). The membranes were
blocked with 5% non-fat dried milk in TBS and Tween-20 for 1 h at
room temperature. The membranes were then incubated overnight at
4°C with the following specific primary antibodies: Anti-CLDN1
(1:1,000; catalog no. ab15098; Abcam, Cambridge, UK), anti-LC3 I/II
(1:1,000; catalog no. 4108S; Cell Signaling Technology, Inc.,
Danvers, MA, USA), anti-P62 (1:1,000; catalog no. 8025S; Cell
Signaling Technology) and rabbit polyclonal anti-GAPDH (1:2,000;
catalog no. sc-25778; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA). Following the primary incubation, membranes were incubated
for 2 h at room temperature with either of the following
horseradish peroxidase-conjugated secondary antibodies: Goat
anti-mouse (1:2,000; catalog no. sc-2005; Santa Cruz Biotechnology,
Inc.) and goat anti-rabbit IgG (1:2,000; catalog no. sc-2004; Santa
Cruz Biotechnology, Inc.). Protein bands were visualized using
ECL-detecting reagent (GE Healthcare, Chicago, IL, USA) and
quantified by densitometry using Quantity One software (version
4.6.6; Bio-Rad Laboratories, Inc., Hercules, CA, USA). Protein
amounts were expressed relative to the internal reference
GAPDH.
RNA interference of CLDN1
Small interference (si)RNAs against CLDN1 (siCLDN1)
and the negative control (siNC) were designed and chemically
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China).
Cell transfections were conducted using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Briefly, 1.5 l 25 pmol/l siCLDN1 or siNC
and 1 µl Lipofectamine® 2000 were added into vials
containing 50 µl Opti Memi medium (Thermo Fisher Scientific, Inc.)
at room temperature. After 5 min, the liquids in the vials were
mixed before standing for 20 min at room temperature, and then the
mixture was added to the cells and incubated for 6 h. Fresh DMEM
supplemented with 10% FBS was provided and cells were grown for 48
h prior to subsequent experiments.
Furthermore, Hep/5FU cells transfected with negative
control siRNA or siRNA against CLDN1 were treated with 50 mg/l 5-FU
for 48 h. Non-silenced Hep/5FU cells were treated with or without 5
mM 3-MA, an autophagy inhibitor, for 48 h. CLDN1-silenced Hep/5FU
cells were treated with or without 10 nM Rapamycin, an autophagy
agonist, for 48 h.
Apoptosis assay
Hep/5FU cells and HepG2 cells were grown to 80%
confluence at room temperature and treated with 50 mg/l 5-FU for 48
h at room temperature. Cells were collected, washed three times
using TBS + 0.1% Tween-20 for 5 min and resuspended in PBS at room
temperature. Apoptosis was analyzed using Annexin V-fluorescein
isothiocyanate (FITC)/propidium iodide (PI) assay (eBioscience,
USA), according to the manufacturer's protocol. Briefly, the cells
were washed three times with PBS + 0.1% Tween-20 and subsequently
incubated for 15 min at room temperature in the dark with 100 µl 1X
Annexin binding buffer containing 5 µl Annexin V-FITC and 2 µl PI.
This detected the amount of phosphatidylserine on the outer surface
of the plasma membrane, a biochemical alteration unique to
membranes of apoptotic cells, and the amount of PI, a dye that
enters dead cells or cells in the late stages of apoptosis and
binds to DNA but does not bind to the plasma membrane of viable
cells. Fluorescence was detected using a flow cytometer and data
were analyzed using CellQuest software version 5.1 (BD
Biosciences). Cells with phosphatidylserine on their surface were
considered to be apoptotic, including early apoptotic cells and
late apoptotic cells.
Transmission electron microscopy
(TEM)
Cells were trypsinized and fixed in 0.1 M cacodylate
buffer (pH 7.4) and 2.5% glutaraldehyde for 2 h at room temperature
and were washed twice using precooled PBS. Subsequently, cells were
post-fixed in 0.1 M cacodylate buffer (pH 7.4) and 1% osmium
tetroxide. The fixed cells were rinsed with PBS, dehydrated with
various doses of ethanol and embedded in epoxy resin. The
ultrastructures of autophagic cells were observed and images were
obtained using an electron microscope (magnification, ×10,000 or
×20,000; JEM-1200; JEOL, Ltd., Tokyo, Japan) at 80 kV.
Statistical analysis
Data were obtained from a minimum of three
experiments. Statistical analysis was performed using SPSS software
(version 13.0; SPSS, Inc., Chicago, IL, USA). Data are presented as
the mean ± standard error of the mean. One-way analysis of variance
was used to assess differences between groups. Duncan's multiple
range test was employed for pairwise comparison, followed by
Bonferroni correction. P<0.05 was considered to indicate a
statistically significant difference.
Results
Development of 5-FU-resistant HepG2
cells
To examine the mechanism underlying 5-FU resistance
in liver cancer, a 5-FU resistant Hep/5FU cell model was first
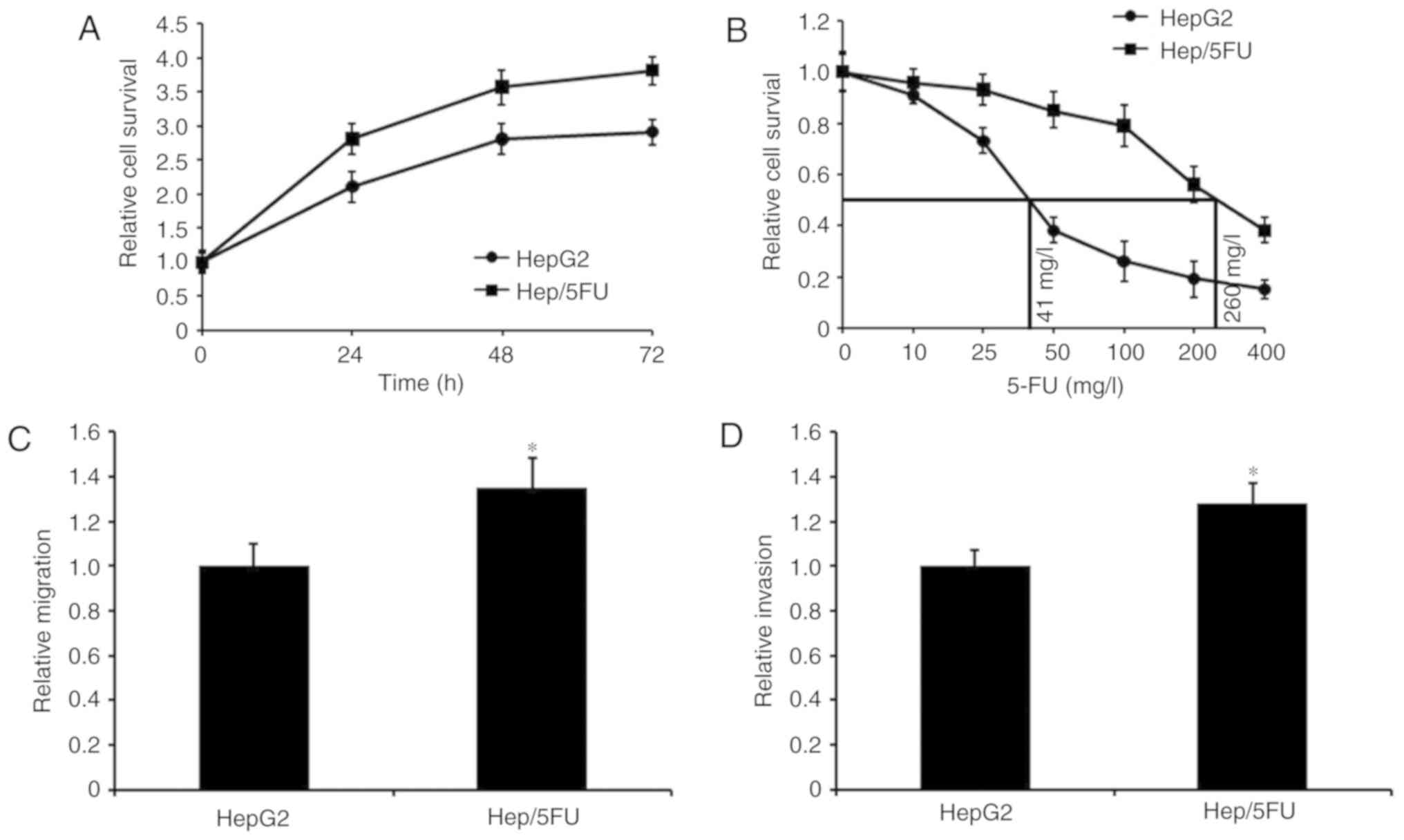
developed. Cell growth was assessed using an MTT assay and growth
curves of Hep/5FU and HepG2 cells are depicted in Fig. 1A. The data demonstrated that Hep/5FU
cells grew faster, compared with HepG2 cells, at the same time
interval. Additionally, the maximal half inhibitory concentration
(IC50) values of 5-FU were determined by exposing
Hep/5FU and HepG2 cells to different concentrations of 5-FU for 48
h. IC50 values of 5-FU in Hep/5FU and HepG2 cells were
calculated to be 260 and 41 mg/l, respectively (Fig. 1B). Additionally, migration and
invasion capabilities of Hep/5FU cells were identified to be
significantly increased, compared with HepG2 cells (Fig. 1C and D). These data indicate that a
5-FU-resistant HepG2 cell line was successfully constructed, which
demonstrated an increased migration and invasion ability, compared
with HepG2 cells.
CLDN1 silencing resensitizes resistant
HepG2 cells to 5-FU
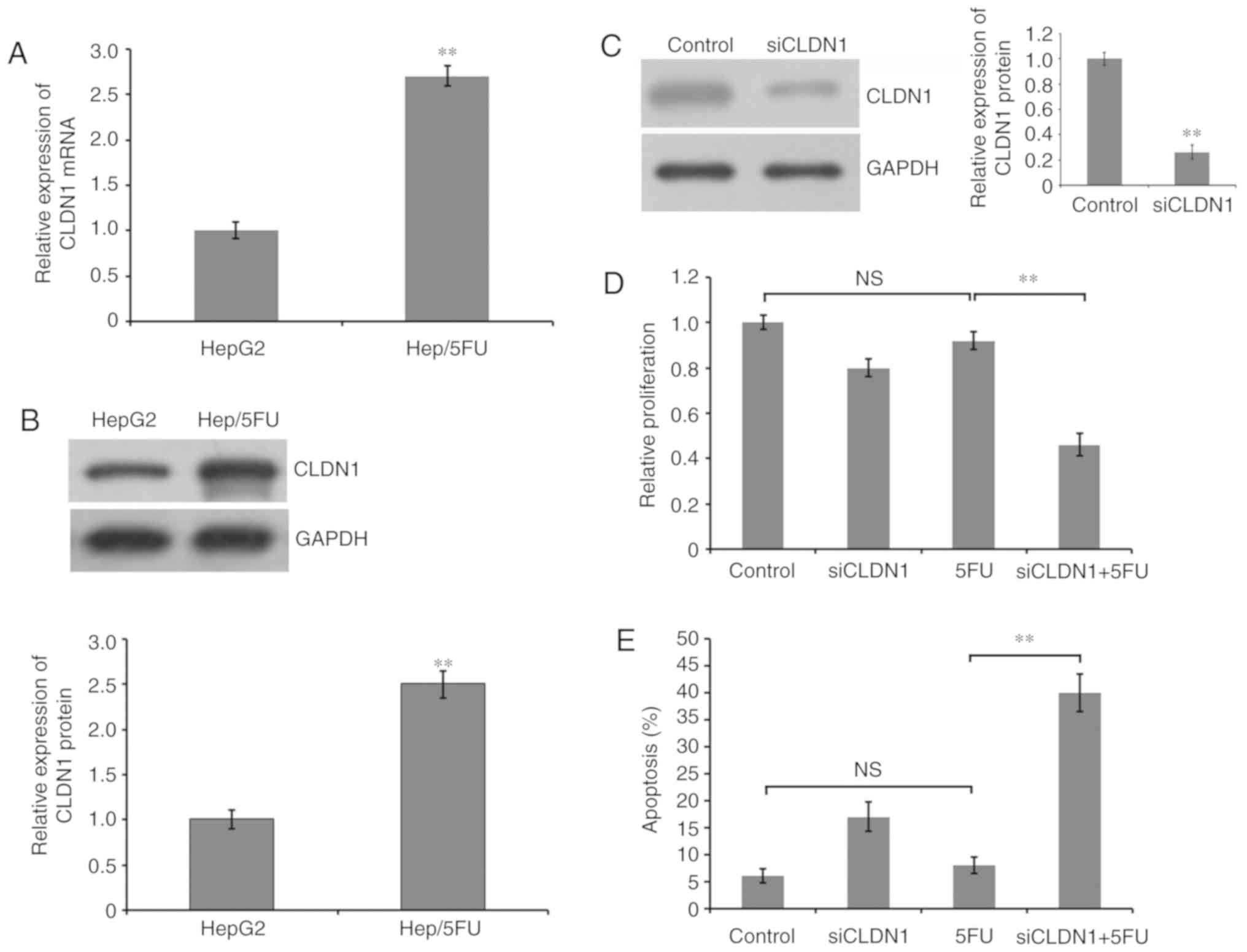
To detect the role of CLDN1 in the generation of
5-FU resistance in HepG2 cells, the expression level of CLDN1 was
examined. RT-qPCR demonstrated that the mRNA expression level of
CLDN1 was significantly increased in 5-FU-resistant HepG2 cells,
compared with HepG2 cells (Fig. 2A).
Additionally, western blot analysis demonstrated that the protein
expression level of CLDN1 was significantly increased in
5-FU-resistant HepG2 cells, compared with HepG2 cells (Fig. 2B). Furthermore, the expression of
CLDN1 was silenced by transfection siRNA and western blot analysis
revealed a significant decrease of CLDN1 expression in
CLDN1-silenced Hep/5FU cells, compared with non-silenced control
cells (Fig. 2C). Additionally,
Hep/5FU cells were transfected with negative control or siRNA
against CLDN1 and cultured in the presence or absence of 50 mg/l
5-FU for 48 h. Cell proliferation was analyzed with an MTT assay. A
slight decrease was observed in the proliferation of CLDN1-silenced
Hep/5FU cells, compared with non-silenced Hep/5FU control cells,
under identical experimental conditions without 5-FU treatment.
Additionally, cell proliferation of CLDN1-silenced Hep/5FU cells
was significantly decreased following 5-FU treatment, compared with
5-FU-treated non-silenced Hep/5FU control cells, and the growth of
non-silenced Hep/5FU control cells was not significantly affected
by 5-FU treatment, compared with untreated control cells (Fig. 2D).
Subsequently, Hep/5FU cells were transfected with
negative control or siRNAs against CLDN1 and cultured in the
presence or absence of 50 mg/l 5-FU for 48 h. Cell apoptosis was
evaluated with an Annexin V-FITC/PI assay. These data indicate that
CLDN1 silencing significantly increased apoptosis following
treatment with 5-FU in Hep/5FU cells, compared with non-silenced
Hep/5FU cells (Fig. 2E). However,
5-FU treatment did not significantly induce apoptosis in
non-silenced Hep/5FU cells, compared with untreated control cells
(Fig. 2E). These results indicate
that CLDN1 is associated with the resistance of HepG2 cells to 5-FU
and CLDN1 silencing may increase the sensitivity of resistant
Hep/5FU cells to 5-FU by enhancing the apoptosis triggered by 5-FU
treatment.
CLDN1 silencing decreases migration
and invasion of Hep/5FU cells
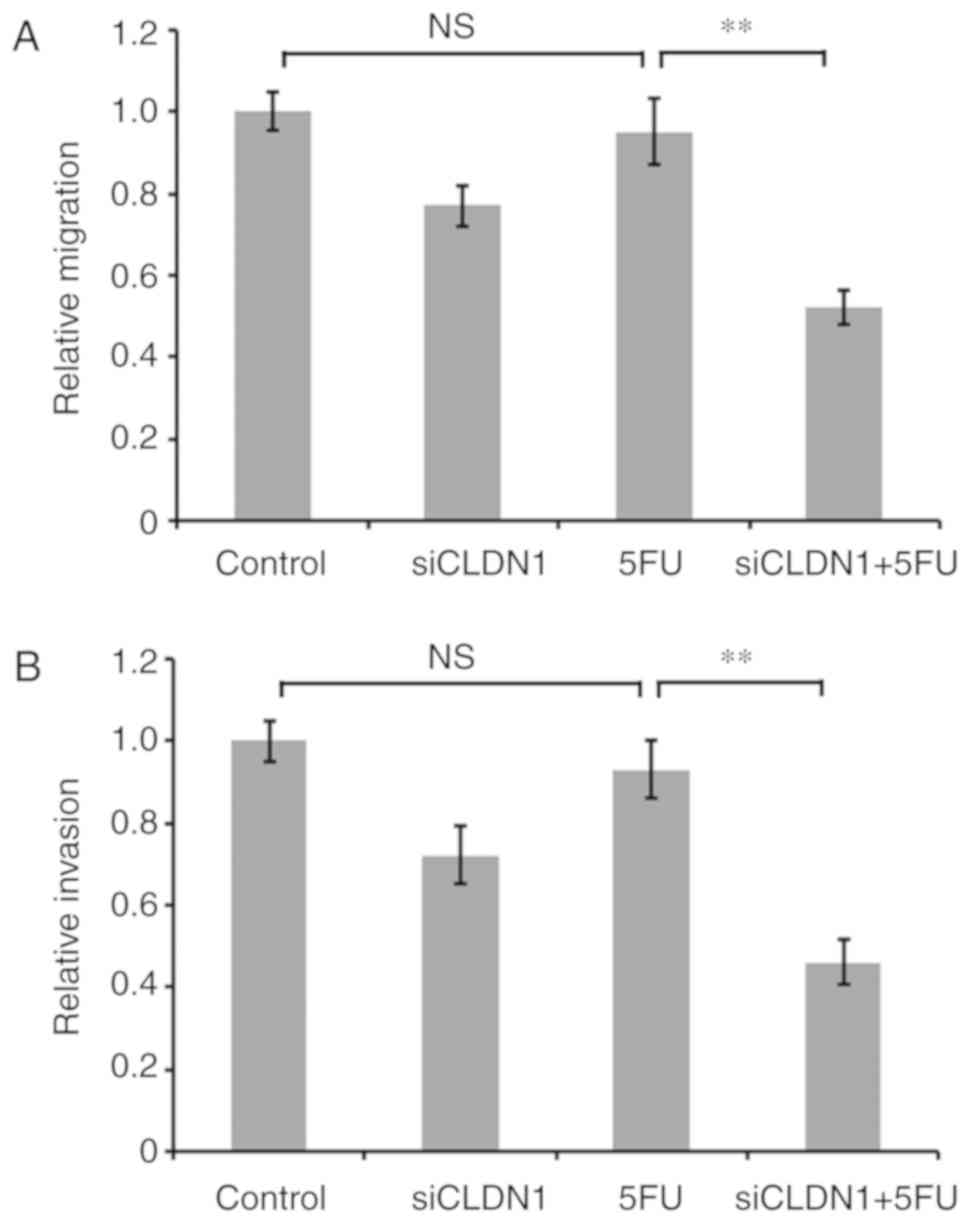
To further investigate the effect of CLDN1 on 5-FU
resistance of Hep/5FU cells, the expression of CLDN1 was
downregulated using siRNA, and cells were cultured in the presence
or absence of 50 mg/l 5-FU for 48 h. Cell migration and invasion
were then evaluated. Transwell and Matrigel assays demonstrated the
migration and invasion abilities, respectively. Following treatment
with 5-FU, the migration and invasion abilities of CLDN1-silenced
Hep/5FU cells were significantly decreased, compared with CLDN1
non-silenced Hep/5FU cells (Fig. 3).
These data indicate that regulation of cell motility by CLDN1 may
be involved in the development of drug resistance in Hep/5FU
cells.
CLDN1 silencing reduces drug
resistance of Hep/5FU cells by inhibiting autophagy
To investigate whether CLDN1 silencing enhances drug
sensitivity of Hep/5FU cells by affecting autophagy, the
ultrastructures of cells were observed and imaged using TEM. The
results demonstrate that an increased number of autophagosomes were
observed in drug-resistant Hep/5FU cells, compared with HepG2 cells
(Fig. 4A). By contrast, CLDN1
silencing reversed the phenomenon and inhibited cell autophagy of
Hep/5FU cells, compared with non-silenced Hep/5FU cells (Fig. 4B). Additionally, it was revealed that
the ratio of LC3B II/I was significantly increased in Hep/5FU
cells, compared with HepG2 cells, while CLDN1 silencing
significantly decreased the ratio of LC3B II/I in Hep/5FU cells,
compared with non-silenced Hep/5FU cells (Fig. 4C). Additionally, a significantly
reduced expression of P62 was observed in Hep/5FU cells, compared
with HepG2 cells, while CLDN1 silencing significantly increased the
expression of P62 in Hep/5FU cells, compared with non-silenced
Hep/5FU cells (Fig. 4C).
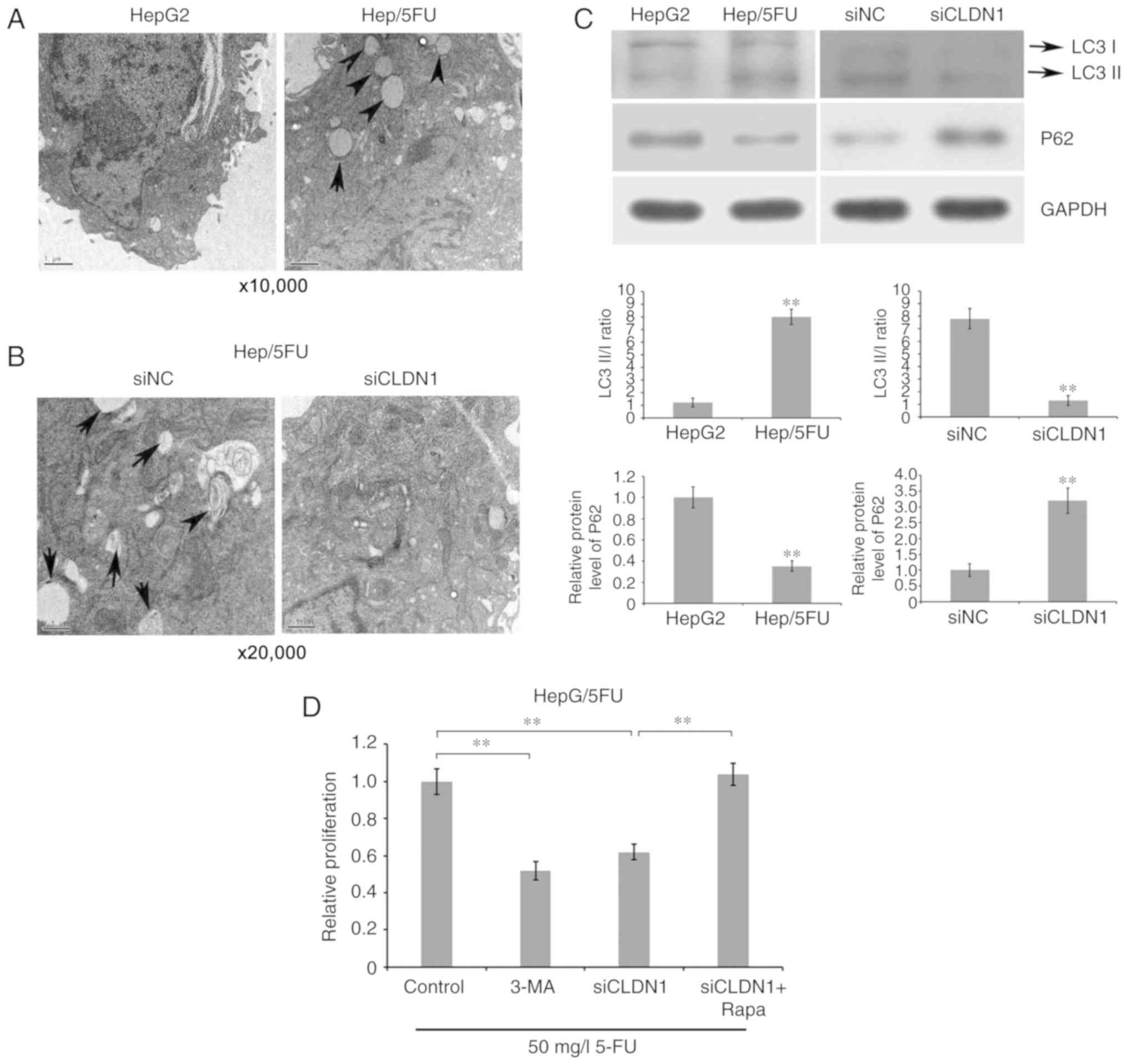 | Figure 4.Association between autophagy and drug
resistance of Hep/5FU cells. (A) HepG2 and Hep/5FU cells were
cultured to 80% confluence and autophagy was detected. The
ultrastructures of cells undergoing autophagy were observed and
imaged using TEM. Magnification, ×10,000. Black arrows indicate
autophagosomes. (B) Autophagy of Hep/5FU cells transfected with
siNC or siCLDN1 was observed using TEM. Magnification, ×20,000.
Black arrows indicate autophagosomes. (C) Protein expression levels
of LC3 I/II and P62 in HepG2 and Hep/5FU cells, as well as Hep/5FU
cells transfected with siNC or siCLDN1 were assessed by western
blot analysis using the corresponding antibodies. Protein bands
were quantified by densitometry and the expression levels were
expressed as a ratio relative to the expression level in HepG2 or
siNC-transfected cells. **P<0.01 vs. HepG2 cells or
siNC-transfected cells. (D) Hep/5FU cells were treated with or
without siCLDN1, 5 mM 3-MA or 10 nM Rapa combined with 50 mg/l 5-FU
for 48 h, and cell proliferation was detected with an MTT assay.
**P<0.01. CLDN1, claudin-1; Hep/5FU, 5-fluorouracil-resistant
HepG2; siCLDN1, small interfering RNA targeting claudin-1; TEM,
transmission electron microscopy; LC3, microtubule-associated
protein 1A/1B-light chain 3; 3-MA, 3-methyladenine; Rapa,
Rapamycin; NC, negative control. |
Subsequently, cell proliferation was detected with
an MTT assay. The results demonstrate that the addition of
autophagy inhibitor 3-MA significantly decreased drug resistance of
Hep/5FU cells, compared with control cells, while incubation with
autophagy agonist Rapamycin significantly increased drug resistance
of CLDN1-silenced Hep/5FU cells, compared with cells not treated
with Rapamycin (Fig. 4D). These data
indicate that CLDN1 silencing resensitizes resistant HepG2 cells to
5-FU, possibly by regulating cell autophagy.
Discussion
5-FU is one of the most common anticancer drugs and
is used as one of the standard chemotherapies for liver cancer
(22). However, cancer cells usually
develop resistance to 5-FU, which is the main cause of treatment
failure (7). Overcoming drug
resistance may be significant to improve prognosis and survival
rates of liver cancer. 5-FU has been demonstrated to irreversibly
inhibit thymidylate synthase and act as an antimetabolite,
resulting in defection of DNA and RNA synthesis, and thus inducing
apoptosis and inhibiting cell growth (23). Therefore, it is important to
investigate the mechanism of 5-FU resistance. The present study
developed Hep/5FU cells and demonstrated that Hep/5FU cells
exhibited enhanced migration, invasion and cell autophagy.
Additionally, the expression of CLDN1 in Hep/5-FU cells was
identified to be significantly increased, compared with 5-FU
sensitive HepG2 cells, and was associated with tumor cell
biological functions. In addition, it was revealed that
downregulation of CLDN1 expression in Hep/5FU cells inhibited cell
autophagy and resensitized drug-resistant Hep/5FU cells to
5-FU.
Tight junctions serve important roles in epithelial
cells by maintaining cell polarity and the epithelial barrier
(8,9). Abnormal expression and distribution of
tight junctions is associated with characteristics of tumor
progression and development, including growth, migration and
invasion (10). CLDN1, a key protein
in tight junctions, is associated with tumor metastasis and
recurrence (14–16,18). It
has been reported that CLDN1 is associated with the prognosis of
colon cancer and enhances the invasion and migration of tumor cells
(24). Additionally, it has been
demonstrated that regulation of osteosarcoma cell motility by CLDN1
is associated with the intracellular localization of CLDN1 protein
(25). It has been identified that
the high expression of CLDN1 indicates a poor prognosis of patients
with non-small-cell lung carcinoma and downregulation of CLDN1
expression hinders cellular migration in breast cancer (26). Furthermore, upregulation of CLDN1 was
determined to stimulate cell proliferation and motility in liver
cancer (27). It has been reported
that CLDN1 expression is elevated in nasopharyngeal carcinoma cells
following treatment with 5-FU and upregulation of CLDN1 expression
confers resistance to 5-FU (23).
However, to the best of our knowledge, the role of CLDN1 in drug
resistance of liver cancer to 5-FU remains unclear.
In the present study, resistant Hep/5FU cells were
constructed, which exhibited resistance to 50 mg/l 5-FU and
demonstrated increased levels of cell proliferation, migration and
invasion. Additionally, the expression level of CLDN1 was
significantly increased in Hep/5FU cells, compared with HepG2
cells, which indicates that CLDN1 may be positively associated with
drug resistance of HepG2 cells to 5-FU. The inhibitory effect of
5-FU on cell proliferation, migration and invasion increased
following silencing of CLDN1 by transfection with siRNA. 5-FU
induced cell apoptosis and therefore reduced drug resistance. These
data indicate that CLDN1 expression increases drug resistance in
Hep/5FU cells by attenuating the inhibition of 5-FU on
proliferation and cell motility. It has previously been identified
that the expression and anti-apoptotic activity of CLDN1 can be
regulated by E-cadherin, which has been associated with the
development of 5-FU resistance in nasopharyngeal carcinoma cells
(28). Whether E-cadherin is
involved in the modulation of CLDN1 and 5-FU resistance in HepG2
cells requires further investigated in future studies.
Autophagy is a homeostatic process that is
responsible for degrading intracellular organelles and damaged
proteins via the delivering of cytoplasmic cargo to the lysosome,
which achieves cell metabolism and renews organelles (29). Autophagy serves an important role in
the progression and development of liver cancer (30). However, the role of autophagy in drug
resistance of liver cancer is not clear. In the present study,
increased levels of autophagy were observed in Hep/5FU cells,
compared with HepG2. Silencing of CLDN1 inhibited autophagy in
Hep/5FU cells and was associated with a downregulation of the ratio
of the LC3 II/I protein and the upregulation of P62. The inhibition
of autophagy in Hep/5FU cells by 3-MA resulted in a decrease in
drug resistance of the cells, while activation of autophagy by
Rapamycin restored drug resistance of CLDN1-silenced Hep/5FU cells
to 5-FU. It has been reported that unc-51 like autophagy activating
kinase 1 (ULK1) serves a critical role in regulating autophagy of
tumor cells and activates autophagy by phosphorylation, which can
be mediated by adenosine monophosphate-activated protein kinase
(AMPK) (30,31). Autophagy-related protein 13 (ATG13),
a target of the target of rapamycin kinase signaling pathway,
modulates autophagy via phosphorylation of ULK1 and ATG13 and the
regulation of the ATG13/ULK1 complex (32,33). The
ATG13/ULK1 complex regulates the kinase activity of cell
proliferation (34). The present
study indicated that CLDN1 modulates the resistance of HepG2 cells
to 5-FU by cell autophagy. However, to the best of our knowledge,
whether AMPK/ULK1/ATG13 signaling is involved in the modulation of
autophagy by CLDN1 remains unknown. Therefore, the underlying
mechanism of CLDN1 regulating autophagy and affecting drug
resistance of HepG2 cells to 5-FU requires further
investigation.
Acknowledgements
The authors would like to thank Dr. Xiaohui Liu
(Ruijin Hospital, Shanghai) for his support and advice during this
project.
Funding
The present study was supported by the Science and
Technology Commission of Shanghai Municipality (grant no.
15411950404).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HT wrote the manuscript and conducted experiments.
TL and WQ designed and helped conduct the experiments. ZZ designed
the study and revised the manuscript. All the authors have reviewed
the manuscript before submission and have approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
American Cancer SocietyCancer Facts &
Figures 2014. Atlanta: American Cancer Society; 2014
|
|
2
|
Krishna R and Mayer LD: Multidrug
resistance (MDR) in cancer: Mechanisms, reversal using modulators
of MDR and the role of MDR modulators in influencing the
pharmacokinetics of anticancer drugs. Eur J Pharm Sci. 11:265–283.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Deng GL, Zeng S and Shen H: Chemotherapy
and target therapy for hepatocellular carcinoma: New advances and
challenges. World J Hepatol. 7:787–798. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Petraccia L, Onori P, Sferra R, Lucchetta
MC, Liberati G, Grassi M and Gaudio E: MDR (multidrug resistance)
in hepatocarcinoma clinical-therapeutic implications. Clin Ter.
154:325–335. 2003.(In Italian). PubMed/NCBI
|
|
5
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohtsu A: Chemotherapy for metastatic
gastric cancer: Past, present, and future. J Gastroenterol.
43:256–264. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Obert E, Strauss R, Brandon C, Grek C,
Ghatnekar G, Gourdie R and Rohrer B: Targeting the tight junction
protein, zonula occludens-1, with the connexin43 mimetic peptide,
aCT1, reduces VEGF-dependent RPE pathophysiology. J Mol Med (Berl).
95:535–552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matsuoka H, Shima A, Uda A, Ezaki H and
Michihara A: The retinoic acid receptor-related orphan receptor a
positively regulates tight junction protein claudin
domain-containing 1 mRNA expression in human brain endothelial
cells. J Biochem. 161:441–450. 2017.PubMed/NCBI
|
|
9
|
Liu W, Mi S, Ruan Z, Li J, Shu X, Yao K,
Jiang M and Deng Z: Dietary tryptophan enhanced the expression of
tight junction protein ZO-1 in intestine. J Food Sci. 82:562–567.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morita K, Furuse M, Fujimoto K and Tsukita
S: Claudin multigene family encoding four-transmembrane domain
protein components of tight junction strands. Proc Natl Acad Sci
USA. 96:511–516. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Furuse M, Fujita K, Hiiragi T, Fujimoto K
and Tsukita S: Claudin-1 and-2: Novel integral membrane proteins
localizing at tight junctions with no sequence similarity to
occludin. J Cell Biol. 141:1539–1550. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim B and Breton S: The MAPK/ERK-signaling
pathway regulates the expression and distribution of tight junction
proteins in the mouse proximal epididymis. Biol Reprod. 94:222016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Akizuki R, Shimobaba S, Matsunaga T, Endo
S and Ikari A: Claudin-5, −7 and −18 suppress proliferation
mediated by inhibition of phosphorylation of Akt in human lung
squamous cell carcinoma. Biochim Biophys Acta. 1864:293–302. 2017.
View Article : Google Scholar
|
|
14
|
Yang Y, Cheon S, Jung MK, Song SB, Kim D,
Kim HJ, Park H, Bang SI and Cho D: Interleukin-18 enhances breast
cancer cell migration via down-regulation of claudin-12 and
induction of the p38 MAPK pathway. Biochem Biophys Res Commun.
459:379–386. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang WN, Li W, Wang XL, Hu Z, Zhu D, Ding
WC, Liu D, Li KZ, Ma D and Wang H: CLDN1 expression in cervical
cancer cells is related to tumor invasion and metastasis.
Oncotarget. 7:87449–87461. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kuo KT, Chen CL, Chou TY, Yeh CT, Lee WH
and Wang LS: Nm23H1 mediates tumor invasion in esophageal squamous
cell carcinoma by regulation of CLDN1 through the AKT signaling.
Oncogenesis. 5:e2392016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kominsky SL: Claudins: Emerging targets
for cancer therapy. Expert Rev Mol Med. 8:1–11. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moldvay J, Fábián K, Jäckel M, Németh Z,
Bogos K, Furák J, Tiszlavicz L, Fillinger J, Döme B and Schaff Z:
Claudin-1 protein expression is a good prognostic factor in
non-small cell lung cancer, but only in squamous cell carcinoma
cases. Pathol Oncol Res. 23:151–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun BS, Yao YQ, Pei BX, Zhang ZF and Wang
CL: Claudin-1 correlates with poor prognosis in lung
adenocarcinoma. Thorac Cancer. 7:556–563. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meena AS, Sharma A, Kumari R, Mohammad N,
Singh SV and Bhat MK: Inherent and acquired resistance to
paclitaxel in hepatocellular carcinoma: Molecular events involved.
PLoS One. 8:e615242013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vu NB, Nguyen TT, Tran LC, Do CD, Nguyen
BH, Phan NK and Pham PV: Doxorubicin and 5-fluorouracil resistant
hepatic cancer cells demonstrate stem-like properties.
Cytotechnology. 65:491–503. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tolba MF and Abdel-Rahman SZ:
Pterostilbine, an active component of blueberries, sensitizes colon
cancer cells to 5-fluorouracil cytotoxicity. Sci Rep. 5:152392015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nakagawa S, Miyoshi N, Ishii H, Mimori K,
Tanaka F, Sekimoto M, Doki Y and Mori M: Expression of CLDN1 in
colorectal cancer: A novel marker for prognosis. Int J Oncol.
39:791–967. 2011.PubMed/NCBI
|
|
25
|
Jian Y, Chen C, Li B and Tian X:
Delocalized Claudin-1 promotes metastasis of human osteosarcoma
cells. Biochem Biophys Res Commun. 466:356–361. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao X, Zou Y, Gu Q, Zhao G, Gray H,
Pfeffer LM and Yue J: Lentiviral vector mediated Claudin1 silencing
inhibits epithelial to mesenchymal transition in breast cancer
cells. Viruses. 7:2965–2979. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fortier AM, Asselin E and Cadrin M:
Keratin 8 and 18 loss in epithelial cancer cells increases
collective cell migration and cisplatin sensitivity through
claudin1 up-regulation. J Biol Chem. 288:11555–7129. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee JW, Hsiao WT, Chen HY, Hsu LP, Chen
PR, Lin MD, Chiu SJ, Shih WL and Hsu YC: Upregulated claudin-1
expression confers resistance to cell death of nasopharyngeal
carcinoma cells. Int J Cancer. 126:1353–1366. 2010.PubMed/NCBI
|
|
29
|
Kim H, Kim Y and Jeoung D: DDX53 promotes
cancer stem cell-like properties and autophagy. Mol Cells.
40:54–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee YJ and Jang BK: The role of autophagy
in hepatocellular carcinoma. Int J Mol Sci. 16:26629–26643. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jang JE, Eom JI, Jeung HK, Cheong JW, Lee
JY, Kim JS and Min YH: Targeting AMPK-ULK1-mediated autophagy for
combating BET inhibitor resistance in acute myeloid leukemia stem
cells. Autophagy. 13:761–762. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nie T, Yang S, Ma H, Zhang L, Lu F, Tao K,
Wang R, Yang R, Huang L, Mao Z and Yang Q: Regulation of ER
stress-induced autophagy by GSK3b-TIP60-ULK1 pathway. Cell Death
Dis. 7:e25632016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim J, Kundu M, Viollet B and Guan KL:
AMPK and mTOR regulate autophagy through direct phosphorylation of
Ulk1. Nat Cell Biol. 13:132–141. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mercer CA, Kaliappan A and Dennis PB: A
novel, human Atg13 binding protein, Atg101, interacts with ULK1 and
is essential for macroautophagy. Autophagy. 5:649–662. 2009.
View Article : Google Scholar : PubMed/NCBI
|