Introduction
Human papillomavirus positive (HPV+)
tonsillar and base of tongue squamous cell carcinoma (TSCC/BOTSCC)
generally have a better outcome than the corresponding HPV negative
(HPV−) cancers and most other head neck squamous cell
carcinoma (HNSCC), and their incidences have increased considerably
(1–15). Most HPV+ TSCC/BOTSCC
patients do therefore not need the intensified treatment, more
recently given to HNSCC patients in general, and which has not
improved survival for patients with HPV+ TSCC/BOTSCC
(5,16–18). To
better individualize treatment, experimental approaches have been
made to find predictive biomarkers (19–27).
Such markers were e.g. high CD8+ tumor infiltrating
lymphocyte counts, HPV16 E2 mRNA expression, absent/low HLA class
I, HLA-A*02, CD44, LMP10 expression, high LRIG1 or CD98 expression,
and combined in mathematical models, they identified ~50% of
patients with high survival probability (16–27).
To identify even more markers for prognostication,
we performed next-generation sequencing (NGS) of hot spot mutations
in 50 cancer related genes and found, similar to others, that
PIK3CA mutations are common in HPV+ TSCC/BOTSCC
(28,29). In our study, however presence of
FGFR3 mutations were also relatively common and correlated to worse
prognosis, and later similar to others we found that FGFR3 was
overexpressed in TSCC/BOTSCC and oropharyngeal cancer (29–31). In
view of the fact, that targeted therapy against FGFR3 and PIK3CA
has recently been initiated in other types of cancers, it could
also be useful to consider for TSCC/BOTSCC, and for reviews see
e.g. (32,33). This would especially be important for
HPV+ TSCC/BOTSCC patients with high risk of a poor
outcome, where today's chemotherapy and radiotherapy is suboptimal
(18,32,33).
Here, the HPV+ cell lines UM-SCC-47 and
UPCI-SCC-154, and the HPV− cell line UT-SCC-60A were
tested for possible presence of the most common FGFR3 and PIK3CA
mutations by competitive allele-specific TaqMan-PCR (CAST-PCR).
They were then treated with FGFR inhibitor AZD4547, or PI3K
inhibitors BEZ235 and BKM120, alone or the former two combined, to
investigate the sensitivity of the cell lines to any of the drugs
alone or combined, and to whether the combination of drugs was
beneficial or antagonistic.
Materials and methods
Cell line characteristics and cell
culture
All cell lines were derived from primary tumors
prior to treatment. HPV+ UM-SCC-47, a squamous cell
carcinoma isolated from the primary tumor of the lateral tongue and
UPCI-SCC-154, a squamous cell carcinoma of the tongue were provided
by Susan Gollin, University of Pittsburgh USA (34,35).
HPV− UT-SSC-60A, a tonsillar squamous cell carcinoma,
was kindly obtained from Reidar Grénman, University of Turku,
Finland (36). All cells were
cultured in Dulbecco's modified Eagle's medium (Gibco), with 10%
fetal bovine serum (FBS; Gibco), 1% L-glutamin, 100 U/ml of
penicillin, and 100 µg/ml streptomycin. All cells were
maintained at 37°C in a humidified incubator with 5%
CO2. Possible presence of FGFR3 or PIK3CA mutations were
analyzed as described below.
Competitive allele-specific TaqMan PCR
(CAST-PCR)
Detection of FGFR3 and PIK3CA mutations was
performed by Competitive Allele-Specific TaqMan® PCR
technology (Thermo Fischer Scientific) as described in detail
earlier (30). Briefly the analysis
was performed in 384-well plates, in 10 µl comprising 5
µl 2X TaqMan Genotyping Mastermix (Thermo Fischer
Scientific), 0.2 µl 50X Exogenous Internal Passive Control
(IPC) template DNA, 1 µl 10X Exogenous IPC mix, µl
Mutation Detection Assay, 1.8 µl deionized water and 20 ng
DNA (in 1 µl). Runs were performed on an Applied Biosystems
7900HT Fast Real-Time PCR System using a following set of reaction
conditions described in detail before (30). The PCR result was analyzed with the
SDS 2.3 software program and Mutation Detector Software 2.0 (Thermo
Fischer Scientific). Ct was determined for exogenous IPC reagents
added to every reaction to evaluate PCR failure or inhibition in a
reaction. The Mutation Detection Assays detecting the reference
FGFR3 gene and its variants p.R248C, p.S249C and p.K650Q have been
described before (30). The same
system was used to detect the reference gene PIK3CA and its
variants p.E542K, p.E545K and p.H1047R. Mutation Detection Assays
were Hs00000822_mu, Hs00000824_mu, Hs00000831_mu, which detects
variants p.E542K, p.E545K and p.H1047R in the PIK3CA gene
respectively, and reference assay Hs00001025_rf was used for
detection of wild-type PIK3CA.
Treatment with FGFR and PI3K inhibitors
after cell seeding and experimental set up
Cell seeding
For most assays, 5,000 cells were seeded out into
80–100 µl medium (without penicillin and streptomycin) per
well of 96 well plates, where the outer wells were not used, but
filled with medium to avoid any edge effects.
Treatment with FGFR and PI3K
inhibitors and experimental set up
The cells were treated with different inhibitors 24
h (h) after seeding. For treatment, the PI3K inhibitors BEZ235,
which is a PI3K and mTOR inhibitor and BKM120, which is a pan-class
I PI3K inhibitor and the selective FGFR inhibitor AZD4547 a potent
selective ATP-competitive receptor tyrosine kinase inhibitor of
FGFR1-3 (all from Selleck Chemicals) were used. The drugs were
diluted in DMSO in order to generate the stock concentrations and
stored at −20°C. These stocks were further diluted with PBS and
added directly to the cells in media to achieve the end
concentrations. For BEZ235 and BKM120 concentrations of 0.25–5
µM were used, while for AZD4547, concentrations of 5–50
µM were used, and in combination experiments the highest
concentrations were generally avoided. All experiments were
repeated at least three times. Overall, after treatment the cells
were incubated for 24–72 h, or up to 120 h, and assays were applied
to investigate cell viability, cytotoxicity and apoptosis for all
cell lines when using one drug at a time. Thereafter, proliferation
tests were performed on HPV+ UPCI-SCC-154 and
HPV− UT-SCC-60A using all three drugs one by one. After
these initial experiments, combinations of AZD4547 and BEZ235 were
tested on HPV+ UPCI-SCC-154 and HPV−
UT-SCC-60A, and viability, cytotoxicity, apoptosis and
proliferation was followed for 24–72 h or up to 120 h. All
experiments were repeated at least three times unless indicated
otherwise. For most figures, a summary of at least three
experiments are presented, however, for proliferation tests,
representative experiments are shown. The ratio-to-control
parameter was calculated by the following formula:
Ratio/Control=(mean experimental values-mean blank values)/(mean
PBS control values-mean blank values), for both the WST-1 and the
cytotoxicity assay. For the apoptosis assay the ratio-to-control
parameter was calculated in comparison to the PBS control.
WST-1 viability assay
Three 96-well plates were seeded and treated the
same way to examine three time-points (for details of seeding see
above). Duplicates were used for each single drug concentration,
and triplicates for drug combinations, PBS was used as a control
and two wells with medium alone, were used to measure the blank
background. After 24, 48 and 72 h, 10 µl of the WST-1
reagent (Roche, Mannheim, Germany) was added. The plates were
incubated for 1 h at 37°C in the incubator and the absorbance was
measured at 450 nm with a Versa Max microplate reader (Molecular
Devices). In the analysis the average blank was subtracted from
every value. The average of the duplicates/triplicates was
calculated. The average values were normalized to the average
control treated with PBS. In addition, the IC50
(inhibitory concentration 50%) of the drugs on different cell lines
were determined from log concentrations-effect curves in GraphPad
Prism (GraphPad Software Version 8) using non-linear regression
analysis.
CellTox Green Cytotoxicity assay
The CellTox Cytotoxicity assay (Promega) was
performed after 48, 72, 96 and 120 h and 5,000 cells were seeded
out in 96-well plates Corning Costar black (Merk) with clear flat
bottom and an end-volume of 80 µl. For each drug
concentration triplicates were prepared, PBS was added to the
negative and positive controls and medium was filled into two blank
wells. Before measurement 4 µl cell lysis solution (Promega)
was added to the positive controls and the plates were incubated
for 30 min (min) at 37°C in the incubator. Then 20 µl
CellTox green dye solution, 1:100 dye to buffer assay (both from
Promega Corporation), was added to treated, blank, and control
wells. The plates were wrapped in tin foil to avoid light
irradiation and incubated on the shaker at RT for 15 min. The read
out was performed on the SPARK 10M multimode microplate plate
reader (Tecan Group Ltd.) with 485 nm excitation and 520 nm
emission.
Caspase-Glo 3/7 apoptosis assay
The Caspase-Glo 3/7 Assay was done in duplicates
immediately afterwards in the plates used for the CellTox Green
Cytotoxicity Assay. 100 µl reagent (Promega Corporation) was
added to every treated well except the positive control of the Cell
Tox Green Cytotoxicity Assay. The plates wrapped in foil were
incubated at RT for one hour on the shaker. Luminescence was
measured with the Centro LB 960 Microplate Luminometer (Berthold
Technologies).
Proliferation assay
Cell proliferation was measured within the
xCELLigence RTCA DP instrument, and by using E-plate VIEW 16 plates
(ACEA Biosciences Inc.). In order to measure the background, 50
µl medium was added into each well and set in the cradle and
starting step one to measure the background. The reads should be
below a cell index of 0.6, where the cell index is defined as
changes in response to cell number and morphology. Afterwards,
5,000 cells in 50 µl medium of the UT-SCC-60A and
UPCI-SCC-154 cell lines were seeded out in the E-Plate VIEW 16 to a
total end volume of 100 µl. The plates were left under the
hood for 30 min at RT to settle and then inserted back into the
reader. Step two was continued with sweeps of the plates initiated
every 2 h during the 72 h experimental run. Step two was paused
after 24 h. The drugs and PBS as a control were added; afterwards
step three was continued for at least 84 h, also with sweeps every
2 h.
Statistical analysis
To evaluate the effects of the single and the
combination treatments a multiple t-test followed by a correction
for multiple comparison of the means according to the Holm-Sidak
method was performed. The combination effects were evaluated
according to the effect-based approach ‘Highest Single Agent’
(37), which reports if the
resulting effect of a drug combination (EAB) is greater
than the effects produced by any of combined individual drugs
(EA and EB). A combination index (CI) was
calculated with the following formula: CI=Max(EA or
B)/EAB. A CI<1 was defined as a positive
combination effect and CI>1 as a negative combination effect.
Two-tailed unpaired Student's t-test was used to examine the
difference in means between two treatments. A P<0.05 was
considered as statistically significant.
Results
Analysis of FGFR3 and PIK3CA mutations
in HPV+ UPCI-SCC-154 and UM-SCC-47 cells, and
HPV− UT-SSC-60A cells by CAST-PCR
DNA was extracted successfully from all three cell
lines, but none of the FGFR3 and PIK3CA mutations, assayed for by
the CAST-PCR, were detected (data not shown).
Treatment of HPV+ UPCI
-SCC-154 and UM-SCC-47 and HPV− UT-SCC-60A with FGFR and
PI3K inhibitors independently
WST-1 viability analysis after
treatment with FGFR and PI3K inhibitors independently on
HPV+ UPCI-SCC-154 and UM-SCC-47 and HPV−
UT-SCC-60A
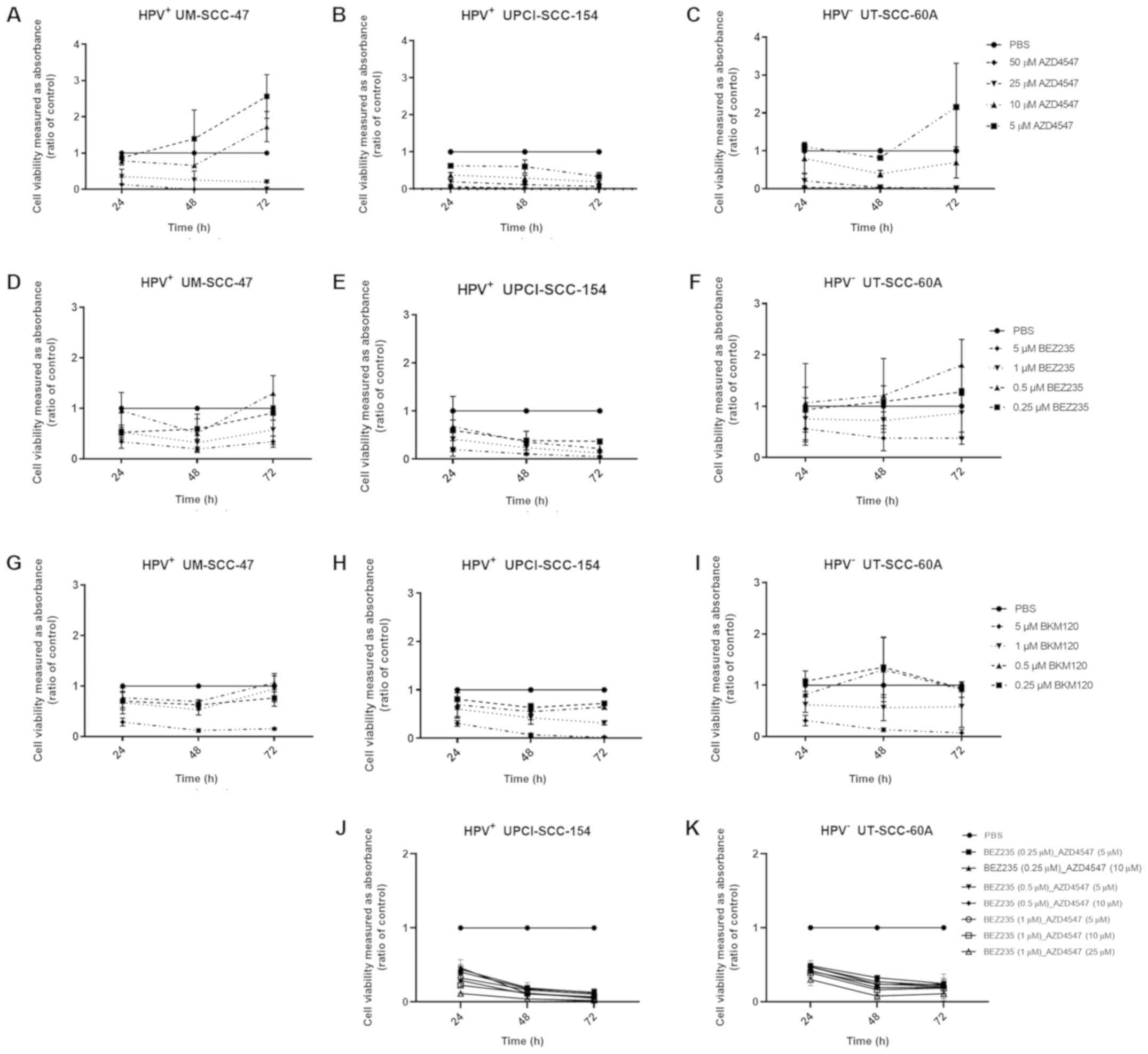
To examine effects of FGFR and PI3K inhibitors on
viablility of TSCC/BOTSCC cell lines, WST-1 viability assays were
performed 24, 48 and 72 h after treatment with FGFR inhibitor
AZD4547 and PI3K inhibitors BEZ235 and BKM120 on HPV+
UM-SCC-47 and UPCI-SCC-154 and HPV− UT-SCC-60A. Data for
three combined experiments using different concentrations of the
drugs are shown in Fig. 1.
In general, all cell lines were sensitive for
treatment with the FGFR and PI3K inhibitors. HPV+
UPCI-SCC-154 was slightly more sensitive than the other two cell
lines to AZD4547, and showed ≥50% decreased absorbance representing
cell viability after treatment with 5 µM AZD4547 after 72 h,
and with 10, 25 and 50 µM AZD4547 after 24, 48 and 72 h (at
least P<0.05 for all) (Fig. 1B).
Similar effects with ≥50% decreased absorption were obtained for
HPV+ UM-SCC-47 and HPV− UT-SCC-60A after
treatment with 25 and 50 µM AZD4547 at 24, 48 and 72 h (at
least P<0.05 for all) (Fig. 1A and
C, respectively).
HPV+ UPCI-SCC-154 was in general also
slightly more sensitive to treatment with BEZ235 and BKM120
compared to the other two cell lines. HPV+ UM-SCC-47 and
UPCI-SCC-154 both showed ≥50% decreased absorbance after treatment
with 1 and 5 µM BEZ235 after 24 and 48 h, and UPCI-SCC-154
also at 72 h (at least P<0.01) (Fig.
1D and E, respectively). Moreover, UPCI-SCC-154 also showed
≥50% decreased absorbance with 0.25 and 0.5 µM BEZ235 at 48
and 72 h (at least P<0.01 for all) (Fig. 1E). For HPV− UT-SCC-60A a
≥50% decrease was obtained only with the highest concentration i.e.
5 µM BEZ235 after 48 and 72 h (at least P<0.05, for all)
(Fig. 1F).
For BKM120, all cell lines reached a significant
decrease for 5 µM BKM120 at 24, 48 and 72 h (at least
P<0.001 for all) (Fig. 1G-I). A
significant decrease was obtained for UPCI-SCC-154 also at 1
µM of BKM120 at 48 and 72 h, while for UM-SCC-47 and
UT-SCC-60A respectively, significance was obtained only
sporadically i.e. after 48 and 24 h respectively, (at least
P<0.05, for all) (Fig. 1G-I).
Finally, the IC50 for each cell line for
each drug were determined (Table I).
Taken together these data confirm that the UPCI-SCC-154 was in
general more sensitive than the other two cell lines to all
drugs.
 | Table I.WST-1 viability analysis following
treatment with FGFR inhibitor AZD4547 and PI3K inhibitors BEZ235
and BKM120 for 24, 48 and 72 h. |
Table I.
WST-1 viability analysis following
treatment with FGFR inhibitor AZD4547 and PI3K inhibitors BEZ235
and BKM120 for 24, 48 and 72 h.
|
|
|
| IC50
(µM) |
|---|
|
|
|
|
|
|---|
| Cell line | Target | Drug | 24 h | 48 h | 72 h |
|---|
| UPCI-SCC-154
(HPV+) | FGFR | AZD4547 | 7.24 | 6.29 | 2.86 |
|
| PI3K | BKM120 | 1.67 | 0.57 | 0.62 |
|
|
| BEZ235 | 0.74 | 0.12 | 0.14 |
| UM-SCC-47
(HPV+) | FGFR | AZD4547 | 18.24 | 16.32 | NA |
|
| PI3K | BKM120 | 1.85 | 0.89 | 2.78 |
|
|
| BEZ235 | 1.90 | 0.44 | 2.71 |
| UT-SCC-60A
(HPV−) | FGFR | AZD4547 | 16.33 | 8.55 | 10.41 |
|
| PI3K | BKM120 | 2.12 | 1.01 | 1.21 |
|
|
| BEZ235 | 5.99 | 3.47 | 4.29 |
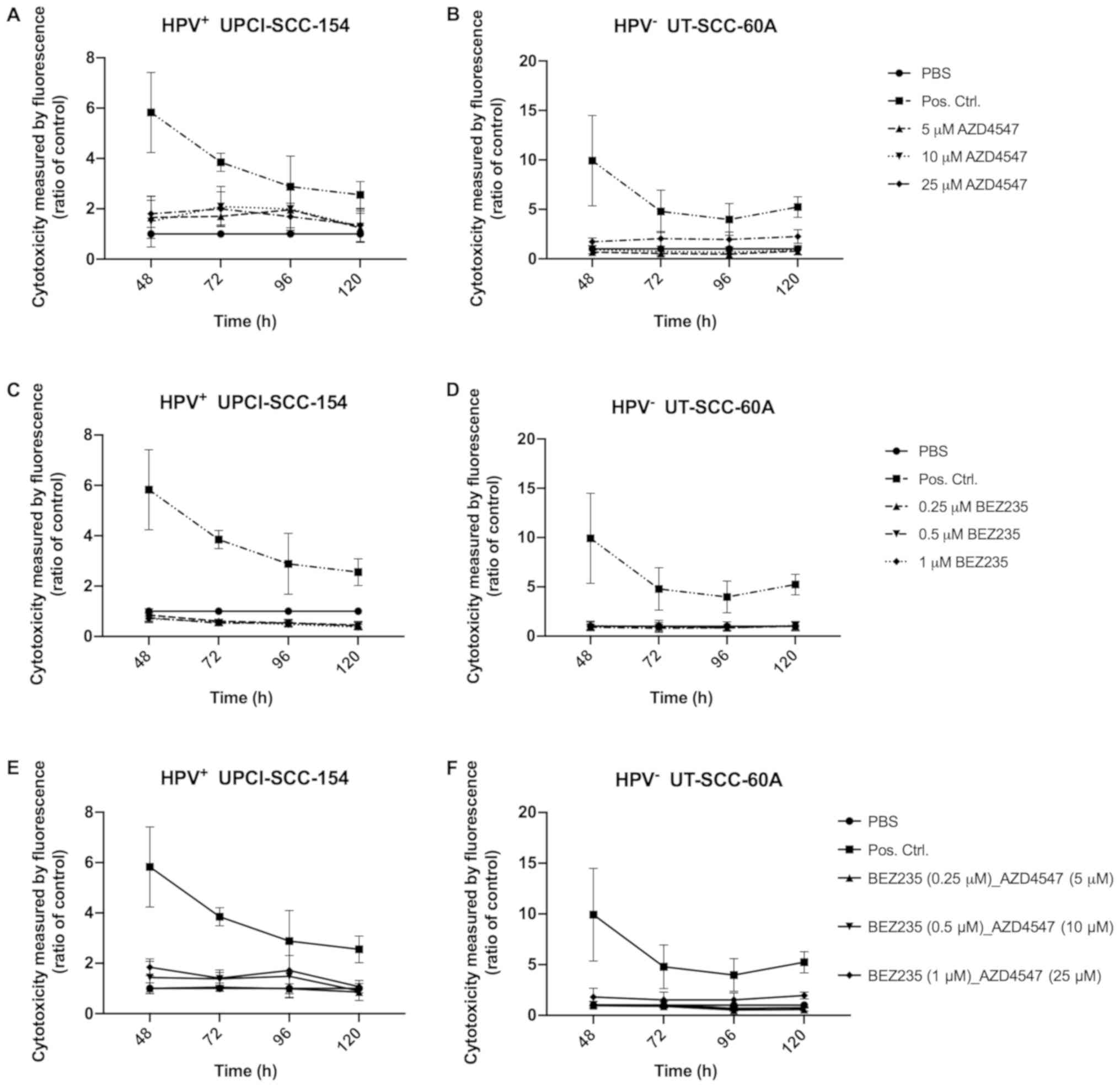
CellTox Green Cytotoxicity Assay with
FGFR and PI3K inhibitors independently on HPV+
UPCI-SCC-154 and HPV− UT-SCC-60A
CellTox Green Cytotoxicity Assays on HPV+
UPCI-SCC-154 and HPV− UT-SCC-60A were performed in four
different experiments 48, 72, 96 and 120 h after treatment with
FGFR inhibitor AZD4547 and PI3K inhibitor BEZ235 to assess
cytotoxic effects in correlation to PBS and lysis buffer
respectively (Fig. 2).
After treatment with 5, 10 and 25 µM AZD4547,
UPCI-SCC-154 showed a trend of increased cytotoxicity as compared
to the PBS control, but this was significant only for 5 µM
at 72 h (P<0.05) (Fig. 2A). For
UT-SCC-60A, the cytotoxic effect seemed lower, however at 48 and
120 h after treatment with 25 µM AZD4547 there was a
significant difference in cytotoxicity when compared to the PBS
control (at least P<0.05) (Fig.
2B). For treatment with BEZ235, there were no significant
increases in cytotoxicity compared to PBS (Fig. 2C and D).
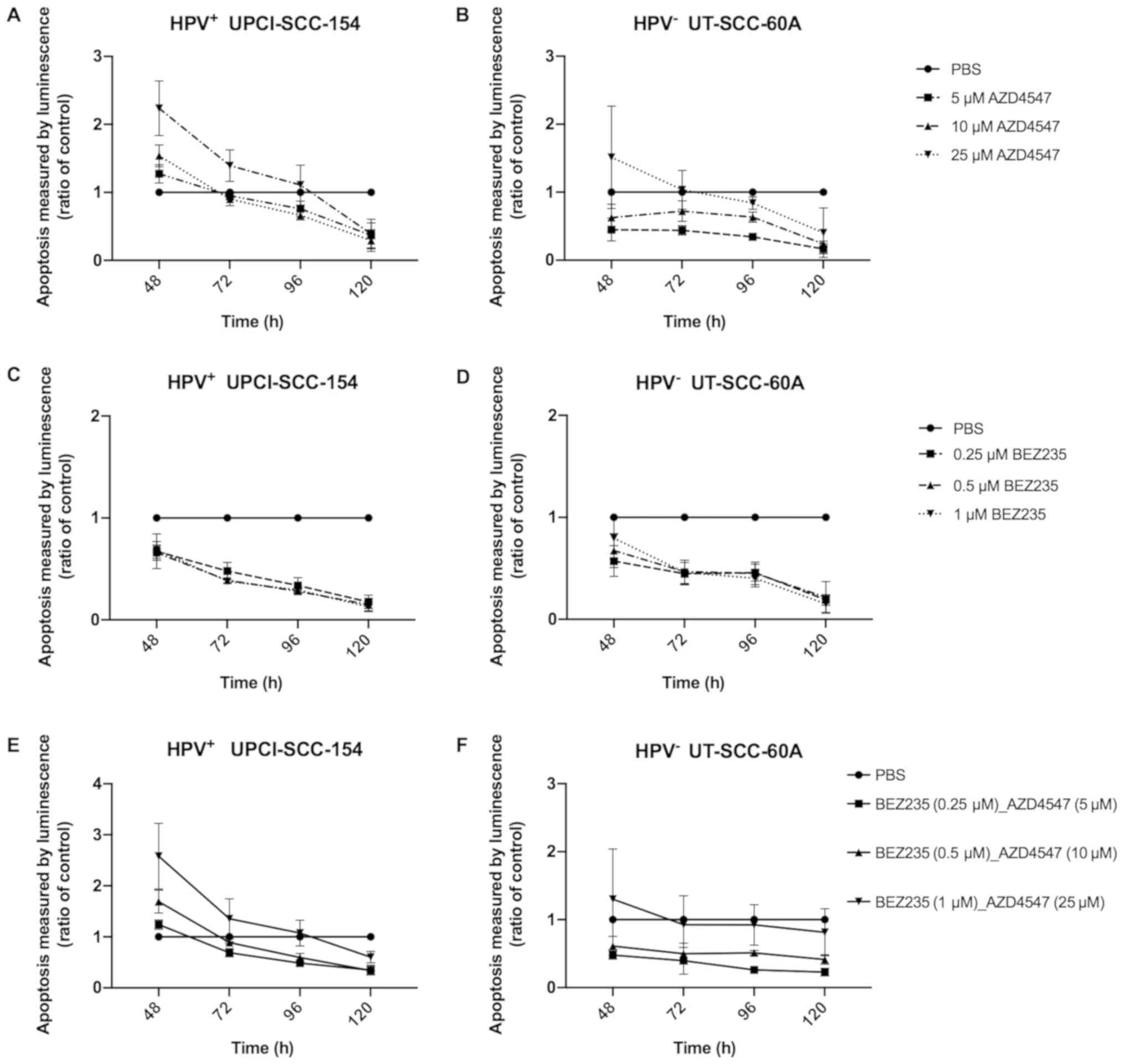
Caspase-Glo 3/7 apoptosis-assay with
FGFR and PI3K inhibitors independently on HPV+
UPCI-SCC-154 and HPV− UT-SCC-60A
To evaluate the effects on viability Caspase-Glo 3/7
apoptosis assays were done 48, 72, 96 and 120 h after treatment
with FGFR inhibitor AZD4547 and PI3K inhibitor BEZ235 at various
concentrations normalized against PBS on HPV+
UPCI-SCC-154 and HPV− UT-SCC-60A. Again, HPV+
UPCI-SCC-154 had a tendency to be more sensitive than
HPV− UT-SCC-60A as shown in Fig. 3.
After 48 h HPV+ UPCI-SCC-154 showed an
increase in apoptosis with 10 and 25 µM AZD4547 (at least
P<0.05) (Fig. 3A). For UT-SCC-60A
a similar weak trend was observed for the 25 µM
concentration, however statistical significance was not reached
(Fig. 3B).
Neither HPV+ UPCI-SCC-154 (Fig. 3C) nor HPV− UT-SCC-60A
(Fig. 3D) showed increased
luminescence representing apoptosis, after treatment with any of
the concentrations of BEZ235.
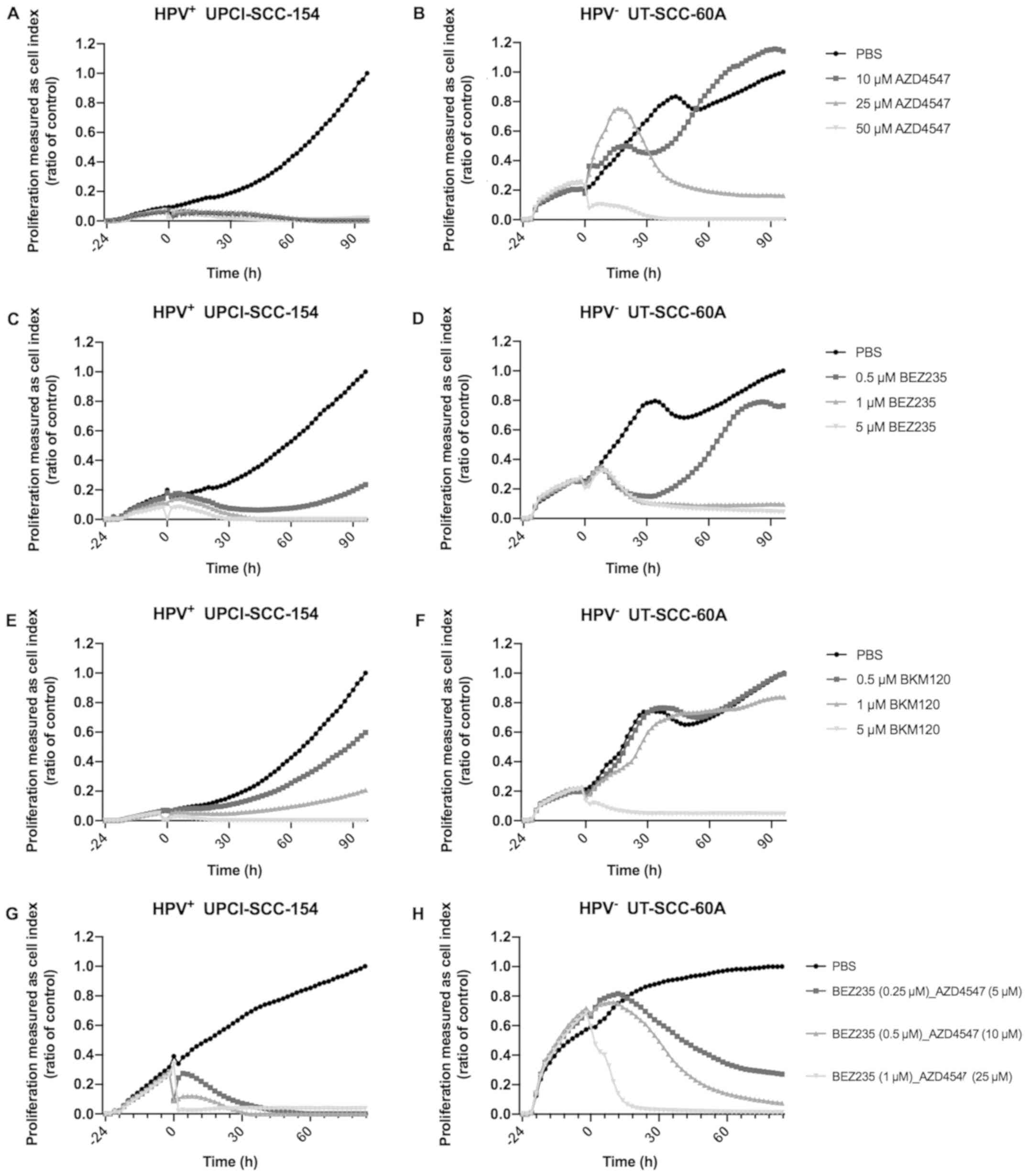
Proliferation assay using the
xCELLigence System after treatment with FGFR and PI3K inhibitors
independently of HPV+ UPCI-SCC-154 and HPV−
UT-SCC-60A
Lastly, to evaluate effects on cell growth,
proliferation assays using the xCELLigence System after treatment
with FGFR inhibitor AZD4547 and PI3K inhibitors BEZ235 and BKM120
on the cell lines HPV+ UPCI-SCC-154 and HPV−
UT-SCC-60A were performed (Fig.
4).
HPV+ UPCI-SCC-154 showed a complete
inhibition of proliferation after treatment with 10, 25 and 50
µM AZD4547 as compared to treatment with PBS during the
whole observation period of 96 h (Fig.
4A). For HPV− UT-SCC-60A treatment with 50 µM
AZD4547 also inhibited proliferation almost throughout the whole
observation of 96 h as compared to treatment with PBS, and in
addition, treatment with 25 µM, but not with 10 µM
AZD4547 eventually inhibited proliferation between 60 and 96 h
(Fig. 4B).
Treatment with 1 and 5 µM of BEZ235, and 5
µM BKM120 almost completely inhibited HPV+
UPCI-SCC-154 proliferation through the 96 h observation period as
compared to PBS, and when treating with lower doses, both BEZ235
and BKM120 induced decreased proliferation when compared to PBS
(Fig. 4C and E, respectively). For
HPV− UT-SCC-60A treatment with 1 and 5 µM BEZ235
and 5 µM BKM120 respectively inhibited proliferation almost
completely as compared to PBS, during most of the 96 h observation
period, while lower concentrations were not as efficient, and
especially not for BKM120 (Fig. 4D and
F).
Treatment of HPV+
UPCI-SCC-154 and HPV− UT-SCC-60A with FGFR and PI3K
inhibitors AZD4547 and BEZ235 combined
Clearly there were inhibitory effects on growth
using FGFR inhibitor AZD4547 and PI3K inhibitors BEZ235 and BKM120
on all the cell lines as shown above. To further investigate their
therapeutic potential we now wanted to examine if lower drug
concentrations could be used when combining two different types of
drugs or whether they had an antagonistic effect. For this purpose,
cell viability, cytotoxicity, apoptosis and proliferation (using
the WST-1 assay, Cell-Tox Green Cytotoxicity Assay, Caspase-Glo 3/7
assay, and the xCELLigence System respectively) of HPV+
UPCI-SCC-154 and HPV− UT-SCC-60A were measured after
combined treatment with the FGFR AZD4547 and the PI3K BEZ235
inhibitors. In this respect, as indicated above HPV+
UPCI-SCC-154 was an example of a generally more sensitive cell
line, while HPV− UT-SCC-60A was an example of a more
resistant cell line.
WST-1 viability analysis after
treatment with FGFR and PI3K inhibitors AZD4547 and BEZ235 combined
on HPV+ UPCI-SCC-154 and HPV− UT-SCC-60A
WST-1 viability assays were performed 24, 48 and 72
h after treatment with FGFR inhibitor AZD4547 at concentrations 5,
10 and 25 µM combined with PI3K inhibitor BEZ235 at
concentrations 0.25, 0.5 and 1 µM on cell lines
HPV+ UPCI-SCC-154 and HPV− UT-SCC-60A (for
details see Fig. 1J and K,
respectively). All combinations decreased viability ≥50% at 24, 48
and 72 h of both cell lines (at least P<0.001 for all) (Fig. 1J and K). Furthermore, for
HPV+ UPCI-SCC-154 viability decreased to ~90% for all
concentrations at 72 h (Fig.
1J).
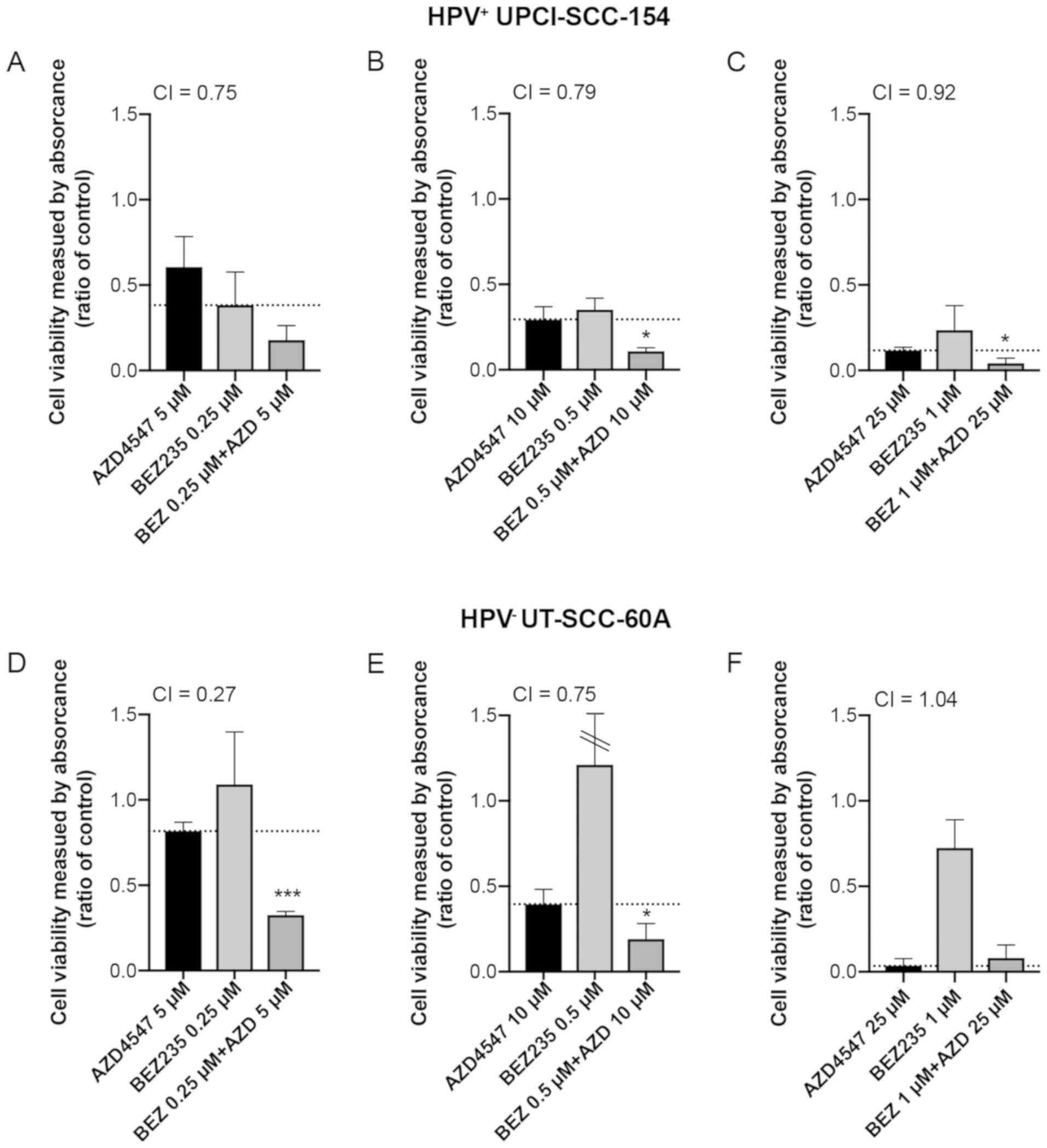
Viability was also illustrated as staple diagrams
for single doses of AZD4547 and BEZ235 as well as combined for
HPV+ UPCI-SCC-154 and HPV− UT-SCC-60A at 48 h
in Fig. 5 (with the same
concentrations of drug combinations shown in Figs. 1–3)
and Fig. S1 (with additional drug
combinations, not tested in the other assays). We also calculated
the combination index (CI) for each combination in order to
investigate whether we had a positive or negative combination
effect, these CIs are displayed in Figs.
5 and S1 as well as summarized
for all time-points in Fig. S2.
At 48 h, a decrease in viability was significant for
HPV+ UPCI-SCC-154 in four out of seven tested
combinations (Fig. 5A-C and S1A-D together) and for HPV−
UT-SCC-60A in six of seven (Figs.
5D-F and S1E-H together).
Especially for HPV− UT-SCC-60A in the lower dose
combinations (AZD4547 5 µM and BEZ235 0.25 or 0.5 µM)
a clear synergistic effect was observed, where the single drugs had
minor (AZD4547) or no effect (BEZ235), while the combination
induced a strong effect on cell viability (Figs. 5D and S1E).
In most cases, with four statistically
non-significant exceptions, the highest concentrations of AZD4547
and BEZ235 in HPV− UT-SCC-60A at all time-points, and
the 10 µM of AZD4547 and 0.25 µM BEZ235 combination
for HPV+ UPCI-SCC-154 at 24 h) the combined treatment
tended to induce greater viability effect than single treatments,
shown by CIs<1 (Fig. S2).
CellTox Green Cytotoxicity Assay with
FGFR and PI3K inhibitors AZD4547 and BEZ235 combined on
HPV+ UPCI-SCC-154 and HPV− UT-SCC-60A
CellTox Green Cytotoxicity assays were done 48, 72,
96 and 120 h after treatment with FGFR inhibitor AZD4547 at
concentrations of 5, 10 and 25 µM combined with PI3K
inhibitor BEZ235 at concentrations of 0.25, 0.5 and 1 µM,
respectively on HPV+ UPCI-SCC-154 and HPV−
UT-SCC-60A. Except for UPCI-SCC-154 at the highest combination at
48 h, and for UT-SCC-60A at the highest combination at 120 h (at
least P<0.05 for both), there were no other significant
increases in induced cytotoxicity with any of the drug combinations
at any of the time-points as compared to PBS (Fig. 2E and F).
Caspase-Glo 3/7 apoptosis-assay with
FGFR and PI3K inhibitors AZD4547 and BEZ235 combined on
HPV+ UPCI-SCC-154 and HPV− UT-SCC-60A
Caspase-Glo 3/7 apoptosis-assays were performed 48,
72, 96 and 120 h after treatment with FGFR inhibitor AZD4547 at
concentrations 5, 10 and 25 µM combined with PI3K inhibitor
BEZ235 at concentrations 0.25, 0.5 and 1 µM on
HPV+ UPCI-SCC-154 and HPV− UT-SCC-60A. An
increase in apoptosis being proportional to measured luminescence
>50% was observed for UPCI-SCC-154 with all drug combinations at
48 h (at least P<0.05 for all) (Fig.
3E). For UT-SCC-60A, possibly a tendency of an increase in
luminescence was observed for the highest drug combination at 48 h
only (P=ns) (Fig. 3F).
Proliferation assay using the
xCELLigence System after combined treatment with FGFR and PI3K
inhibitors AZD4547 and BEZ235 of HPV+ UPCI-SCC-154 and
HPV− UT-SCC-60A
Proliferation assays were performed up to 84 h
after combined treatment with FGFR inhibitor AZD4547 at
concentrations 5, 10 and 25 µM and PI3K inhibitor BEZ235 at
concentrations 0.25, 0.5 and 1 µM, respectively, on
HPV+ UPCI-SCC-154 and HPV− UT-SCC-60A
(Fig. 4G and H). Compared to PBS
alone, treatment with all combination treatments inhibited
proliferation of HPV+ UPCI-SCC-154 during most of the
observation period of 84 h (Fig.
4G).
Likewise, compared to PBS alone, treatment with all
combinations inhibited the proliferation of HPV−
UT-SCC-60A, however the lower doses of 5 and 10 µM AZD4547
and 0.25 and 0.5 µM of BEZ235, were not as efficient as the
highest dose (Fig. 4H).
Discussion
This study has investigated the sensitivity of
HPV+ and HPV− TSCC/BOTSCC cell lines
UPCI-SCC-154, UM-SCC-47 and UT-SCC-60A to FGFR inhibitor AZD4547,
and PI3K inhibitors BEZ235 and BKM120 and disclosed that they were
more or less sensitive to all inhibitors. Furthermore, taken
together the data indicated that HPV+ UPCI-SCC-154 was
generally more sensitive than the HPV+ UM-SCC-47 and
HPV− UT-SCC-60A cell lines to these afore mentioned
drugs. In addition, combination treatments with the FGFR inhibitor
AZD4547 and PI3K inhibitor BEZ235, allowed for the possibility to
decrease the concentrations of the respective FGFR and PI3K
inhibitors, especially for the less sensitive HPV−
UT-SCC-60A cell line.
The above was illustrated e.g. by the fact that
viability for HPV+ UPCI-SCC-154 was decreased for all
doses of each drug at all three time-points 24, 48 and 72 h. In
contrast, for cell lines HPV+ UM-SCC-47 and
HPV− UT-SCC-60A, viability went down early on at 24 h
and then increased considerably for the two lower doses of AZD4547
and BEZ235 at later time-points 48 and 72 h, although at higher
AZD4547 and BKM120 concentrations both latter cell lines did also
respond. We also analyzed the combinations according to the Highest
Single Agent approach. It is a simple, but yet clinically relevant
method which provides evidence of the superiority of the drug
combination compared to its single agents. However, it offers
limited evidence of synergy except when at least one drug is
inactive as a single treatment, which was the case for several
combinations in the resistant cell line HPV− UT-SCC-60A.
Likewise, in the WST-1 viability assay, HPV+
UPCI-SCC-154 was sensitive at all doses at all time-points 24–72 h
in contrast to the two other cell lines, where sensitivity was only
observed at 24 and 72 h only with the highest doses.
Cytotoxicity measured by CellTox Green Cytotoxicity
assay was not really obvious for any of the cell lines, and
apoptosis was observed at 48 h only for the most efficient
concentrations of the AZD4547. For BEZ235 none of the
concentrations showed efficient increases in luminescence for
HPV+ UPCI-SSC-154 nor HPV− UT-SCC-60A,
suggesting that apoptosis was not observed during the 48–120 h
observation period. The proliferation assays however, showed a
similar profile to the viability assays, with a very strong
inhibition of HPV+ UPCI-SCC-154 with the majority of the
used concentrations of AZD4547 and BEZ235, while the proliferation
curves were more diverse for HPV− UT-SCC-60A.
Together the data indicated a sensitivity of these
cell lines to the above-mentioned drugs. Furthermore, when
combining FGFR and PI3K inhibitors, we found that the positive
effects were mainly seen on the more resistant cell line, since the
sensitive one was more or less sensitive to all doses. Moreover,
the different assays complemented each other quite well, since e.g.
viability (WST-1 assay), apoptosis (Caspase-Glo 3/7
apoptosis-assay) and proliferation (xCELLigence System) for each
cell line treated with a specific drug and dose seemed to correlate
relatively well. Nonetheless, the cytotoxicity assay (CellTox Green
Cytotoxicity Assay) did not indicate that much, which we interpret
as that the obtained effect, was an inhibition of proliferation,
rather than a cytotoxic crisis.
The use of these types of drugs have frequently
been tested on urothelial cancer cell lines in vitro and
synergistic effects have also been reported, as has the
documentation that using one drug alone often can result in the
gain of resistance to that drug (38–40). The
same type of drugs have also been used on head and neck cancer cell
lines in vitro, and in some cases on HPV+ and
HPV− head and neck squamous cell carcinoma lines and
similar drug concentrations were used (41–43).
To our knowledge this is the first time, that drugs
against FGFR and PI3K were used in combination for treatment of
HPV+ and HPV− TSCC/BOTSCC cell lines,
although the combination of AZD4547 and an MTOR inhibitor has been
used on a nasopharyngeal carcinoma cell line (44). Furthermore, to our knowledge, these
drugs have not been used under physiological conditions as single
treatments on patients with HPV+ TSCC/BOTSCC or combined
treatments for patients with HPV+ or HPV−
TSCC/BOTSCC. Nevertheless, when comparing the doses to those used
on urinary bladder cell lines, where data are more frequently
available, similar doses are used, but our cell lines are possibly
slightly more resistant than the urinary bladder cancer cell lines
(38). Then on the other hand, in
contrast to the urinary bladder cancer cell lines, our cell lines
do to our knowledge not contain any modifications/fusion products
of the most commonly mutated FGFR or PI3K gene variants. Such cell
lines remain to be tested. However, it has also been reported in a
phase I study, that the presence of PI3K mutations or not, does not
always influence the sensitivity to a clinical response (45). Nevertheless, in other situations PI3K
mutations have been reported to influence the responses of FGFR
inhibitors used to combat cervical tumors with FGFR3-TACC3-fusion
genes (46). In that paper, the
authors therefore suggest the use of combination treatments.
There are limitations in this study, since only
three cell lines were tested. On the other hand, the cell lines
that have been tested are representative of the types of cell lines
used in these circumstances (34–36).
Furthermore, several types of assays were used and they all
indicated that the UPCI-SCC-154 cell line was most likely more
sensitive than the UM-SCC-47 or the UT-SCC-60A. For future studies,
other cell lines with FGFR or PI3K mutations could be of use and
such cell lines could very well be more sensitive to FGFR and PI3K
targeted therapy. On the other hand, several publications have
indicated that this is the case in some, but not all circumstances
(47). Definitely, more knowledge on
the subject will be required.
There are also other limitations, in that
resistance can arise to either FGFR and PI3K inhibitors when used
as single treatments and that these treatments can also result in
serious side effects (38,47–49).
However, if lower doses can inhibit both the development of
resistance and induce fewer side effects this would definitely be
of benefit for the patients.
Finally, one last limitation in this study was that
detailed mechanistic studies were not performed. It is known that
BEZ235 is both a PI3K and MTOR inhibitor, this allows for multiple
effects. In this study, we only were aware of that our cells did
not exhibit PIK3CA mutations, and we do not know if MTOR was
deregulated. Therefore, it is possible that BEZ235 exhibited
multiple effects on our cell lines. Likewise, AZD4547 can target
more than one tyrosine kinase, and we have on our cell lines
focused on FGFR3 mutations, therefore also here AZD4547 could also
have more than one effect. To scrutiny such mechanisms in more
detail, additional studies with more selective drugs, and more
detailed information on the nature of targets of the drugs utilized
in the studied cell lines could be of interest.
To conclude, FGFR inhibitor AZD4547 and PI3K
inhibitors BEZ235 and BKM120 were tested alone on HPV+
UM-SCC-47, and UPCI-SCC-154 and HPV− UT-SCC-60A, and
AZD4547 and BEZ235 combined on the latter two cell lines. All cell
lines were found more or less sensitive to treatment with these
drugs, and especially for the latter two cell lines when used in
combination. Nonetheless, more studies also testing TSCC/BOTSCC
cell lines with FGFR3 and PIK3CA mutations should be useful in the
future.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Swedish
Cancer Foundation (grant no. 18 0440), the Stockholm Cancer Society
(grant no. 181053), the Swedish Cancer and Allergy Foundation
(grant no. 190), the Royal Swedish Academy of Sciences (grant no.
2017-2018), the Stockholm City Council (grant no. 20180037),
Karolinska Institutet (grant no. 2018:0007) and Stiftelsen Clas
Groschinsky Minnesfond (grant no. M19376).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
SH and ONK performed the majority of the
experiments, interpreted the data, calculated the statistics and
contributed to the writing of the manuscript. AO initiated the
experiments and the interpretation of the initial experiments and
contributed to the writing of the material and methods section. CB
supervised AO, and planned the initial experiments and performed
the CAST-PCR. BKAL and TA collaborated with SH and ONK and
performed several experiments. BKAL contributed to the writing of
the manuscript together with SH and ONK. TR assisted AO when
necessary and contributed to the calculation of the statistics. MW
assisted TA with the planning of some experiments, and performed
final interpretation and presentation of the data and the
statistics of the manuscript. TD made substantial contributions to
conception and design, acquisition of data, analysis and
interpretation of data and has been involved in drafting the
manuscript and revising it critically for important intellectual
content. TD has also given final approval of the version to be
published and has agreed to be accountable for all aspects of the
work in ensuring that questions related to the accuracy and
integrity of any part of the work are appropriately investigated
and resolved. All authors critically read and approved the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mellin H, Friesland S, Lewensohn R,
Dalianis T and Munck-Wikland E: Human papillomavirus (HPV) DNA in
tonsillar cancer: Clinical correlates, risk of relapse, and
survival. Int J Cancer. 89:300–304. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dahlgren L, Dahlstrand HM, Lindquist D,
Högmo A, Björnestål L, Lindholm J, Lundberg B, Dalianis T and
Munck-Wikland E: Human papillomavirus is more common in base of
tongue than in mobile tongue cancer and is a favorable prognostic
factor in base of tongue cancer patients. Int J Cancer.
112:1015–1019. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Attner P, Du J, Näsman A, Hammarstedt L,
Ramqvist T, Lindholm J, Marklund L, Dalianis T and Munck-Wikland E:
Human papillomavirus and survival in patients with base of tongue
cancer. Int J Cancer. 128:2892–2897. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sivars L, Näsman A, Tertipis N, Vlastos A,
Ramqvist T, Dalianis T, Munck-Wikland E and Nordemar S: Human
papillomavirus and p53 expression in cancer of unknown primary in
the head and neck region in relation to clinical outcome. Cancer
Med. 3:376–384. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dalianis T: Human papillomavirus and
oropharyngeal cancer, the epidemics, and significance of additional
clinical biomarkers for prediction of response to therapy (review).
Int J Oncol. 44:1799–1805. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marklund L, Näsman A, Ramqvist T, Dalianis
T, Munck-Wikland E and Hammarstedt L: Prevalence of human
papillomavirus and survival in oropharyngeal cancer other than
tonsil or base of tongue cancer. Cancer Med. 1:82–88. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Robinson KL and Macfarlane GJ:
Oropharyngeal cancer incidence and mortality in scotland: Are rates
still increasing? Oral Oncol. 39:31–36. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hammarstedt L, Lindquist D, Dahlstrand H,
Romanitan M, Dahlgren LO, Joneberg J, Creson N, Lindholm J, Ye W,
Dalianis T and Munck-Wikland E: Human papillomavirus as a risk
factor for the increase in incidence of tonsillar cancer. Int J
Cancer. 119:2620–2623. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sturgis EM and Cinciripini PM: Trends in
head and neck cancer incidence in relation to smoking prevalence:
An emerging epidemic of human papillomavirus-associated cancers?
Cancer. 110:1429–1435. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Näsman A, Attner P, Hammarstedt L, Du J,
Eriksson M, Giraud G, Ahrlund-Richter S, Marklund L, Romanitan M,
Lindquist D, et al: Incidence of human papillomavirus (HPV)
positive tonsillar carcinoma in Stockholm, Sweden: An epidemic of
viral-induced carcinoma? Int J Cancer. 125:362–366. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Braakhuis BJ, Visser O and Leemans CR:
Oral and oropharyngeal cancer in the Netherlands between 1989 and
2006: Increasing incidence, but not in young adults. Oral Oncol.
45:e85–89. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Attner P, Du J, Näsman A, Hammarstedt L,
Ramqvist T, Lindholm J, Marklund L, Dalianis T and Munck-Wikland E:
The role of human papillomavirus in the increased incidence of base
of tongue cancer. Int J Cancer. 126:2879–2884. 2010.PubMed/NCBI
|
|
13
|
Chaturvedi AK, Engels EA, Pfeiffer RM,
Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M,
Cozen W, et al: Human papillomavirus and rising oropharyngeal
cancer incidence in the United States. J Clin Oncol. 29:4294–4301.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Näsman A, Nordfors C, Holzhauser S,
Vlastos A, Tertipis N, Hammar U, Hammarstedt-Nordenvall L, Marklund
L, Munck-Wikland E, Ramqvist T, et al: Incidence of human
papillomavirus positive tonsillar and base of tongue carcinoma: A
stabilisation of an epidemic of viral induced carcinoma? Eur J
Cancer. 51:55–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Haeggblom L, Attoff T, Yu J, Holzhauser S,
Vlastos A, Mirzae L, Ährlund-Richter A, Munck-Wikland E, Marklund
L, Hammarstedt-Nordenvall L, et al: Changes in incidence and
prevalence of human papillomavirus in tonsillar and base of tongue
cancer during 2000–2016 in the Stockholm region and Sweden. Head
Neck. 41:1583–1590. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mirghani H, Amen F, Blanchard P, Moreau F,
Guigay J, Hartl DM and Lacau St Guily J: Treatment de-escalation in
HPV-positive oropharyngeal carcinoma: Ongoing trials, critical
issues and perspectives. Int J Cancer. 136:1494–1503. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Näsman A, Bersani C, Lindquist D, Du J,
Ramqvist T and Dalianis T: Human papillomavirus and potentially
relevant biomarkers in tonsillar and base of tongue squamous cell
carcinoma. Anticancer Res. 37:5319–5328. 2017.PubMed/NCBI
|
|
18
|
Bersani C, Mints M, Tertipis N, Haeggblom
L, Sivars L, Ährlund-Richter A, Vlastos A, Smedberg C, Grün N,
Munck-Wikland E, et al: A model using concomitant markers for
predicting outcome in human papillomavirus positive oropharyngeal
cancer. Oral Oncol. 68:53–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ang KK, Harris J, Wheeler R, Weber R,
Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C,
et al: Human papillomavirus and survival of patients with
oropharyngeal cancer. N Engl J Med. 363:24–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Näsman A, Andersson E, Marklund L,
Tertipis N, Hammarstedt-Nordenvall L, Attner P, Nyberg T, Masucci
GV, Munck-Wikland E, Ramqvist T and Dalianis T: HLA class I and II
expression in oropharyngeal squamous cell carcinoma in relation to
tumor HPV status and clinical outcome. PLoS One. 8:e770252013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Näsman A, Nordfors C, Grün N,
Munck-Wikland E, Ramqvist T, Marklund L, Lindquist D and Dalianis
T: Absent/weak CD44 intensity and positive human papillomavirus
(HPV) status in oropharyngeal squamous cell carcinoma indicates a
very high survival. Cancer Med. 2:507–518. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tertipis N, Haeggblom L, Nordfors C, Grün
N, Näsman A, Vlastos A, Dalianis T and Ramqvist T: Correlation of
LMP10 expression and clinical outcome in human papillomavirus (HPV)
positive and HPV-negative tonsillar and base of tongue cancer. PLoS
One. 9:e956242014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lindquist D, Näsman A, Tarján M,
Henriksson R, Tot T, Dalianis T and Hedman H: Expression of LRIG1
is associated with good prognosis and human papillomavirus status
in oropharyngeal cancer. Br J Cancer. 110:1793–1800. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tertipis N, Villabona L, Nordfors C,
Näsman A, Ramqvist T, Vlastos A, Masucci G and Dalianis T: HLA-A*02
in relation to outcome in human papillomavirus positive tonsillar
and base of tongue cancer. Anticancer Res. 34:2369–2375.
2014.PubMed/NCBI
|
|
25
|
Nordfors C, Grün N, Tertipis N,
Ährlund-Richter A, Haeggblom L, Sivars L, Du J, Nyberg T, Marklund
L, Munck-Wikland E, et al: CD8+ and CD4+
tumour infiltrating lymphocytes in relation to human papillomavirus
status and clinical outcome in tonsillar and base of tongue
squamous cell carcinoma. Eur J Cancer. 49:2522–2530. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ramqvist T, Mints M, Tertipis N, Näsman A,
Romanitan M and Dalianis T: Studies on human papillomavirus (HPV)
16 E2, E5 and E7 mRNA in HPV-positive tonsillar and base of tongue
cancer in relation to clinical outcome and immunological
parameters. Oral Oncol. 51:1126–1131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rietbergen MM, Martens-de Kemp SR,
Bloemena E, Witte BI, Brink A, Baatenburg de Jong RJ, Leemans CR,
Braakhuis BJ and Brakenhoff RH: Cancer stem cell enrichment marker
CD98: A prognostic factor for survival in patients with human
papillomavirus-positive oropharyngeal cancer. Eur J Cancer.
50:765–773. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tinhofer I, Budach V, Saki M, Konschak R,
Niehr F, Jöhrens K, Weichert W, Linge A, Lohaus F, Krause M, et al:
Targeted next-generation sequencing of locally advanced squamous
cell carcinomas of the head and neck reveals druggable targets for
improving adjuvant chemoradiation. Eur J Cancer. 57:78–86. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bersani C, Sivars L, Haeggblom L,
DiLorenzo S, Mints M, Ährlund-Richter A, Tertipis N, Munck-Wikland
E, Näsman A, Ramqvist T and Dalianis T: Targeted sequencing of
tonsillar and base of tongue cancer and human papillomavirus
positive unknown primary of the head and neck reveals prognostic
effects of mutated FGFR3. Oncotarget. 8:35339–35350. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bersani C, Haeggblom L, Ursu RG, Giusca
SE, Marklund L, Ramqvist T, Näsman A and Dalianis T: Overexpression
of FGFR3 in HPV-positive tonsillar and base of tongue cancer is
correlated to outcome. Anticancer Res. 38:4683–4690. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koole K, van Kempen PM, Swartz JE, Peeters
T, van Diest PJ, Koole R, van Es RJ and Willems SM: Fibroblast
growth factor receptor 3 protein is overexpressed in oral and
oropharyngeal squamous cell carcinoma. Cancer Med. 5:275–284. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ghedini GC, Ronca R, Presta N and
Giacomini A: Future applications of FGF/FGFR inhibitors in cancer.
Expert Rev of Anticancer Ther. 18:861–872. 2018. View Article : Google Scholar
|
|
33
|
Arafeh R and Samuels Y: PIK3CA in cancer:
The past 30 years. Semin Cancer Biol. 2019.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Brenner JC, Graham MP, Kumar B, Saunders
LM, Kupfer R, Lyons RH, Bradford CR and Care TE: Genotyping of 73
USCC head and neck squamous cell carcinoma cell lines. Head Neck.
32:417–426. 2010.PubMed/NCBI
|
|
35
|
White JS, Weissfeld JL, Ragin CCR, Rossie
KM, Martin CL, Shuster M, Ishwad CS, Law JC, Myers EN, Johnson JT
and Gollin SM: The influence of clinical and demographic risk
factors on the establishment of head and neck squamous cell
carcinoma cell lines. Oral Oncol. 43:701–712. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lange MJ, Lasiter JC and Misfeldt ML:
Toll-like receptros in tonsillar epithelial cells. Int J Pediatr
Otorhinolaryngol. 73:613–621. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Foucquier J and Guedj M: Analysis of drug
combinations: Current methodological landscape. Pharmacol Res
Perspect. 3:e001492015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang L, Šuštić T, Leite de Oliveira R,
Lieftink C, Halonen P, van de Ven M, Beijersbergen RL, van den
Heuvel MM, Bernards R and van der Heijden MS: A functional genetic
screen identifies the phosphinositide 3-kinas pathway as a
determinant of resistance to fibroblast grown factor receptor
inhibitors in FGFR mutant urothelial cell carcinoma. Eur Urol.
71:858–862. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang J, Mikse O, Liao RG, Li Y, Tan L,
Janne PA, Gray NS, Wong KK and Hammerman PS: Ligand-associated
ERBB2/3 activation confers acquired resistance to FGFR inhibition
in FGFR3-dependent cancer cells. Oncogene. 34:2167–2177. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Herrera-Abreu MT, Pearson A, Campbell J,
Shnyder SD, Knowles MA, Ashworth A and Turner NC: Parallel RNA
interference screens identify EGFR activation as an escape
mechanism in FGFR3-mutant cancer. Cancer Discov. 3:1058–1071. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Brands RC, Knierim LM, De Donno F,
Steinacker V, Hartmann S, Seher A, Kübler AC and Müller-Richter
UDA: Targeting VEGFR and FGFR in head and neck squamous cell
carcinoma in vitro. Oncol Rep. 38:1877–1885. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ma BB, Lui VW, Hui CW, Lau CP, Wong CH,
Hui EP, Ng MH, Cheng SH, Tsao SW, Tsang CM, et al: Preclinical
evaluation of the mTOR-PI3K inhibitor BEZ235 in nasopharyngeal
models. Cancer Lett. 343:24–32. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Koole K, Brunen D, van Kempen PM, Noorlag
R, de Bree R, Lieftink C, van Es RJ, Bernards R and Willems SM:
FGFR1 is a potential prognostic biomarker and therapeutic target in
head and neck squamous cell carcinoma. Clin Cancer Res.
22:3884–3893. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Singleton KR, Hinz TK, Kleczko EK, Marek
LA, Kwak J, Harp T, Kim J, Tan AC and Heasley LE: Kinome RNAi
screens reveal synergistic targeting of MTOR and FGFR1 pathways for
treatment of lung cancer and HNSCC. Cancer Res. 75:4398–4406. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Munster P, Aggarwal R, Hong D, Schellens
JH, van der Noll R, Specht J, Witteveen PO, Werner TL, Dees EC,
Bergsland E, et al: First-in-human phase I study of GSK2126458, an
oral pan-class I phosphatidylinositol-3-kinase inhibitor, in
patients with advanced solid tumor malignancies. Clin Cancer Res.
22:1932–1939. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tamura R, Yoshihara K, Saito T, Ishimura
R, Martínez-Ledesma JE, Xin H, Ishiguro T, Mori Y, Yamawaki K, Suda
K, et al: Novel therapeutic strategy for cervical cancer harboring
FGFR3-TACC3 fusions. Oncogenesis. 7:42018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Esposito A, Viale G and Curigliano G:
Safety, tolerability, and management of toxic effects of
phosphatidylinositol 3-kinase inhibitor treatment in patients with
cancer: A review. JAMA Oncol. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Schram AM, Voss MH and Hyman DM:
Genome-driven paradigm for the development of selective fibroblast
growth factor receptor inhibitors. J Clin Oncol. 35:131–134. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
de Weerdt I, Koopmans SM, Kater AP and van
Gelder M: Incidence and management of toxicity associated with
ibrutinib and idelalisib: A practical approach. Haematologica.
102:1629–1639. 2017. View Article : Google Scholar : PubMed/NCBI
|