Introduction
Breast cancer is one of the most common types of
cancer worldwide. In recent years, in China, the mortality rate of
patients with breast cancer has continuously increased. As such,
breast cancer is a significant cause of mortality in females
(1). As the result of
multidisciplinary collaborations, there have been significant
advances in the treatment of patients with breast cancer, of which
radiotherapy is a key strategy (2,3).
Postoperative radiotherapy decreases the lymph node recurrence rate
by approximately two-thirds. Clinical data has demonstrated that
breast-conserving surgery combined with radiotherapy decreases the
local recurrence rate from 18–35 to 2–10% (4); however, certain patients do experience
relapse, suggesting that their tumors may be resistant to
radiotherapy. Therefore, it is important to examine the mechanism
underlying this phenomenon and determine the intrinsic
radiosensitivity of cells and the cellular microenvironment
(5,6). Studies regarding radiosensitivity have
progressed through 3 different stages; from the tissue to the
cellular level and subsequently to the molecular level. The third
stage concerns how biological processes may affect cell
radiosensitivity, including hypoxia, cell cycle distribution, cell
proliferation, apoptosis, DNA damage repair and more (7–9), and
these mechanisms involve genetic variation and epigenetic
modification. Therefore, it may be possible to determine the effect
of cell radiosensitivity by examining the regulatory mechanisms
underlying these processes.
Invasion and metastasis are important behavioral
characteristics of cancer cells, and acquisition of these processes
endows cells with a variety of properties. For example,
epithelial-mesenchymal transition (EMT) is associated with cell
invasion and metastasis, cell stemness, and radiotherapy and
chemotherapy tolerance (10).
Numerous transcription factors regulate EMT, including Zinc finger
E-box-binding homeobox protein, Snail, Slug, and Twist. These
factors downregulate epithelial cadherin (E-cadherin), which is
pivotal in the EMT process (11).
E-cadherin participates in the regulation of cell adhesion, which
serves a critical role in cancer cell invasion and metastasis
(12). Our previous study indicated
that human epidermal growth factor receptor 2 (HER2) decreased
breast cancer cell radiosensitivity by activating focal adhesion
kinase in vitro and in vivo (13). Therefore, cell adhesion processes,
and invasion and metastatic processes may be associated with the
response to radiotherapy. To determine whether there was an
association between migration and metastasis, radiosensitivity,
daughter cell lines with differing migratory capabilities from 2
parent cell lines were established in the present study using
Transwell chambers in a 24-well plate. There was a negative
association between migration and radiosensitivity and this may be
associated with the expression of cell adhesion molecules and/or
EMT.
Materials and methods
Cell lines and cell culture
The breast cancer MDA-MB-231 and ZR-75–30 cell lines
were purchased from the American Type Culture Collection and
maintained in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.), penicillin (100 units/ml),
streptomycin (100 µg/ml) 2 mM l-glutamine and 1 mM sodium pyruvate.
All cells were incubated in a 37°C in a humidified atmosphere with
5% CO2.
Migration assays
The migration assay was performed as previously
described (13,14). Briefly, a total of 2×104
of MDA-MB-231 or ZR-75–30 were placed in the upper chamber of a
Transwell chamber (BD Biosciences) with an 8-µm pore filter between
the chambers. The cells were allowed to migrate at 37°C for 8 h
toward the chamber of medium supplemented with 2.5% fetal bovine
serum. Non-migrating cells on the upper side of the insert were
removed and the migrated cells on the lower side of the insert were
fixed with ice-cold methanol for 10 min at room temperature,
stained with 0.1% crystal violet for 20 min at room temperature,
imaged and counted at magnification ×200 under a light microscope.
The assay was repeated 3 times in duplicate.
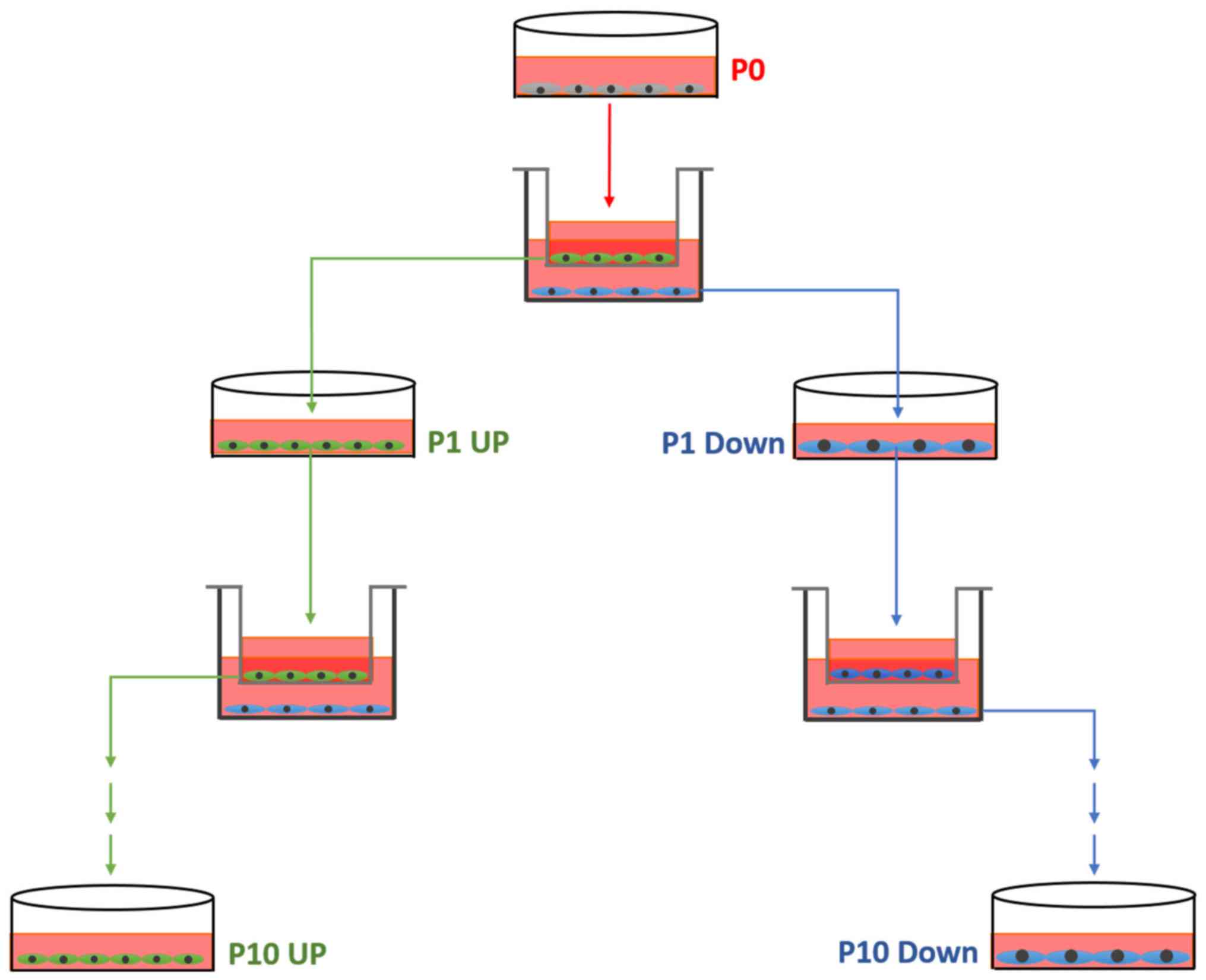
Establishment of the cell model
From each cell line, a pair of cell lines differing
in migratory ability was established, according to the schematic
diagram in Fig. 1. Initially, a
migration assay was performed as aforementioned (named P0). The
cells from the upper chamber, which had not migrated, were
collected and cultured in new dishes, termed 231 Down-1 (P1). The
cells which had migrated through the insert after 8 h were also
collected and cultured. These cells were termed 231 UP-1 (P1). This
process was repeated 10 times, each time using the collected cells
that had or had not migrated, until the MDA-MB-231 UP-10,
MDA-MB-231 Down-10, ZR-75-30-UP-10 and ZR-75-30-Down-10 cell
cultures were established.
Genes microarray
Total RNA was isolated from 2×106 target
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and was treated with DNase I to remove any
contaminating genomic DNA. Microarray analysis was used to screen
changes in genome-wide gene expression patterns in the MDA-MB-231
UP-10 and MDA-MB-231 Down-10 cells, the ZR-75–30 UP-10 and ZR-75–30
Down-10 cell line. The changes in human gene expression patterns
were assessed using Affymetrix gene microarrays (CapitalBio
Technology Co., Ltd.) and 3 replicates were used for microarrays
analysis. Gene ontology (GO) enrichment analysis (14,15) and
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
(16–18) were performed to analyze the pathways
involved.
Irradiation and clone formation
assay
MDA-MB-231 UP-10, MDA-MB-231 Down-10 cells, ZR-75–30
UP-10 and ZR-75–30 Down-10 cells were seeded into 6 cm cell culture
dishes at a density of 1×106 cells. A total of 24 h
later, the cells were irradiated with a single dose of X-rays (0,
2, 4, 6 and 8 Gy), using a linear accelerator (Varian Medical
Systems) at 3 Gy/min and then seeded into 6 cm petri dishes and
incubated for 10–14 days to allow colonies to form. The cell
inoculum of the single cell suspension was determined for each
sample according to the expected number of colony formation
(30–100). The colonies were fixed with 4% paraformaldehyde for 20
min at room temperature and stained with 0.1% crystal violet for 20
min at room temperature. The number of colonies with ≥50 cells were
counted under a microscope. The plating efficiency (PE) and
survival fraction (SF) were calculated using the following
equations: PE = number of colonies formed without irradiation/the
number of cells inoculated ×100%; and SF = colonies counted/cells
seeded × PE. All assays were performed independently at least 3
times.
Western blot analysis
MDA-MB-231 UP-10, MDA-MB-231 Down-10 cells, ZR-75–30
UP-10 and ZR-75–30 Down-10 cells were seeded into 10-cm culture
dishes. Cell lysates were harvested at 75% confluence using 500 µl
cell lysis buffer (25 mmol/l Tris-HCl at pH 7.6, 150 mmol/l NaCl,
1.0% Nonidet P-40, 1.0% sodium deoxycholate and 0.1% SDS) with a 30
min incubation on ice. The supernatant was collected under rotation
and was centrifuged at 13,000 × g for 30 min at 4°C. The protein
concentrations were determined using a BCA Protein Assay Reagent
(Thermo Fisher Scientific, Inc.). Western blot analyses were
performed as described previously (19). The primary antibodies included
anti-fibronectin 1 (FN1; cat. No sc69681; 1:1,000; Santa Cruz
Biotechnology, Inc.), anti-β-actin (cat. no. 3700s; 1:1,000; Cell
Signaling Technology, Inc.), anti-tight junction protein ZO-1
(ZO-1; cat. no. 8193s; 1:1,000; Cell Signaling Technology, Inc.),
anti-focal adhesion kinase (FAK; cat. no. 9330T; 1:1,000; Cell
Signaling Technology, Inc.), anti-epidermal growth factor receptor
(EGFR; cat. no. 4267s; 1:1,000; Cell Signaling Technology, Inc.),
anti-pEGFR (cat. nos. Y1068 and 3777s; 1:1,000; Cell Signaling
Technology, Inc.), anti-pEGFR (cat. nos. Y1173 and 4407s; 1:1,000;
Cell Signaling Technology, Inc.) and transcription factor SOX-9
(SOX9; cat. no. 82630s; 1:1,000; Cell Signaling Technology, Inc.).
Horseradish peroxidase-conjugated secondary antibodies included
donkey anti-mouse immunoglobulin (cat. no. NA931; 1:2,000; GE
Healthcare) and donkey anti-rabbit immunoglobulin (cat. no. NA9340;
1:2,000; GE Healthcare).
Statistical analysis
Statistical analysis was performed using the SPSS
Statistics version 22.0 (IBM Corp.). Quantitative data were
presented as the mean ± standard deviation. Comparisons between the
means of two groups were conducted using a unpaired Student's
t-test. The experimental data were analyzed and plotted using
GraphPad Prism (v6.0; GraphPad Software, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
Establishment and verification of the
cell model
To accurately determine whether there was any
association between migratory capacity and radiosensitivity, the
differences in genetic background between different cells must be
removed. Therefore, cell models from the same parent cell line that
exhibited different degrees of migratory capabilities was
established. In this model, the Transwell chamber served as a
‘selection’ tool, which separated MDA-MB-231 or ZR-75–30 cells into
two groups of cells based on their migratory capacities. For each
parent cell line, 2 daughter cell lines were established,
MDA-MB-231 UP, MDA-MB-231 Down cell line, ZR-75–30 UP and ZR-75–30
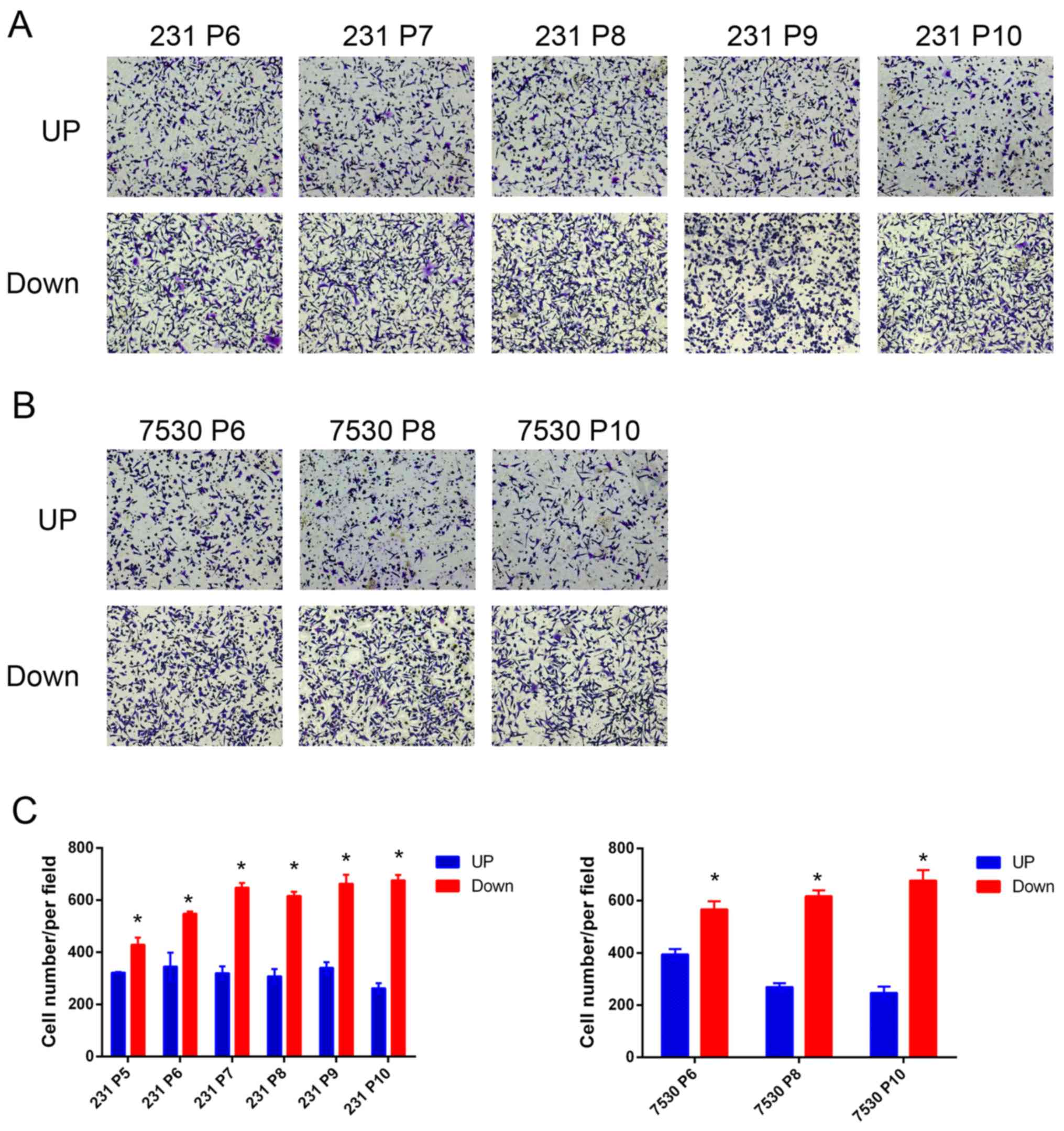
Down cell line. To examine the model, a migration assay was
performed and the number of cells that had migrated through the
membrane was quantified. The results indicated that a greater
number of MDA-MB-231 Down cells migrated through the membrane
compared with the MDA-MB-231 UP cells after the 5th repetition
(P5). A similar effect was observed in the ZR-75–30 daughter cell
lines. Furthermore, the cells in the later generations tended to
exhibit increased migratory capabilities (Fig. 2). Following the 10th repetition
(P10), the resultant daughter cell lines were used for further
study.
A high migratory capacity is
associated with increased radio-resistance
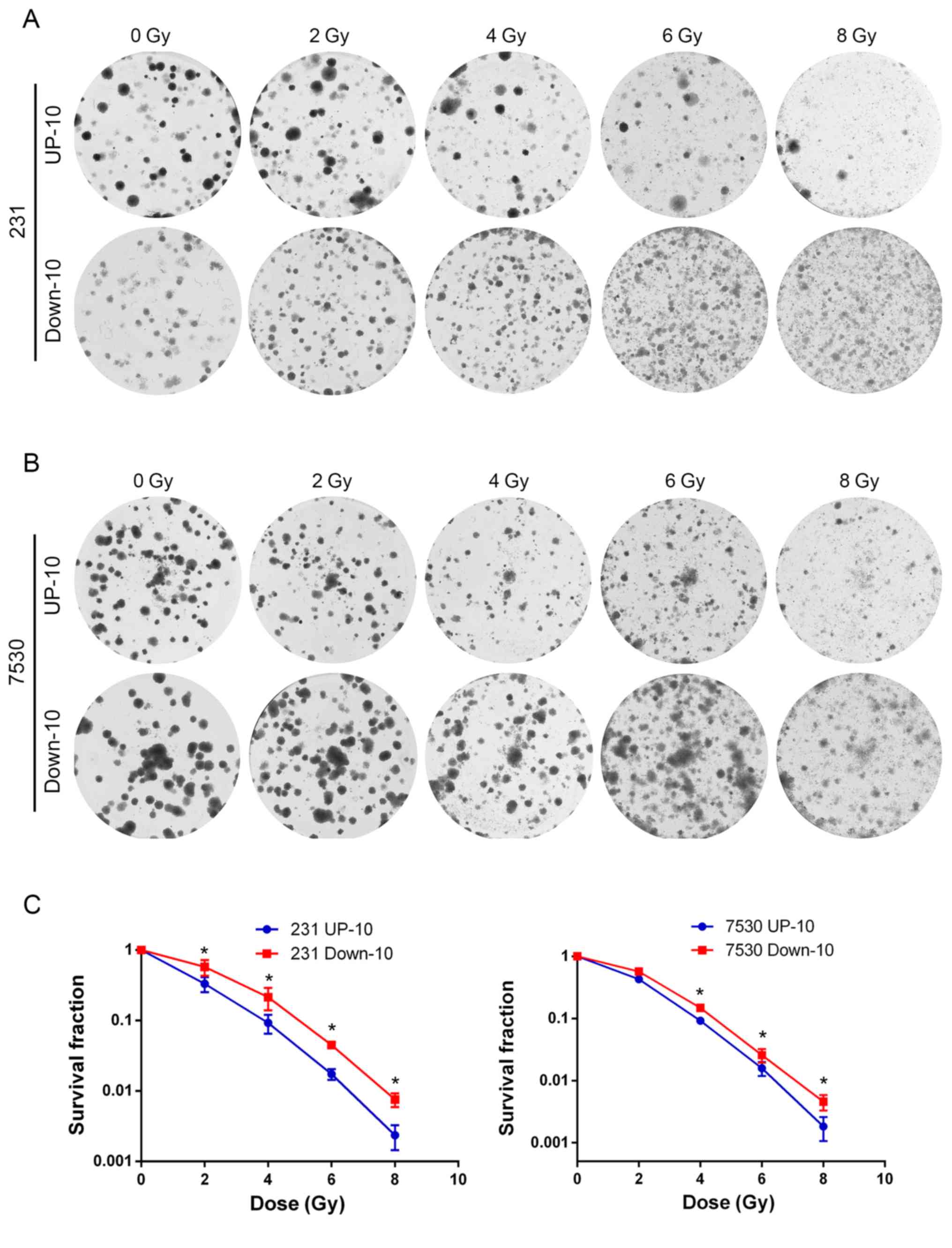
To investigate the association between the migratory
capacity and radiosensitivity, irradiation and colony formation
assays were performed to determine the response to irradiation in
the 4 daughter cell lines. The survival curve indicated that the
MDA-MB-231 Down-10 and ZR-75–30 Down-10 cells were less sensitive
to radiation compared with the corresponding UP-10 cells when
treated with a dose between 2–8 Gy (Fig.
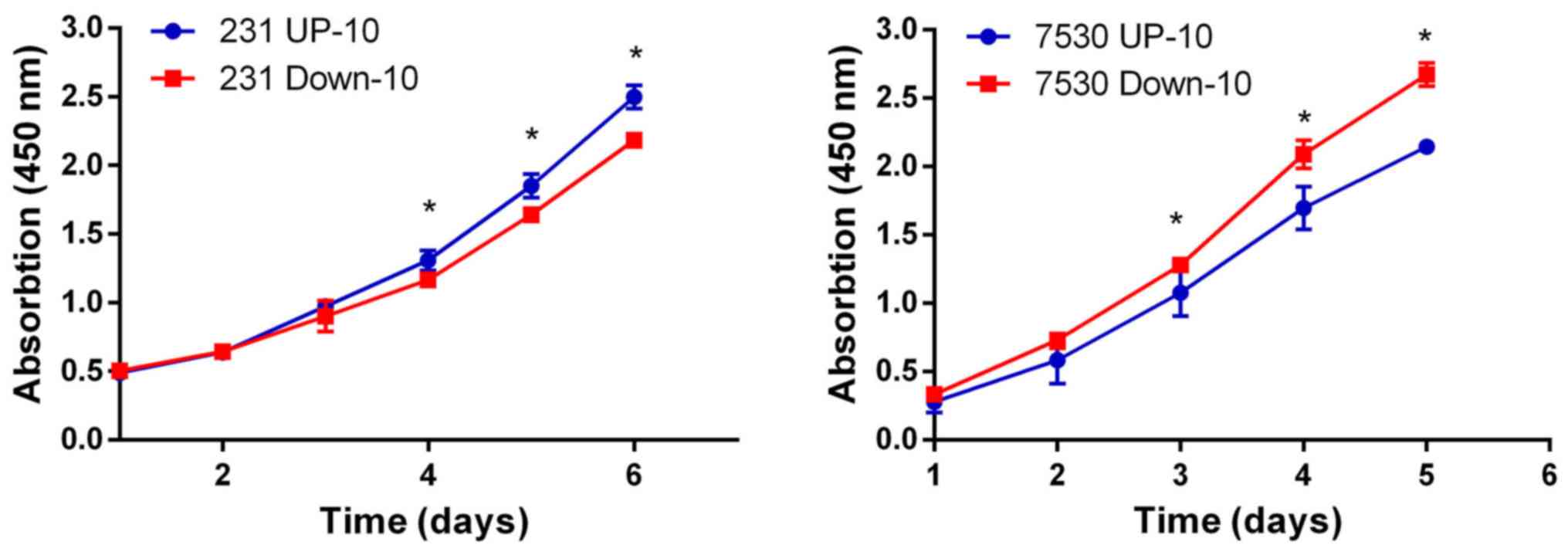
3). Although the MDA-MB-231 Down-10 cells exhibited a greater
migratory capacity compared with the MDA-MB-231 UP-10 cells, the
proliferative capacity of the MDA-MB-231 UP-10 cells was increased.
However, the proliferative capacity was lower in the ZR-75–30 UP-10
compared with the ZR-75–30 Down-10 cells (Fig. 4).
Bioinformatics analysis
To examine the mechanism underlying cells with an
increased migratory capacity increased radio-resistance,
genome-wide gene profiling was performed to determine the
differentially expressed genes in the MDA-MB-231 UP-10 compared
with MDA-MB-231 Down-10 cells, and in the ZR-75–30 UP-10 compared
with the ZR-75–30 Down-10 cells. Affymetrix gene microarrays
indicated that numerous genes were upregulated or downregulated in
the UP-10 cells compared with the Down-10 cells. GO and KEGG
pathway assays demonstrated that a number of pathways were
involved. Amongst the 10 of the most significantly altered
pathways, both GO and KEGG pathway assays suggested that the cell
adhesion pathway was involved (Fig. 5A
and B). Our previous study demonstrated that Fak-associated
genes were important in HER2-mediated radio-resistance (13). Therefore, the expression of Fak was
determined. Western blot analysis results demonstrated that total
Fak expression levels were similar in the UP-10 cells and Down-10
cells. However, the expression of (pFak), including Y397, Y576 and
Y925 sites, were significantly increased in the Down-10 cells
compared with the UP-10 cells. Furthermore, EGFR, pEGFR, ZO-1, FN1
and SOX9 were also expressed robustly in the Down-10 cells
(Fig. 5C).
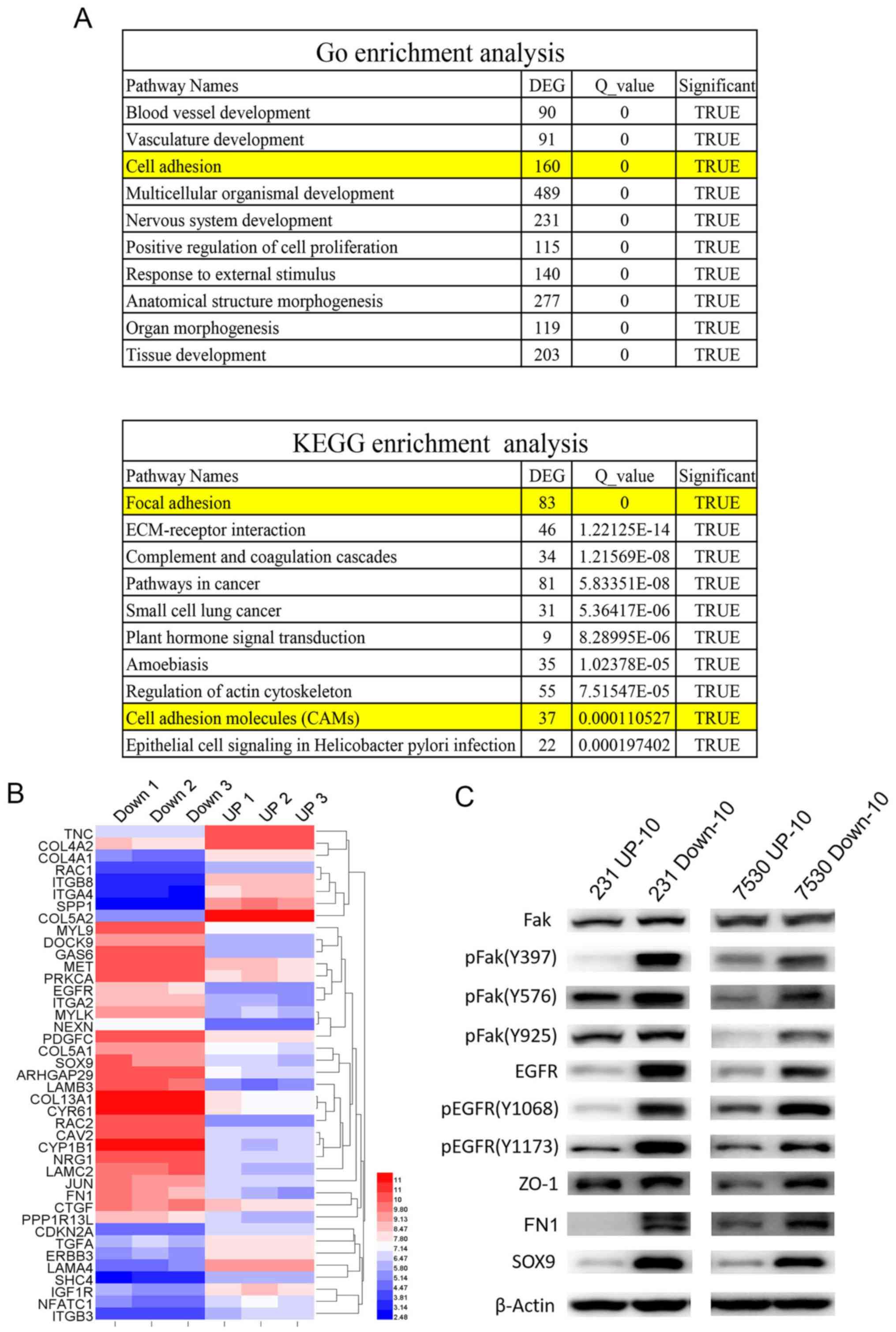 | Figure 5.Cell adhesion pathway is associated
with radio-resistance of cells with high migratory capacities. (A)
GO and KEGG pathway analyses indicated that the cell adhesion
pathway was involved in radio-resistance of cells with an increased
migratory capacity (231 Down-10 and 7530 Down-10 cells). (B)
Cluster map of microarray-identified DEGs, demonstrating a notable
change in cell adhesion associated genes. The left panel represents
the gene expression ratio of 231 UP-10 cells with 231 Down-10
cells. Right panel represents the gene expression ratio of 7530
UP-10 cells with 7530 Down-10 cells. (C) Western blot analysis of
molecules associated with cell adhesion signaling pathway and the
epithelial-mesenchymal transition. Fak, pFak, EGFR, pEGFR, ZO-1,
FN1 SOX9 were protein expression levels were measured. β-actin was
used as the loading control. DEG, differentially expressed genes;
231, MDA-MB-231; 7530, ZR-75-30; Fak, focal adhesion kinase 1; p,
phosphorylated; EGFR, epidermal growth factor receptor; ZO-1, tight
junction protein ZO-1; FN1, fibronectin 1; SOX9, transcription
factor SOX-9; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes
and Genomes. |
Discussion
In recent years, as a result of multidisciplinary
research approaches, there has been a significant advancement in
the diagnosis and treatment of breast cancer, and the overall
survival of patients continues to improve. The contribution of
radiotherapy to this has been indispensable. However, certain
patients may experience relapse following radiotherapy, suggesting
that these tumors may be radio-resistant. The intrinsic sensitivity
of radiotherapy is important for the radio-response, therefore,
examining the mechanism of radiosensitivity of breast cancer cells
may improve the radiotherapeutic effect. A number of biological
processes may affect cell radiosensitivity, including hypoxia, cell
cycle distribution, cell proliferation, apoptosis, DNA damage
repair and others. Invasion and metastasis are important
characteristics of cancer cells and acquisition of these processes
endow cells with a variety of properties. For example, EMT is
associated with cell invasion and metastasis (10). A number of studies have demonstrated
an association between EMT and the treatment tolerance of cancer,
which have provided evidence to support the investigation of the
association between cell invasiveness and radiosensitivity
(20–22). Certain studies have demonstrated that
genes regulating cell invasiveness can affect cell radiosensitivity
(23–25). However, the direct evidence
concerning the effects of cell invasiveness on radiosensitivity
remain scarce. The present study focused on the association between
cell migration and radiosensitivity. To investigate this issue, a
cell model was established from the same parent cell line, with
different migratory capacities, by using Transwell chambers as a
selection pressure. The results indicated that, following 5 rounds
of screening, the ‘Down’ groups of cells exhibited increased levels
of migration compared with the ‘Up’ groups of cells. The effect was
more evident as the selection process continued. Therefore, cells
obtained following the 10th round of selection were used for
further investigation. The result of the clone formation assay
verified the hypothesis that cells with higher migratory capacities
were associated with increased levels of radio-resistance. The
proliferative capacity of the MDA-MB-231 UP-10 cells was increased
compared with MDA-MB-231 Down-10 cells, but was reduced in the
ZR-75–30 UP-10 compared with the ZR-75–30 Down-10 cells, suggesting
that the proliferation process was not associated with the change
in radiosensitivity.
As cell invasion was associated with
radio-resistance, the underlying molecular mechanisms governing
this association were examined. By doing so, it is possible to
improve the understanding of the underlying mechanism regulating
cell invasion and radiosensitivity. Additionally, it may unveil
potential therapeutic strategies that simultaneously solve multiple
issues (metastases and radio-resistance). Therefore, genome-wide
gene profiling was performed to screen differentially expressed
genes between the UP-10 and respective Down-10 cells. Numerous
genes were differentially expressed and a number of associated
pathways were identified. Analysis of these pathways revealed that
the cell adhesion pathway was highly importance. Our previous study
demonstrated that HER2 decreased the radiosensitivity of breast
cancer cells by activating Fak in vitro and in vivo,
and by inhibiting Fak activity using a Fak inhibitor (PF-562281),
the radiosensitivity in HER2-overexpressing breast cancer cells was
restored (13). As Fak serves a
central role in the cell adhesion process, the expression of Fak in
the cells lines in the present study was determined and it was
identified that pFak, including Y397, Y576 and Y925 sites, were
significantly increased in the Down-10 cells compared with the
respective UP-10 cells. The expression Fak-associated genes was
also identified. The results indicated that the expression of cell
adhesion-associated genes, including EGFR, pEGFR, ZO-1, FN1 and
SOX9 were significantly increased in the Down-10 cells compared
with the respective UP-10 cells.
Fak is a 125 kD protein that is recruited as a
participant in focal adhesion dynamics between cells, and has a
role in motility and cell survival (26,27). It
is typically located at structures known as focal adhesions, which
are multi-protein structures that are connected to the
extracellular matrix. It has been demonstrated that when Fak was
inhibited, breast cancer cells became less metastatic as a result
of a decrease in mobility (28).
Numerous previous studies have indicated that Fak may participate
in regulating cell proliferation, migration, anchoring and
apoptosis (29,30). Certain studies have indicated that
Fak is also involved in radiosensitivity regulation, although there
are conflicting data on this issue: For example, Cordes et
al (23) described a type of
cell adhesion-mediated radio-resistance. Such a phenomenon was
associated with the cell adhesion network regulated by integrin and
Fak. Furthermore, a second study indicated that overexpression of
Fak in the lung cancer A549 cell line resulted in cell
radio-resistance, while knockdown of Fak in pancreatic cancer cells
led to radiosensitization (24,25). A
recent study identified that Fak may mediate the radioresistance of
HIEC cells. Fak knockdown markedly increased the radiosensitivity
of HIEC cells, and mice treated with FAK inhibitors exhibited
aggravated mice rectal damage (31).
In addition, Zhang et al (32) identified that β1-integrin-blocking
antibody and FAK inhibitor-enhanced radiation induced apoptosis in
esophageal cancer cells. In addition, Fak is associated with cell
stemness and may initiate the EMT process. These phenotypes have
been suggested to increase cell radio-resistance (33). However, contradictory results have
been described by Graham et al (34), who revealed that the loss of Fak
expression led to cell radio-resistance, and increasing expression
of Fak in these cells restored radiosensitivity.
In conclusion, invasion and metastasis are hallmarks
of cancer cells and these processes infer a variety of properties
in cells. The association between migration and radiosensitivity is
not fully understood. A cell model was established in which
daughter cell lines with either increased or decreased migratory
capacities were derived from the same parent cell line. Cell lines
with an increased migratory capacity were more radio-resistant,
potentially due to the upregulation of factors involved in cell
adhesion and EMT. Additionally, Fak may serve a central role in
radiosensitization and may be an important potential therapeutic
target. A limitation of the present study was that the conclusion
was based on only two pairs of cell lines. Additional cell lines
are required to validate the conclusion and may provide an improved
understanding of the specific pathways and components involved.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Nature Science Foundation of China (grant nos. 81702640 and
81660439) and The Guizhou provincial science and technology project
[grant nos. (2017)1114 and (2017)1104].
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the author on reasonable request.
Authors' contributions
JH, LL, HZ, HC, NW and MD designed and performed
experiments, prepared the figures and interpreted the data. LL and
HZ drafted the results section of the manuscript. NW verified the
statistical analysis and revised the manuscript. XG, QN and JH
conceived and designed the study, interpreted the data, wrote and
revised the manuscript and obtained funding to support the
research. All authors have read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li H, Zheng RS, Zhang SW, Zeng HM, Sun XK,
Xia CF, Yang XZ, Chen WQ and He J: Incidence and mortality of
female breast cancer in china, 2014. Zhonghua Zhong Liu Za Zhi.
40:166–171. 2018.PubMed/NCBI
|
|
2
|
Early Breast Cancer Trialists'
Collaborative Group (EBCTCG), ; Darby S, McGale P, Correa C, Taylor
C, Arriagada R, Clarke M, Cutter D, Davies C, Ewertz M, et al:
Effect of radiotherapy after breast-conserving surgery on 10-year
recurrence and 15-year breast cancer death: Meta-analysis of
individual patient data for 10,801 women in 17 randomised trials.
Lancet. 378:1707–1716. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
EBCTCG (Early Breast Cancer Trialists'
Collaborative Group), ; McGale P, Taylor C, Correa C, Cutter D,
Duane F, Ewertz M, Gray R, Mannu G, Peto R, et al: Effect of
radiotherapy after mastectomy and axillary surgery on 10-year
recurrence and 20-year breast cancer mortality: Meta-analysis of
individual patient data for 8135 women in 22 randomised trials.
Lancet. 383:2127–2135. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fisher B, Anderson S, Bryant J, Margolese
RG, Deutsch M, Fisher ER, Jeong JH and Wolmark N: Twenty-year
follow-up of a randomized trial comparing total mastectomy,
lumpectomy, and lumpectomy plus irradiation for the treatment of
invasive breast cancer. N Engl J Med. 47:1233–1241. 2002.
View Article : Google Scholar
|
|
5
|
Joubert A and Foray N: Intrinsic
radiosensitivity and DNA double-strand breaks in human cells.
Cancer Radiother. 11:129–142. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barker HE, Paget JT, Khan AA and
Harrington KJ: The tumour microenvironment after radiotherapy:
Mechanisms of resistance and recurrence. Nat Rev Cancer.
15:409–425. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marples B and Collis SJ: Low-dose
hyper-radiosensitivity: Past, present, and future. Int J Radiat
Oncol Biol Phys. 70:1310–1318. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhivotovsky B, Joseph B and Orrenius S:
Tumor radiosensitivity and apoptosis. Exp Cell Res. 248:10–17.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pawlik TM and Keyomarsi K: Role of cell
cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol
Biol Phys. 59:928–942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meng J, Li P, Zhang Q, Yang Z and Fu S: A
radiosensitivity gene signature in predicting glioma prognostic via
EMT pathway. Oncotarget. 5:4683–4693. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Anzai E, Hirata K, Shibazaki M, Yamada C,
Morii M, Honda T and Yamaguchi N and Yamaguchi N: FOXA1 induces
E-cadherin expression at the protein level via suppression of slug
in epithelial Breast cancer cells. Biol Pharm Bull. 40:1483–1489.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Canel M, Serrels A, Frame MC and Brunton
VG: E-cadherin-integrin crosstalk in cancer invasion and
metastasis. J Cell Sci. 126:393–401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hou J, Zhou Z, Chen X, Zhao R, Yang Z, Wei
N, Ni Q, Feng Y, Yu X, Ma J and Guo X: HER2 reduces breast cancer
radiosensitivity by activating focal adhesion kinase in vitro and
in vivo. Oncotarget. 7:45186–45198. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. Nat
Genet. 25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
The Gene Ontology Consortium: The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou J, Wang Z, Xu H, Yang L, Yu X, Yang Z,
Deng Y, Meng J, Feng Y, Guo X and Yang G: Stanniocalicin 2
suppresses breast cancer cell migration and invasion via the
PKC/claudin-1-mediated signaling. PLoS One. 10:e1221792015.
|
|
20
|
Santamaría PG, Moreno-Bueno G and Cano A:
Contribution of Epithelial plasticity to therapy resistance. J Clin
Med. 8:6762019. View Article : Google Scholar
|
|
21
|
Xie P, Yu H, Wang F, Yan F and He X:
Inhibition of LOXL2 enhances the radiosensitivity of
castration-resistant prostate cancer cells associated with the
reversal of the EMT process. BioMed Res Int. 27:40125902019.
|
|
22
|
Fischer KR, Durrans A, Lee S, Sheng J, Li
F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al:
Epithelial-to-mesenchymal transition is not required for lung
metastasis but contributes to chemoresistance. Nature. 527:472–476.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cordes N and Meineke V: Cell
adhesion-mediated radioresistance (CAM-RR). Extracellular
matrix-dependent improvement of cell survival in human tumor and
normal cells in vitro. Strahlenther Onkol. 179:337–344. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cordes N, Frick S, Brunner TB, Pilarsky C,
Grützmann R, Sipos B, Klöppel G, McKenna WG and Bernhard EJ: Human
pancreatic tumor cells are sensitized to ionizing radiation by
knockdown of caveolin-1. Oncogene. 26:6851–6862. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beinke C, Van Beuningen D and Cordes N:
Ionizing radiation modules of the expression and tyrosine
phosphorylation of the focal adhesion-associated proteins focal
adhesion kinase (FAK) and its substrates p130cas and paxillin in
A549 human lung carcinoma cells in vitro. Int J Radiat Biol.
79:721–731. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Braren R, Hu H, Kim YH, Beggs HE,
Reichardt LF and Wang R: Endothelial FAK is essential for vascular
network stability, cell survival, and lamellipodial formation. J
Cell Biol. 172:151–162. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hanks SK, Ryzhova L, Shin NY and Brábek J:
Focal adhesion kinase signaling activities and their implications
in the control of cell survival and motility. Front Biosci.
8:d982–d996. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chan KT, Cortesio CL and Huttenlocher A:
FAK alters invadopodia and focal adhesion composition and dynamics
to regulate breast cancer invasion. J Cell Biol. 185:357–370. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han EK, Mcgonigal T, Wang J, Giranda VL
and Luo Y: Functional analysis of focal adhesion kinase (FAK)
reduction by small inhibitory RNAs. Anticancer Res. 24:3899–3905.
2004.PubMed/NCBI
|
|
30
|
Sulzmaier FJ, Jean C and Schlaepfer DD:
FAK in cancer: Mechanistic findings and clinical applications. Nat
Rev Cancer. 14:598–610. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li JJ, Tu WZ, Chen XM, Ying HY, Chen Y, Ge
YL, Wang J, Xu Y, Chen TF, Zhang XW, et al: FAK alleviates
radiation-induced rectal injury by decreasing apoptosis. Toxicol
Appl Pharmacol. 360:131–140. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang C, Deng X, Qiu L, Peng F, Geng S,
Shen L and Luo Z: Knockdown of C1GalT1 inhibits radioresistance of
human esophageal cancer cells through modifying β1-integrin
glycosylation. J Cancer. 9:2666–2677. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cicchini C, Laudadio I, Citarella F,
Corazzari M, Steindler C, Conigliaro A, Fantoni A, Amicone L and
Tripodi M: TGFbeta-induced EMT requires focal adhesion kinase (FAK)
signaling. Exp Cell Res. 314:143–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Graham K, Moran-Jones K, Sansom OJ,
Brunton VG and Frame MC: FAK deletion promotes p53-mediated
induction of p21, DNA-damage responses and radio-resistance in
advanced squamous cancer cells. PLoS One. 6:e278062011. View Article : Google Scholar : PubMed/NCBI
|