Introduction
L-asparaginase (L-asp) is a fundamental drug in the
systemic treatment of childhood B cell precursor acute
lymphoblastic leukemia (BCP-ALL). The introduction of L-asp
improved the 5-year event-free survival (EFS) by 20% and boosted
treatment response rates by 60% (1,2).
However, in genetic subtypes of ALL with a high risk of relapse,
L-asp treatment did not meet expectations, with drug resistance
being one of the major mechanisms contributing to treatment failure
(2–6). Although L-asp resistance is
multifaceted and still not fully explained, several potential
mechanisms have been proposed, such as hydrolysis of asparaginyl
bonds and enzymatic degradation of the drug particle itself,
immunological response against the drug triggered by presentation
of L-asp to dendritic cells through MHC class II complex and
increased production of asparagine as a response to its depletion
in the blast environment during L-asp treatment (7–9).
Previous studies have suggested that the genes involved in the
degradation of L-asp and subsequent minimization of its
effectiveness, may be upregulated by primary genetic abnormalities
and copy number variations (CNVs) identified in BCP-ALL, such as
hypodiploidy, breakpoint cluster region and Abelson murine leukemia
viral oncogene homolog 1 (BCR-ABL1) fusion, lack of ETS
variant 6 and runt-related transcription factor 1
(ETV6-RUNX1) fusion, BCR-ABL1-like phenotype and
ikaros family zinc finger protein 1 (IKZF1) deletion
(1,10,11).
Of the genes identified to be involved in the
mechanisms of resistance to L-asp treatment, three key examples are
asparagine synthetase (ASNS), legumain (LGMN) and
cathepsin B (CTSB). ASNS synthesizes asparagine to reverse
the antineoplastic action of L-asp, which depletes the surrounding
leukemic cells of asparagine (12,13).
LGMN is a cysteine protease that inactivates drugs by specifically
hydrolyzing asparaginyl bonds; it is also involved in the major
histocompatibility complex (MHC) class II antigen presentation
process, which may account for the allergic reactions to L-asp
(14). CTSB encodes a
peptidase that degrades L-asp (9).
Although the aforementioned genes act in opposition to L-asp, no
data on functional associations between them are currently
available.
The role of the aforementioned genes in high-risk
genetic subtypes of childhood BCP-ALL has not been fully
elucidated. Studies of ASNS expression in groups with a
number of genetic alterations are ambiguous (2,5,15). In addition, poor response to L-asp
treatment is associated with ASNS overexpression at the
protein, but not mRNA level (5,12,15,16).
LGMN has been demonstrated to be upregulated in the
intrachromosomal amplification of chromosome 21 (iAMP21) subtype of
ALL, which is associated with high risk of relapse when treated
with a low or standard risk protocol (14). No studies on the relevance of
CTSB expression levels in BCP-ALL patients are currently
available in the literature to the best of our knowledge, and a key
aim of the present study was to determine the expression levels of
the selected candidate genes underlying L-asp resistance in
patients with different genetic types of BCP-ALL.
Materials and methods
Study group
A group of 52 patients ≤18 years were recruited
retrospectively from three pediatric oncology centers in Poland:
Lodz, Zabrze and Lublin. The patients were diagnosed with BCP-ALL
between December 2005 and January 2016 and treated with the
Berlin-Frankfurt-Münster backbone protocols with L-asp as a core
regimen (ALL IC, 2002 and 2009) (17,18).
Matched DNA and RNA samples from the bone marrow aspiration at the
point of diagnosis, as well as clinical data, were acquired.
BCR-ABL1 fusion, ETV6-RUNX1 fusion, hypodiploidy and
hyperdiploidy were assessed routinely by fluorescence in
situ hybridization and karyotyping performed in a certified
laboratory (Genetic Laboratory at Collegium Medicum; Bydgoszcz,
Poland) at the time of diagnosis. Each patient and/or their parents
provided signed informed consent in writing prior to inclusion in
the study.
Multiplex ligation-dependent probe
amplification (MLPA)
For all collected bone marrow samples, MLPA analysis
with P327-B1 iAMP21-ERG probe mix was performed according to the
manufacturer's protocol (MRC-Holland BV). Additionally, MLPA P329
CRLF2-CSF2RA-IL3RA and P335 ALL-IKZF1 (MRC-Holland BV) probe mixes
for cytokine receptor-like factor 1, IKZF1, IKZF2, IKZF3,
cyclin-dependent kinase inhibitor (CDKN)
2A/2B, retinoblastoma 1 (RB1), PAX5 and
ETV6 amplification or deletion status detection were used.
Absolute fluorescence was normalized by comparing peak patterns of
DNA in the sample of interest with a DNA isolated from blood
samples from age- and gender-matched healthy individuals who
provided written consent to participate in the study. The relative
probe ratio of the tested samples was compared with the average
relative probe ratio in the reference samples to calculate the
dosage quotient. Data analysis and interpretation were conducted
using GeneMarker v2.7.4 software (Softgenetics, LLC) according to
the manufacturer's protocol.
Gene expression measurement
To obtain cDNA for quantitative PCR, all available
RNA samples that were extracted using the TRIzol reagent (Thermo
Fisher Scientific, Inc.) were reverse transcribed using a
High-Capacity cDNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.). The thermal conditions were 30 min at 50°C, 15
min at 95°C, followed by storage at 4°C. TaqMan Gene Expression
assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) for
ASNS (cat. no. Hs04186194_m1), LGMN (cat. no.
Hs00271599_m1) and CTSB (cat. no. Hs00947433_m1) were used;
GAPDH (cat. no. Hs02786624_g1) was selected as a control.
The thermocycling conditions were as follows: 50°C for 2 min and
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C
for 1 min. All samples were tested in duplicate and the
2−ΔΔCq value was calculated (19) with GAPDH used for
normalization. The relative mRNA expression was used for further
comparisons.
Transcription factor binding sites
search
The search for PAX5 binding sites throughout
the human and mouse genome matrix was conducted using the HOCOMOCO
v11, TRANSFAC database version 2018.3 (GeneXplain, GmbH). This is
an online engine for searches through multiple module databases and
was used as previously described (20).
Statistical analysis
Statistica 12.0 software (TIBCO Software, Inc.) was
used for all analyses. Nominal variables are presented as
percentages; differences between the groups were evaluated using
the χ2 test. Continuous variables are presented as
medians with interquartile range (IQR); differences between groups
were evaluated using the Kruskal-Wallis test and post-hoc Tukey's
HSD test or the Mann-Whitney U test for paired groups were used to
establish groups with significant differences. Correlations between
continuous variables were calculated with the Spearman's
rank-correlation coefficient. Kaplan-Meier curves were used to
present relapse-free survival (RFS) time, and hazard ratios (HRs)
were calculated using Cox proportional hazards. P<0.05 was
considered to indicate a statistically significant difference. For
the graphical presentation of result, GraphPad Prism 7.05 software
(GraphPad Software, Inc.) was used.
Results
Clinical and genetic
characteristics
A total of 52 children with BCP-ALL were enrolled in
the study with a median age at diagnosis of 6.54 years (IQR,
2.97–12.38 years). The group was predominantly male (60.40%). The
median white blood cell (WBC) count at diagnosis was
1.40×104 cells/µl (IQR, 0.41–9.68×104
cells/µl) and the median leukemic cell percentage in bone marrow
collected at diagnosis assessed by flow cytometry was 93.10% (IQR,
83.50–96.70%). The mean follow-up time was 4.43 years (range,
0.24–9.6 years).
The patient cohort was divided into groups based on
the major genetic abnormalities detected in leukemic cells that
have prognostic value within the used treatment protocols: The
BCR-ABL1 fusion-positive group (n=5; 9.62% of all patients),
hyperdiploid group (n=7; 13.46%), hypodiploid (n=2; 3.85%),
ETV6-RUNX1 fusion (n=3; 5.77%) and other BCP-ALL group with
no clear primary genetic aberrations (n=35; 67.31%) (Table I). The subgroups differed
significantly in age at diagnosis, with the BCR-ABL1 fusion
group being the oldest at diagnosis (median, 13.76 years; IQR,
6.76–13.90 years; P=0.04).
 | Table I.Clinical characteristics of the
analyzed cohort of 52 patients with BCP-ALL. |
Table I.
Clinical characteristics of the
analyzed cohort of 52 patients with BCP-ALL.
| Characteristic | BCR-ABL1,
n=5 | ETV6-RUNX1,
n=3 | Hyperdiploid,
n=7 | Hypodiploid,
n=2 | Other BCP-ALL,
n=35 | P-value |
|---|
| Male, % | 60.00 | 66.67 | 71.43 | 50.00 | 57.14 | 0.89 |
| Mean age at
diagnosis, years | 13.76 |
8.54 |
2.97 |
1.62 | 7.03 | 0.04 |
| WBC, ×104
cells/µl |
5.27 |
0.81 |
0.62 |
5.70 | 2.35 | 0.33 |
| Blast count, % | 88.00 | 83.25 | 96.80 | 94.00 | 94.50 | 0.64 |
| Poor steroid
response, % | 40.00 | 33.33 | 0 | 50.00 | 10.71 | 0.23 |
| MRD at day 15,
% |
0.40 | 11.34 |
0.71 | 29.45 | 2.23 | 0.51 |
| Relapses, % | 0 | 0 | 0 | 0 | 22.86 | 0.04 |
| Deaths, % | 0 | 0 | 0 | 0 | 20.59 | 0.07 |
The incidence of CNVs frequently observed in
childhood ALL varied between the groups. All patients in the
ETV6-RUNX1 fusion group carried IKZF1 and ETV6
deletions in the leukemic clone, and 2 out of the 3 patients
displayed concomitant CDKN2A/2B deletion. The majority of
patients with BCR-ABL1 fusion exhibited IKZF1 and
PAX5 deletions (both were present in 3 out of 5 patients)
and CDKN2A/2B, CRLF2, ETV6, RB1 and transcriptional
regulator ERG (ERG) deletions were detected in 1 out of 7
patients each. In the patients from the hyperdiploid group,
IKZF1 (3 out of 7), CRLF2 (2 out of 7),
CDKN2A/2B (1 out of 7), PAX5 (1 out of 7) and Janus
kinase 2 (JAK2; 1 out of 7) deletions were identified. The
other BCP-ALL group was the most heterogenic among all groups; all
patients exhibited CNVs in at least one of the tested genes with an
incidence of 29 for IKZF1, 2 for IKZF2, 2 for
IKZF3, 17 for CDKN2A/2B, 8 for PAX5, 4 for
CRLF2, 4 for JAK2, 9 for ETV6, 4 for
ERG and 6 for RB1 out of 35 patients in this group
(Table SI).
Expression analysis
Expression levels of ASNS, LGMN and
CTSB were measured in diagnostic bone marrow samples from
the 52 patients. The median relative expression value for
ASNS was 2.57 (IQR, 2.26–3.26), for LGMN was 1.48
(IQR, 1.21–2.14) and for CTSB was 1.14 (IQR, 0.79–1.77). The
median relative mRNA expression of ASNS correlated
significantly with LGMN and CTSB (r=0.59 and r=0.62,
respectively; P<0.05, data not shown. No correlations were
identified between ASNS, LGMN and CTSB expression
levels and minimal residual disease (MRD) on day 15 of the
treatment protocol or RFS time (data not shown). In addition, no
relevant differences in the median relative mRNA expression levels
of the evaluated genes between the distinct molecular groups of
BCP-ALL were identified (Table SII;
Fig. 1A-C).
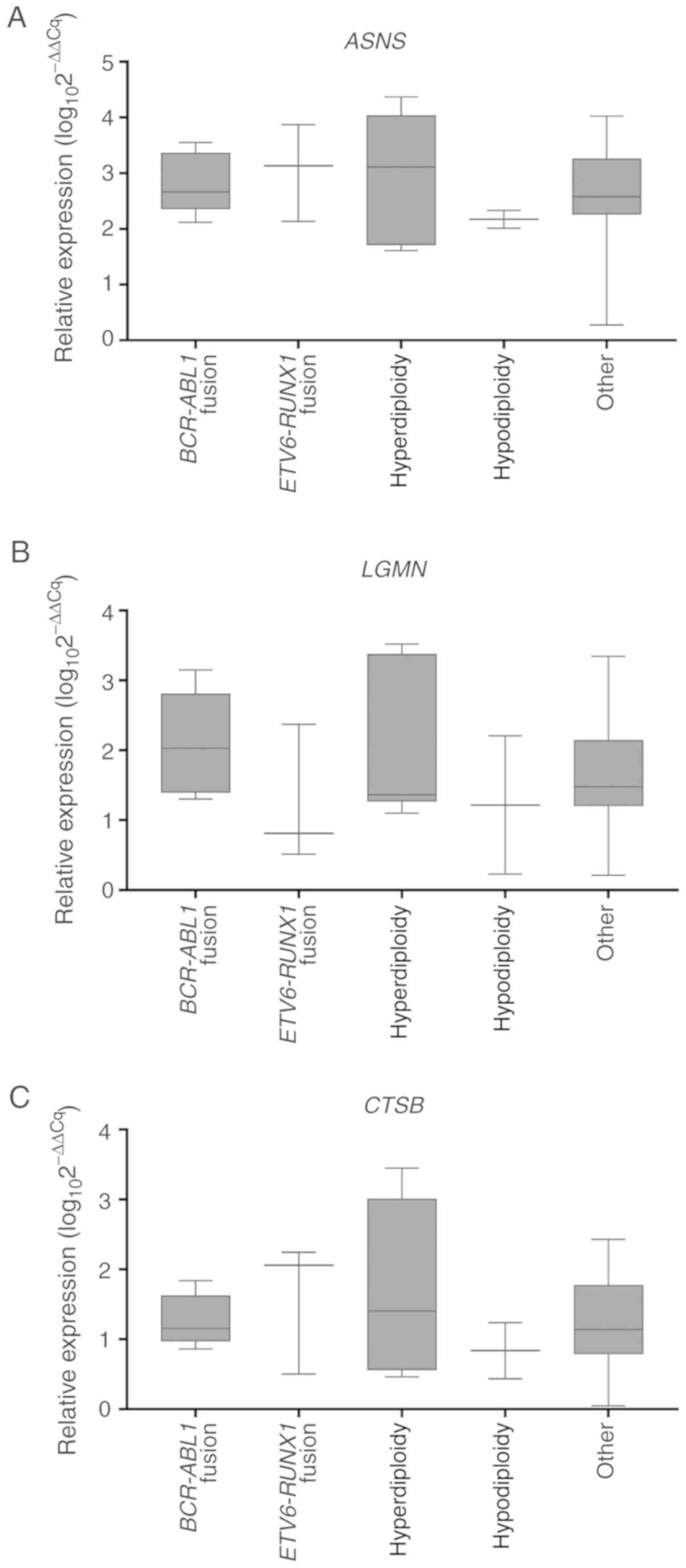 | Figure 1.ASNS, LGMN and CTSB
expression levels in patients with BCP-ALL with various genetic
aberrations. Expression of (A) ASNS, (B) LGMN and (C)
CTSB in every group according to the primary genetic
abnormality identified in the leukemic clone of BCP-ALL. Sample
sizes: BCR-ABL1 fusion, N=5; hyperdiploidy, N=7;
hypodiploidy, N=2; ETV6-RUNX1 fusion, N=3; other, N=35. Data
were obtained by reverse transcription-quantitative PCR and are
presented as relative mRNA expression. Horizontal line, median;
whiskers, minimum to maximum; boxes, interquartile range. ASNS,
asparagine synthetase; LGMN, legumain; CTSB,
cathepsin B; BCP-ALL, B cell precursor acute lymphoblastic
leukemia; BCR-ABL1, breakpoint cluster region and Abelson
murine leukemia viral oncogene homolog 1 fusion; ETV6-RUNX1,
ETS variant 6 and runt-related transcription factor 1 fusion. |
Analysis of the gene expression profiles based on
common CNVs revealed a significantly higher median LGMN
relative expression level in patients with PAX5 deletion
compared with patients with wild-type PAX5 in all BCP-ALL
subgroups (median, 1.93 and 1.34, respectively; P=0.0282; Table SIII). Patients with PAX5
deletion also exhibited a significant correlation of CTSB
median expression level with ASNS and LGMN (r=0.73
and r=0.64, respectively; P<0.05, data not shown). By contrast,
deletion of IKZF1, IKZF2, IKZF3, CDKN2A/2B, CRLF2, JAK2, ETV6,
RB1 or ERG did not exhibit predictive significance for
ASNS, LGMN and CTSB median expression levels as a
single factor (Table SIII).
Differences in ASNS, LGMN and CTSB
expression levels in the other BCP-ALL group were identified when
the patients were divided into subgroups based on PAX5
deletion status; 8/35 (24%) patients carried PAX5 deletions,
whereas 27/35 (76%) did not. Patients with PAX5 deletions
exhibited a higher relative expression level of ASNS
compared with the wild-type PAX5 group (3.14 vs. 2.47;
P=0.0117; Fig. 2A). Similar results
were observed for LGMN expression levels, which were higher
in patients with PAX5 deletion compared with patients with
wild-type PAX5 (2.24 vs. 1.45; P=0.0029; Fig. 2A). In addition, median CTSB
expression levels were higher in patients with
PAX5-deletions compared with patients with wild-type
PAX5 (2.08 vs. 1.04; P<0.0001; Fig. 2A). No significant associations
between the median expression levels of the assessed genes were
identified when the other BCP-ALL patient group was divided based
on IKZF1 deletion status (82.86% of the group carried
deletions; Fig. 2B). However, in
cases with concomitant deletions of IKZF1 and PAX5
(14.29% of the group-5 patients carried both deletions),
significant differences were observed in ASNS and
CTSB expression levels (P=0.0334 and P=0.0358, respectively)
but not in LGMN expression levels (P=0.0735) (Table SIV; Fig.
2C).
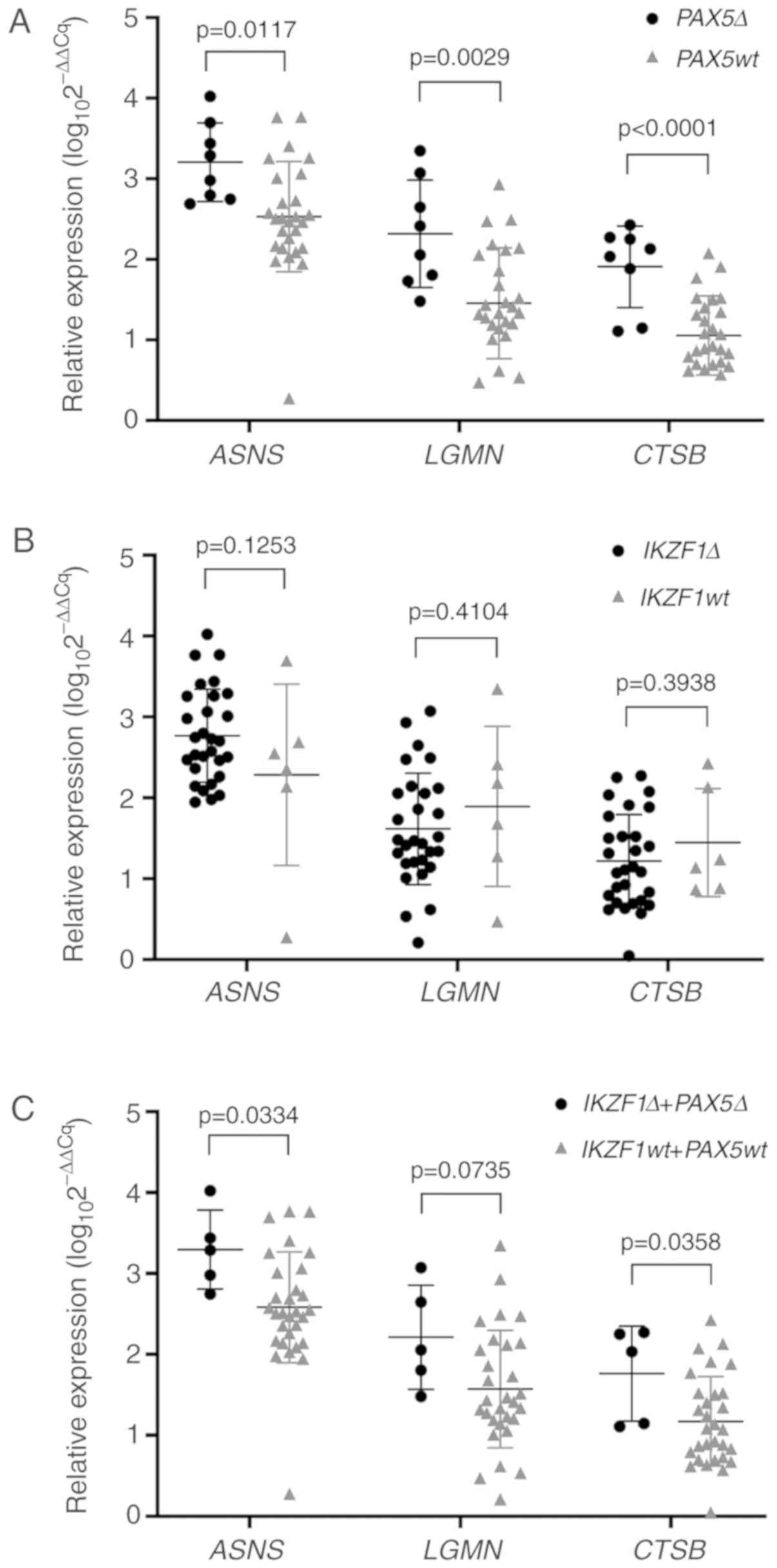 | Figure 2.Expression of ASNS, LGMN and
CTSB in patients carrying PAX5 and IKZF1
deletions. (A) Patients from the other BCP-ALL group with no
identified primary genetic abnormality were separated into
subgroups based on PAX5 deletion (N=8) or wild-type (N=27).
Relative expression of ASNS (3.21 vs. 2.50), LGMN
(2.32 vs. 1.43) and CTSB (1.91 vs. 1.03) in patients with
PAX5 deletions was significantly higher compared with that
in patients with wild-type PAX5. (B) Patients from the other
BCP-ALL group were divided according to the IKZF1 deletion
status (IKZF1 deletions, N=29; IKZF1 wild-type, N=6).
No significant differences were observed in the relative expression
levels of ASNS, LGMN and CTSB. (C) Relative
expression levels of ASNS, LGMN and CTSB in patients
from the other BCP-ALL group with concomitant IKZF1 and
PAX5 deletions (N=5). Middle horizontal line, median value;
whiskers, interquartile range. ASNS, asparagine synthetase;
LGMN, legumain; CTSB, cathepsin B; PAX5,
paired box 5; IKZF1, ikaros family zinc finger protein 1;
wt, wild-type; Δ, deletion; BCP-ALL, B cell precursor acute
lymphoblastic leukemia. |
Of the 12 BCP-ALL patients with PAX5
deletion, four deletions were partial and heterozygous and eight
affected the whole gene: Seven were heterozygous and one was
homozygous. Additionally, TRANSFAC software (GeneXplain, GmbH) was
used to test the PAX5 transcription factor binding sites
(TFBS) for a possible association with ASNS, LGMN or
CTSB. A search through human and mouse databases revealed
that the three genes of interest were not directly regulated by
PAX5.
Outcome description
In the analyzed cohort of 52 patients, eight
relapsed. Median time to relapse was 2.27 years (IQR, 1.37–3.04
years). All relapses occurred in patients in the other BCP-ALL
group. RFS did not differ significantly when patients in the other
BCP-ALL group were divided according to PAX5 deletion
status; median RFS was 5.14 in cases with PAX5 deletion and
5.10 in PAX5 wild-type cases (P=0.4540; HR, 1.56; 95% CI,
0.24–10.20). Out of the eight relapses, six (75%) occurred in
patients with wild-type PAX5, whereas two (25%) occurred in
patients with PAX5 deletions (P=0.0002). When the impact of
PAX5 deletion on RFS was analyzed in the entire cohort, no
significant differences were identified (median RFS for wild-type
PAX5, 4.69 vs. 4.26 years in cases with PAX5
deletion; P=0.7084; HR, 1.50; 95% CI, 0.22–10.03).
The cohort was divided based on the expression
levels of ASNS, LGMN and CTSB, with the cut-off value
set as the median expression for each gene (ASNS, 2.57;
LGMN, 1.48; CTSB, 1.14). When median RFS was analyzed
in patients with high and low ASNS expression, high
ASNS expression was associated with a longer time to relapse
(P=0.0315; HR, 0.19; 95% CI, 0.04–0.86; Fig. 3A). The 5-year RFS rate of patients
with high ASNS expression was 90.15% (95% CI, 87.90–92.40%),
and the 5-year RFS rate of patients with low ASNS expression
was 73.34% (95% CI, 69.40–77.30%) (P=0.0164). No significant
differences in RFS between patients with high and low LGMN
or CTSB expression were observed (P=0.2878; HR, 2.36; 95%
CI, 0.54–10.37; and P=0.7714; HR, 1.25; 95% CI, 0.28–5.49,
respectively; Fig. 3B and C).
Patients with high LGMN expression exhibited a 5-year RFS
rate of 88.46% (95% CI, 85.70–91.20%), and those with low
LGMN expression exhibited a 5-year RFS rate of 77.54% (95%
CI, 74.30–80.80%). Similarly, the 5-year RFS rate was 84.71% (95%
CI, 81.70–87.70%) for patients with high expression of CTSB
and 80.54% (95% CI, 77.40–83.70%) for those with low CTSB
expression.
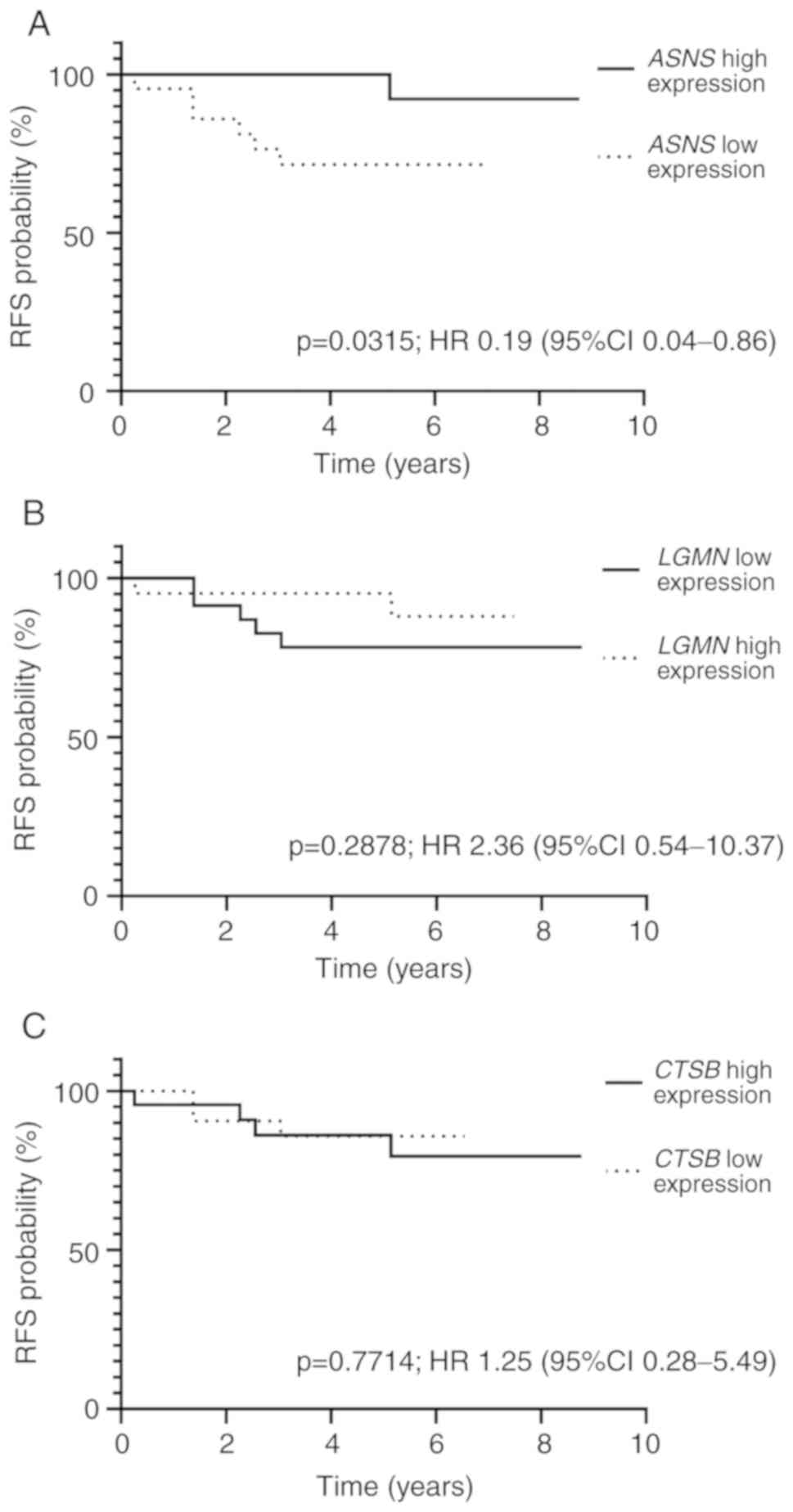 | Figure 3.Kaplan-Meier curves for RFS based on
ASNS, LGMN and CTSB expression levels. (A) For
ASNS, the 5-year RFS rate was 90.15% (95% CI, 87.90–92.40%;
N=24) in the high expression group and 73.34% (95% CI,
69.40–77.30%; N=25) in the low expression group. (B) For
LGMN, the 5-year RFS rate in the high and low LGMN
expression groups was 88.46% (95% CI, 85.70–91.20%; N=23) and
77.54% (95% CI, 74.30–80.80%; N=25), respectively. (C) In the high
and low CTSB expression groups, 5-year RFS rate was 84.71% (95% CI,
81.70–87.70%; N=23) and 80.54% (95% CI, 77.40–83.70%; N=26),
respectively. ASNS, asparagine synthetase; LGMN,
legumain; CTSB, cathepsin B; RFS, relapse-free survival; HR,
hazard ratio; CI, confidence interval. |
Discussion
To evaluate the differences in ASNS, LGMN and
CTSB gene expression in different types of BCP-ALL with
possible additional effects of PAX5 and/or IKZF1 gene
deletions, the patient cohort was grouped according to the major
genetic abnormalities identified in leukemic cells such as
BCR-ABL1 fusion, ETV6-RUNX1 fusion, hypodiploidy and
hyperdiploidy (21). A large
proportion of patients in the current study were classified into
the other BCP-ALL group, in which none of the aforementioned
aberrations were identified. However, considering the high
incidence of IKZF1 and PAX5 deletions in these
patients, it is likely that the group mostly comprised the
BCR-ABL1-like type BCP-ALL (3). However, due to a lack of data on the
gene expression profile, it is impossible to conclusively confirm
this hypothesis, which is one of the limitations of the present
study. Additionally, the small number of patients in the
ETV6-RUNX1 fusion group substantially limited the
comparisons between the remaining groups; therefore, the clinical
characteristics of the group may not fully reflect the favorable
profile described in previous reports (21,22).
The median expression of ASNS did not
significantly differ between the groups; however, relative mRNA
expression of ASNS was the highest in the ETV6-RUNX1
fusion group, which was consistent with previous observation
(5). Previous studies have suggested
the existence of an association between L-asp resistance and the
upregulation of ASNS protein rather than mRNA (5,12,15,16);
however, the data obtained in the present study were insufficient
to verify this hypothesis, which was an additional limitation of
this study. Despite this, longer RFS was observed in patients with
high ASNS gene expression compared with those with low
ASNS expression, which corresponded with the aforementioned
findings (5).
The only previous study of LGMN gene
expression in patients with BCP-ALL reported significantly higher
expression in patients with iAMP21-positive BCP-ALL across all
major genetic variants (14), which
was associated with poor prognosis if not treated aggressively. Due
to the lack of iAMP21-positive cases in the cohort analyzed in the
present study, it was not possible to support those conclusions. No
significant differences in LGMN expression were observed
between the BCP-ALL subgroups; however, the lowest level of
amplification was identified in patients with ETV6-RUNX1
fusion.
To the best of our knowledge, the results of the
present study are the first to describe the CTSB expression
profile in patients with childhood ALL. Despite the lack of
statistical significance, the findings suggest that high
CTSB expression may be associated with positive prognostic
factors such as ETV6-RUNX1 fusion and hyperdiploidy. These
findings contradict our initial hypothesis of the association
between high ASNS, CTSB and LGMN expression levels
and an inferior outcome, although it is in line with the high
ASNS expression observed in cases with ETV6-RUNX1
fusion, as well as the longer time to relapse noted in a previous
study (5).
The results of the present study also suggested an
association between PAX5 deletions and increased ASNS,
LGMN and CTSB expression in the other BCP-ALL patient
group, which has not previously been reported. ASNS, CTSB
and LGMN expression was significantly higher in patients
with PAX5 deletion compared with those with concomitant
IKZF1 deletion, which suggested that PAX5 may
influence the expression of L-asp resistance genes as an
independent factor. Although these comparisons were statistically
significant, further experiments including in vitro PAX5
silencing, as well as larger study cohorts are needed to support
these results.
Although the outcome of PAX5 deletion varies
depending on its location in the front, middle or end of the gene
(23), any PAX5 deletion,
regardless of its location and extent of deleted region, results in
the dysfunction of the PAX5 protein. A small number of this
subgroup enables to statistically connect the region of PAX5
deletion with gene expression levels or outcome. Thus, patients
with PAX5 CNVs were analyzed as a single cohort,
irrespective of the type of PAX5 deletion. This was a
limitation of the present study, and in vitro tests (such as
gene silencing) are required for further confirmation.
TRANSFAC analysis did not provide any information
on the direct influence of PAX5 TFBS on ASNS, LGMN or
CTSB and could not explain the results of the present study.
This suggested that PAX5 may have an indirect effect on
ASNS, LGMN or CTSB expression, and additional
functional studies are required for clarification.
RFS was used for outcome assessment, as it is
superior to overall survival when investigating the influence of
gene expression on resistance to treatment; hence, relapse was the
most accurate end point. High expression of the genes considered to
be responsible for L-asp resistance did not correspond with shorter
RFS or decreased 5-year RFS in this cohort, which rejected the
initial hypothesis of the present study. In addition, higher 5-year
RFS was observed in patients with high ASNS expression,
although the results for CTSB and LGMN were not
significant. Despite the differences in the median RFS in the
subgroup analysis not being significant, patients in the other
BCP-ALL group carrying PAX5 deletions exhibited a longer
time to relapse compared with those with wild-type PAX5.
The main limitation of the present study was a
small number of patients and the lack of BCR-ABL1-like
subtype confirmation, which resulted in challenges in achieving
significant differences between variables. The present study is the
first to report CTSB expression among different genetic
subtypes of BCP-ALL, and the findings encourage further research.
The results of the present study confirmed the association between
PAX5 deletions and the gene expression levels of ASNS,
LGMN and CTSB, as well as between ASNS expression
and 5-year RFS. The latter result, however, requires further study
in a larger cohort for a more thorough explanation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the PRELUDIUM grant
from the National Science Center (grant. no. 2014/13/N/NZ5/03660)
and the National Center of Research and Development PersonALL
project (grant. no. STRATEGMED3/304586/5/2017).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
EW, WM and AP designed the study. ML, JK, LS and TS
obtained clinical data and bone marrow samples, and reviewed the
manuscript. EW, JM and AP conducted the experiments. EW, JM, JJ and
BP collected the data and interpreted the results. EW performed
statistical analyses. EW and WM prepared a final manuscript for
publication.
Ethics approval and consent to
participate
All procedures performed in this retrospective
study involving human participants were in accordance with the
ethical standards of the institutional and national research
committee and with the 1964 Helsinki Declaration and its later
amendments, and were approved by the Bioethical Committee of the
Medical University of Lodz (approval no. RNN/155/13/KE). Informed
consent was obtained from all individual participants and/or their
parents included in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pieters R, Appel I, Kuehnel HJ,
Tetzlaff-Fohr I, Pichlmeier U, Van Der Vaart I, Visser E and
Stigter R: Pharmacokinetics, pharmacodynamics, efficacy, and safety
of a new recombinant asparaginase preparation in children with
previously untreated acute lymphoblastic leukemia: A randomized
phase 2 clinical trial. Blood. 112:4832–4838. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Appel IM, Den Boer ML, Meijerink JP,
Veerman AJ, Reniers NC and Pieters R: Up-regulation of asparagine
synthetase expression is not linked to the clinical response to
L-asparaginase in pediatric acute lymphoblastic leukemia. Blood.
107:4244–4249. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Den Boer ML, van Slegtenhorst M, De
Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ,
Beverloo HB, Van der Spek PJ, Escherich G, et al: A subtype of
childhood acute lymphoblastic leukaemia with poor treatment
outcome: A genome-wide classification study. Lancet Oncol.
10:125–134. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Richards NG and Kilberg MS: Asparagine
synthetase chemotherapy. Annu Rev Biochem. 75:629–654. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Su N, Pan YX, Zhou M, Harvey RC, Hunger SP
and Kilberg MS: Correlation between asparaginase sensitivity and
asparagine synthetase protein content, but not mRNA, in acute
lymphoblastic leukemia cell lines. Pediatr Blood Cancer.
50:274–279. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Accordi B, Galla L, Milani G, Curtarello
M, Serafin V, Lissandron V, Viola G, te Kronnie G, De Maria R,
Petricoin EF III, et al: AMPK inhibition enhances apoptosis in
MLL-rearranged pediatric B-acute lymphoblastic leukemia cells.
Leukemia. 27:1019–1027. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dimitriou H, Choulaki C, Perdikogianni C,
Stiakaki E and Kalmanti M: Expression levels of ASNS in mesenchymal
stromal cells in childhood acute lymphoblastic leukemia. Int J
Hematol. 99:305–310. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chien WW, Le Beux C, Rachinel N, Julien M,
Lacroix CE, Allas S, Sahakian P, Cornut-Thibaut A, Lionnard L,
Kucharczak J, et al: Differential mechanisms of asparaginase
resistance in B-type acute lymphoblastic leukemia and malignant
natural killer cell lines. Sci Rep. 5:80682015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Patel N, Krishnan S, Offman MN, Krol M,
Moss CX, Leighton C, van Delft FW, Holland M, Liu J, Alexander S,
et al: A dyad of lymphoblastic lysosomal cysteine proteases
degrades the antileukemic drug L-asparaginase. J Clin Invest.
119:1964–1973. 2009.PubMed/NCBI
|
|
10
|
Hermanova I, Zaliova M, Trka J and
Starkova J: Low expression of asparagine synthetase in lymphoid
blasts precludes its role in sensitivity to L-asparaginase. Exp
Hematol. 40:657–665. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Krejci O, Starkova J, Otava B, Madzo J,
Kalinova M, Hrusak O and Trka J: Upregulation of asparagine
synthetase fails to avert cell cycle arrest induced by
L-asparaginase in TEL/AML1-positive leukemic cells. Leukemia.
18:434–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He Y, Li B, Luo C, Shen S, Chen J, Xue H,
Tang J and Gu L: Asparagine synthetase is partially localized to
the plasma membrane and upregulated by L-asparaginase in U937
cells. J Huazhong Univ Sci Technol Med Sci. 31:159–163. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lomelino CL, Andring JT, McKenna R and
Kilberg MS: Asparagine synthetase: Function, structure, and role in
disease. J Biol Chem. 292:19952–19958. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Strefford JC, van Delft FW, Robinson HM,
Worley H, Yiannikouris O, Selzer R Richmond T, Hann I, Bellotti T,
Raghavan M, et al: Complex genomic alterations and gene expression
in acute lymphoblastic leukemia with intrachromosomal amplification
of chromosome 21. Proc Natl Acad Sci USA. 103:8167–8172. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stams WA, Den Boer ML, Beverloo HB,
Meijerink JP, Stigter RL, Van Wering ER, Janka-Schaub GE, Slater R
and Pieters R: Sensitivity to L-asparaginase is not associated with
expression levels of asparagine synthetase in t(12;21)+pediatric
ALL. Blood. 101:2743–2747. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aslanian AM, Fletcher BS and Kilberg MS:
L-asparaginase resistance in MOLT-4 human leukaemia cells. Biochem
J. 357:321–328. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
International BFM Study Group, : A
randomized trial of the I-BFM-SG for the management of childhood
ALL IC-BFM. 2009.
|
|
18
|
Stary J, Zimmermann M, Campbell M,
Castillo L, Dibar E, Donska S, Gonzalez A, Izraeli S, Janic D,
Jazbec J, et al: Intensive chemotherapy for childhood acute
lymphoblastic leukemia: Results of the randomized intercontinental
trial ALL IC-BFM 2002. J Clin Oncol. 32:174–184. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wingender E: TheTRANSFAC project as an
example of framework technology that supports the analysis of
genomic regulation. Brief Bioinform. 9:326–332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wenzinger C, Williams E and Gru AA:
Updates in the pathology of precursor lymphoid neoplasms in the
revised fourth edition of the WHO classification of tumors of
hematopoietic and lymphoid tissues. Curr Hematol Malig Rep.
13:275–288. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Moorman AV: New and emerging prognostic
and predictive genetic biomarkers in B-cell precursor acute
lymphoblastic leukemia. Haematologica. 101:407–416. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schwab C, Nebral K, Chilton L, Leschi C,
Waanders E, Boer JM, Žaliová M, Sutton R, Öfverholm II, Ohki K, et
al: Intragenic amplification of PAX5: A novel subgroup in B-cell
precursor acute lymphoblastic leukemia? Blood Adv. 1:1473–1477.
2017. View Article : Google Scholar : PubMed/NCBI
|

















