Introduction
Breast cancer is a heterogeneous disease that can be
divided into different subtypes based on pathological markers. One
of the most important markers is the estrogen receptor (ER).
Estrogen receptor-positive (ER+) and estrogen
receptor-negative (ER−) tumors differ in their
recurrence patterns. Patients with ER− tumors exhibit
higher recurrence risks over the first 5 years following diagnosis.
Thereafter, the risk of recurrence decreases rapidly. However,
patients with ER+ tumors experience late recurrences
more frequently. The biological differences between ER+
and ER− tumors are also evident. Several studies have
revealed that different chromosomal and gene expressional patterns
are present in patients with different ER statuses (1–3).
The majority of breast cancer tumors (75–80%) are
ER+ and of these, ~75% are also progesterone
receptor-positive (PR+) (4,5).
ER+/PR− breast cancer is associated with a
more aggressive phenotype (6),
larger tumors, axillary lymph node metastases and higher S-phase
fractions compared with ER+/PR+ tumors
(7). Differences between these
subgroups have also been observed at the DNA level.
ER+/PR− tumors exhibit a more unstable
genetic profile and possess twice as many copy number gains or
losses as ER+/PR+ tumors (8). Thus, separate analyses for subgroups
based on hormone receptor status is appropriate.
The aim of the current study was to identify new
prognostic factors for ER+ breast cancer via gene copy
number analysis and to investigate if these factors had prognostic
value in subgroups created based on PR status.
The present study identified the gene RAB6C,
located on chromosome 2q21.1, which encodes a 254 amino acid
protein. It was determined to be a prognostic marker following
analysis and was subsequently investigated for prognostic value in
a second independent data set from a study with systemically
untreated patients. To the best of our knowledge, this is the first
study to determine the prognostic value of RAB6C in clinical breast
cancer tissue. However, experimental studies have demonstrated that
RAB6C interacts with p53, which is frequently mutated in breast
cancer (9–12). RAB6C has also been revealed to
inhibit cell proliferation, invasion and metastases, leading to the
hypothesis that it may function as a tumor suppressor (13).
Materials and methods
Patients of the public dataset
The current study utilized a public data set
obtained from the tumor bank of the Erasmus Medical Center, which
included 313 patients with breast cancer. Data is available at the
NCBI GEO website (accession no. 10099) and includes information
pertaining to gene copy number, clinical data and information on
distant recurrences. The patients were treated between 1980 and
1995, and all were determined to be lymph node negative. Patients
also did not receive any adjuvant systemic therapy. Gene copy
number was previously analyzed using the Affymetrix GeneChip Human
Mapping 100K Array. Additionally, Affymetrix Chromosome Copy Number
Tool 3.0 software was used to generate a value representing the
copy number of each probe set. The previously determined data was
determined by Zhang et al (3). From this cohort, the current study
selected ER+ tumors (n=199) for further analyses.
Hormone receptors and grade of the
public dataset
Protein levels of ER and PR were measured using a
ligand binding assay, an enzyme immunoassay or immunohistochemistry
(IHC) for a selection of tumors. The cut-off value for the
classification of patients as positive or negative for ER and PR
was 10 fmol/mg protein or 10% positive tumor cells (14). Grade was assessed by regional
pathologists and reflected the current practice of clinicians over
the years that the tumor samples were collected (3).
Patients of the independent
cohort
To investigate the prognostic value of RAB6C at the
protein level and to analyze a cohort comparable with GSE10099,
systemically untreated patients were selected from two randomized
studies conducted by the Stockholm breast cancer group between 1976
and 1990 (15,16). One study included postmenopausal
‘low-risk’ patients (tumor size ≤30 mm and lymph node-negative) and
the other included premenopausal ‘high-risk’ patients (tumor size
>30 mm and/or lymph node positive). After selecting systemically
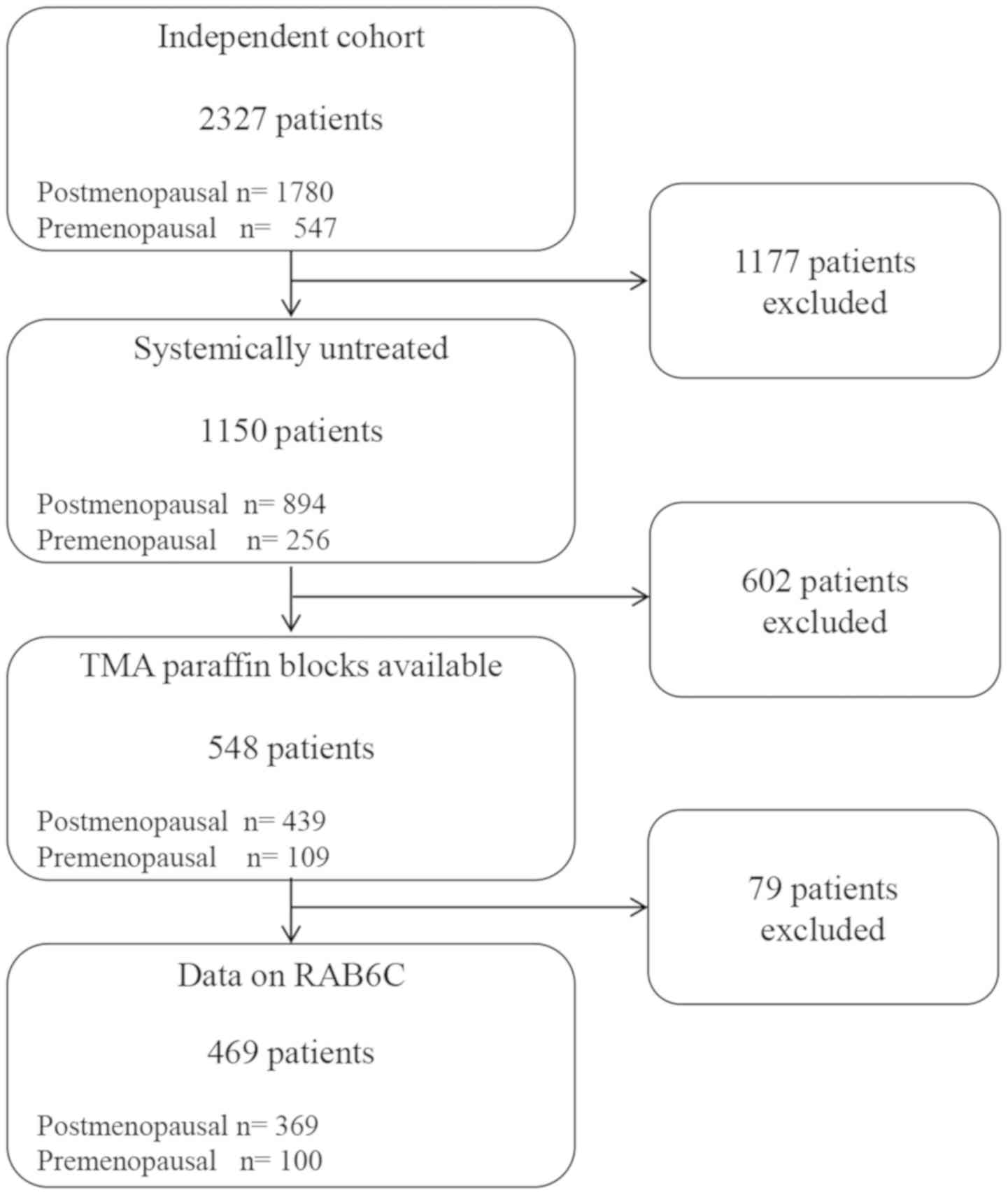
untreated patients, the cohort of the current study consisted of
1,150 patients, where tissue microarrays were available for 548
tumors. Of these, RAB6C expression could be evaluated in a total of
469 patients (Fig. 1).
Hormone receptors, HER2 status, Grade
and RAB6C of the independent cohort
ER, PR and HER2 data were collected from previous
studies. For postmenopausal patients, ER and PR status was assessed
retrospectively via IHC using the Ventana® automated
slide stainer (Ventana Medical Systems, S.A.). CONFIRM™ mouse
anti-ER primary monoclonal antibodies (clone 6F11) and CONFIRM™
mouse anti-PR antibodies (clone 16) were obtained from Ventana
Medical Systems. The cut-off level of positively stained tumor cell
nuclei was set to 10% (17). HER2
was analyzed via IHC as previously described (17). For premenopausal patients, the ER and
PR status that was determined during clinical routine practice was
assessed with a cut-off level of 0.05 fmol/µg DNA (18). HER2 was analyzed via IHC using the
same antibody as for postmenopausal patients. Grade was analyzed
retrospectively in both cohorts using the same investigator for all
tumor samples within each.
The protein expression of RAB6C was analyzed via IHC
and the staining pattern was evaluated independently by three
investigators (JS, TB and HF). Polyclonal rabbit antibodies (cat.
no. ab200396; Abcam) were used. The intensity of RAB6C in the
nucleus was analyzed and scored as 0, 1, 2 or 3. If the nuclei
exhibited an intensity ≥2, the tumor was considered to highly
express RAB6C (RAB6C+). Otherwise, it was considered to
exhibit a low RAB6C expression (RAB6C−). Fig. S1 presents examples of tumors that
were graded as RAB6C+ and RAB6C−,
respectively. RAB6C expression in the cytoplasm was also evaluated
and scored as 0, 1, 2 or 3.
Statistical analysis
The interquartile range was calculated for each SNP
and the 20 most varied were selected and analyzed separately. For
each SNP, the patients were divided into two groups based on their
gene copy number, with the median value as a cut-off. To compare
the association between RAB6C and clinical characteristics, the
Pearson χ2 test was utilized.
Cumulative distant-recurrence risk was estimated
using the Kaplan-Meier method. In the public data set, distant
recurrence was calculated as previously described by Zhang et
al (3). In the independent
cohort, the end-point was defined as the first distant recurrence
from the patient's primary breast tumor as described by Rutqvist
and Johansson (15,16). In this cohort, 3 patients died from
breast cancer, but no date of distant recurrence was recorded. For
these patients, the date of death was used as date of distant
recurrence. Patients were censored at the last follow-up or at
death due to causes other than breast cancer. Hazard ratios (HRs)
and 95% confidence intervals (CIs) were estimated using Cox's
proportional hazards model. P-values were obtained from two-sided
Wald tests and the patients were followed up until 10 years after
diagnosis. The microarray data was processed using R 2.14.1 and the
statistical analyses were performed with Stata/SE 13.1
software.
Results
Public data set results
The 20 SNPs that varied the most between patients
were mapped as MECT1, UBN1 (3 SNPs), SKI, GALNT1,
FLJ35424, FCGR1A, WIG1, RAB6C, MUM1L1, 7p22.3, POU5FLC20,
ZNF195, LOC200030, LOC122618, ODF1 and OSR2 (3 SNPs;
Table SI). When analyzing the
distant recurrence risk of each SNP after applying Bonferroni's
correction, only RAB6C had a statistically significant
impact on prognostic value. Furthermore, since previous studies
indicated that RAB6C might be a favorable prognostic and predictive
factor, this gene was selected for further analysis (11,13,19,20).
The second most significant P-value was obtained for
the MUM1L1 gene (HR, 0.55; 95% CI, 0.35–0.86; P=0.01). The
protein that the MUM1L1 gene encodes has been previously
revealed to be highly expressed in several types of cancer.
According to the protein atlas, the expression of MUM1L1 is
not detectable in normal breast tissue and is expressed in low
levels in breast cancer tissue (21). However, due to a non-significant
P-value after Bonferroni's correction, no further analysis was
performed on MUM1L1 in the current study.
The patients that were subdivided into groups
according to their tumor copy number being above or below the
median value of 3.6 were denoted as the RAB6C+
and RAB6C− groups, respectively. The HR of the
patients that exhibited a RAB6C+ tumor compared
with those that exhibited a RAB6C− tumor was 0.44
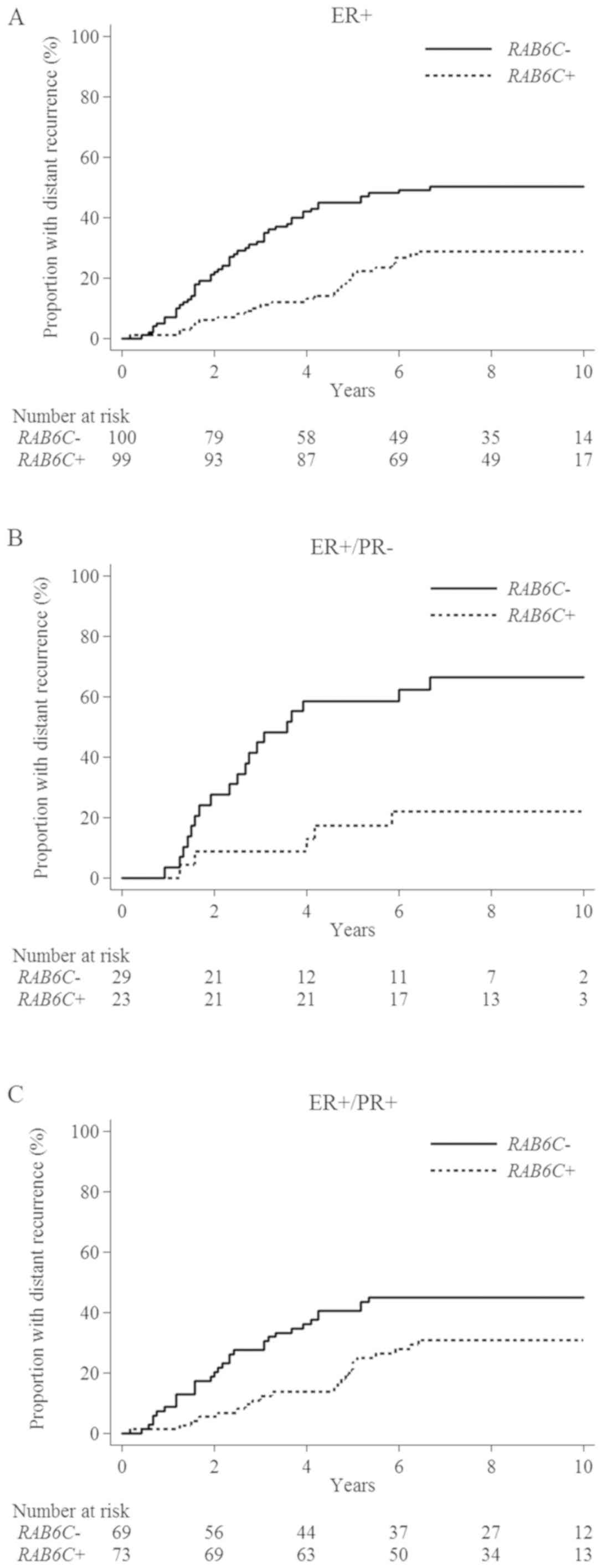
(95% CI, 0.28–0.71; P=0.001; Fig.
2A). The prognostic value of RAB6C was determined by
performing a multivariable analysis with adjustments for age, tumor
stage, grade and PR. Of these factors, RAB6C had the
strongest prognostic value (HR, 0.45; 95% CI, 0.28–0.72; P=0.001;
Tables I and SII).
 | Table I.Distant recurrence rate in the data
set GSE10099 for RAB6C+ compared with
RAB6C− stratified by hormone receptor status. |
Table I.
Distant recurrence rate in the data
set GSE10099 for RAB6C+ compared with
RAB6C− stratified by hormone receptor status.
|
|
|
| Univariable |
|
Multivariablea |
|
|---|
|
|
|
|
|
|
|
|
|---|
|
| Number of
patients/events | HR (95% CI) |
| HR (95% CI) |
|
|---|
|
|
|
|
|
|
|
|---|
| Hormone receptor
status |
RAB6C+ |
RAB6C− |
RAB6C+ vs.
RAB6C− | P-value |
RAB6C+ vs.
RAB6C− | P-value |
|---|
| ER+ | 99/28 | 100/50 | 0.44
(0.28–0.71) | 0.001 | 0.45
(0.28–0.72) | 0.001 |
|
ER+/PR+ | 73/22 | 69/31 | 0.55
(0.32–0.96) | 0.034 | 0.55
(0.32–0.95) | 0.033 |
|
ER+/PR− | 23/5 | 29/19 | 0.23
(0.08–0.61) | 0.003 | 0.15
(0.05–0.46) | 0.001 |
The patients were further divided into two subgroups
based on PR status. In both subgroups, RAB6C had a favorable
prognostic value, which was more evident among
ER+/PR− patients. In these patients, there
was a 77% risk reduction (HR, 0.23; 95% CI, 0.08–0.61; P=0.003) in
tumors that were RAB6C+ compared with those that
were RAB6C− (Fig.
2B). For ER+/PR+ tumors, the risk
reduction was 45% (HR, 0.55; 95% CI, 0.32–0.96; P=0.03; Fig. 2C). Based on PR status, two separate
multivariable analyses were performed. RAB6C+
revealed a statistically significant improvement in the prognosis
of both subgroups. However, a stronger independent prognostic value
was identified among ER+/PR− tumors (HR,
0.15; 95% CI, 0.05–0.46 vs. HR, 0.55; 95% CI, 0.32–0.95; Tables I and SIII).
A higher gene copy number of RAB6C was
associated with favorable prognostic factors.
RAB6C+ tumors were more frequently of T1 size (56
vs. 49%), PR+ (76 vs. 70%) or of grade I or II (32 vs.
26%) when compared with RAB6C− tumors. However,
none of these associations were statistically significant. In the
ER+/PR− subgroup, RAB6C+
tumors were more commonly grade I or II (P=0.008; Table II).
 | Table II.Patient characteristics for the data
set GSE10099, stratified by hormone receptor status. |
Table II.
Patient characteristics for the data
set GSE10099, stratified by hormone receptor status.
|
| ER+, n
(%) |
|
ER+/PR+, n (%) |
|
ER+/PR−, n (%) |
|
|---|
|
|
|
|
|
|
|
|
|---|
|
Characteristics |
RAB6C− |
RAB6C+ | P-value |
RAB6C− |
RAB6C+ | P-value |
RAB6C− |
RAB6C+ | P-value |
|---|
| Total no. of
patients | 100 | 99 |
| 69 | 73 |
| 29 | 23 |
|
| Age, years |
|
|
|
|
|
|
|
|
|
|
≤50 | 46 (46) | 38 (38) | 0.28 | 37 (54) | 30 (41) | 0.14 | 9 (31) | 7 (30) | 0.96 |
|
>50 | 54 (54) | 61 (62) |
| 32 (46) | 43 (59) |
| 20 (69) | 16 (70) |
|
| Tumour stage |
|
|
|
|
|
|
|
|
|
| T1 | 49 (49) | 55 (56) | 0.36 | 32 (46) | 41 (56) | 0.24 | 16 (55) | 11 (48) | 0.60 |
|
T2-T4 | 51 (51) | 44 (44) |
| 37 (54) | 32 (44) |
| 13 (45) | 12 (52) |
|
| PR status |
|
|
|
|
|
|
|
|
|
|
Negative | 29 (30) | 23 (24) | 0.38 |
|
|
|
|
|
|
|
Positive | 69 (70) | 73 (76) |
|
|
|
|
|
|
|
|
Unknown | 2 | 3 |
|
|
|
|
|
|
|
| Grade |
|
|
|
|
|
|
|
|
|
|
I–II | 17 (26) | 22 (32) | 0.40 | 14 (35) | 11 (23) | 0.23 | 3 (13) | 9 (50) | <0.01 |
|
III | 49 (74) | 46 (68) |
| 26 (65) | 36 (77) |
| 21 (88) | 9 (50) |
|
|
Unknown | 34 | 31 |
| 29 | 26 |
| 5 | 5 |
|
Independent cohort results
The prognostic value of RAB6C based on protein
expression was tested in two different cohorts that included
premenopausal ‘high-risk’ patients and postmenopausal ‘low-risk’
patients, respectively. Since statistical analysis revealed similar
results in both cohorts, they were merged into a single cohort to
increase the statistical power. Cytoplasmic RAB6C expressions were
observed in almost all cases (96%) with staining intensities of 2
or 3 in 71%. The results of nuclear staining were more varied, with
scores of 0, 1, 2 and 3 in 34, 23, 25 and 19% of cases,
respectively. The statistical analysis of cytoplasmic RAB6C
expression did not demonstrate any prognostic value and as such,
all results included in the current study were based on the nuclear
expression of RAB6C.
The dataset contained both ER+ and
ER− tumors, which revealed a strong positive correlation
between RAB6C and ER status (P=0.001). However, a negative
correlation with borderline significance was observed between RAB6C
and HER2 (P=0.057). RAB6C+ tumors were also frequently
of a lower grade than RAB6C− tumors (P<0.001;
Table III). The current study
subsequently analyzed the association between RAB6C and the three
individual components of grading (pleomorphism, tubule formation
and mitosis). The results revealed a negative correlation between
RAB6C positivity and each of the components (P<0.001). Among the
ER+ tumors, no statistically significant associations
between RAB6C and the prognostic factors presented in Table III were detected, but HER2 was
negativity associated with RAB6C+ tumors (P=0.07).
ER+/PR− tumors with high RAB6C levels were
frequently of a lower grade (P=0.016; Table IV).
 | Table III.Patient characteristics for the
independent cohort. |
Table III.
Patient characteristics for the
independent cohort.
|
| All patients, n
(%) |
| ER+, n
(%) |
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics |
RAB6C− |
RAB6C+ | P-value |
RAB6C− |
RAB6C+ | P-value |
|---|
| Total no. of
patients | 265 | 204 |
| 157 | 153 |
|
| Age, years |
|
|
|
|
|
|
|
≤50 | 61 (23) | 44 (22) | 0.71 | 38 (24) | 34 (22) | 0.68 |
|
>50 | 204 (77) | 160 (78) |
| 119 (76) | 119 (78) |
|
| Tumour size,
mm |
|
|
|
|
|
|
|
<30 | 218 (85) | 181 (90) | 0.14 | 134 (87) | 137 (90) | 0.39 |
|
≥30 | 37 (15) | 20 (10) |
| 20 (13) | 15 (10) |
|
|
Unknown | 10 | 3 |
| 3 | 1 |
|
| Lymph node
status |
|
|
|
|
|
|
| N0 | 214 (81) | 169 (83) | 0.56 | 124 (79) | 127 (83) | 0.37 |
|
N+ | 51 (19) | 35 (17) |
| 33 (21) | 26 (17) |
|
| ER status |
|
|
|
|
|
|
|
Negative | 68 (30) | 30 (16) | 0.001 |
|
|
|
|
Positive | 157 (70) | 153 (84) |
|
|
|
|
|
Unknown | 40 | 21 |
|
|
|
|
| PR status |
|
|
|
|
|
|
|
Negative | 111 (51) | 78 (45) | 0.24 | 50 (35) | 51 (37) | 0.76 |
|
Positive | 105 (49) | 94 (55) |
| 92 (65) | 87 (63) |
|
|
Unknown | 49 | 32 |
| 15 | 15 |
|
| HER2 |
|
|
|
|
|
|
|
Negative | 197 (84) | 166 (90) | 0.057 | 134 (91) | 138 (96) | 0.07 |
|
Positive | 38 (16) | 18 (10) |
| 14 (9) | 6 (4) |
|
|
Unknown | 30 | 20 |
| 9 | 9 |
|
| NHG |
|
|
|
|
|
|
| I | 38 (17) | 50 (28) | <0.001 | 32 (23) | 45 (32) | 0.10 |
| II | 119 (52) | 106 (60) |
| 86 (61) | 81 (58) |
|
|
III | 73 (32) | 21 (12) |
| 23 (16) | 14 (10) |
|
|
Unknown | 35 | 27 |
| 16 | 13 |
|
 | Table IV.Patient characteristics for the
independent cohort stratified by hormone receptor status. |
Table IV.
Patient characteristics for the
independent cohort stratified by hormone receptor status.
|
|
ER+/PR+, n (%) |
|
ER+/PR−, n (%) |
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics |
RAB6C− |
RAB6C+ | P-value |
RAB6C− |
RAB6C+ | P-value |
|---|
| Total no. of
patients | 92 | 87 |
| 50 | 51 |
|
| Age, years |
|
|
|
|
|
|
|
≤50 | 22 (24) | 14 (16) | 0.19 | 9 (18) | 11 (22) | 0.65 |
|
>50 | 70 (76) | 73 (84) |
| 41 (82) | 40 (78) |
|
| Tumour size,
mm |
|
|
|
|
|
|
|
<30 | 79 (87) | 79 (91) | 0.40 | 41 (85) | 46 (92) | 0.30 |
|
≥30 | 12 (13) | 8 (9) |
| 7 (15) | 4 (8) |
|
|
Unknown | 1 | 0 |
| 2 | 1 |
|
| Lymph node
status |
|
|
|
|
|
|
| N0 | 70 (76) | 76 (87) | 0.05 | 45 (90) | 43 (84) | 0.39 |
|
N+ | 22 (24) | 11 (13) |
| 5 (10) | 8 (16) |
|
| HER2 |
|
|
|
|
|
|
|
Negative | 85 (97) | 84 (99) | 0.33 | 38 (79) | 42 (89) | 0.17 |
|
Positive | 3 (3) | 1 (1) |
| 10 (21) | 5 (11) |
|
|
Unknown | 4 | 2 |
| 2 | 4 |
|
| NHG |
|
|
|
|
|
|
| I | 21 (26) | 25 (30) | 0.55 | 6 (12) | 16 (36) | 0.02 |
| II | 47 (59) | 50 (60) |
| 33 (67) | 24 (55) |
|
|
III | 12 (15) | 8 (10) |
| 10 (20) | 4 (9) |
|
|
Unknown | 12 | 4 |
| 1 | 7 |
|
Since the analyses performed using the public
dataset were based on ER+ tumors, the independent cohort
was divided into two groups based on ER status. Among these
subgroups, no statistically significant differences were determined
in distant recurrence rates between RAB6C+ and
RAB6C− cases, independently of ER status. For patients
with ER+ tumors, the HR was 0.77 (95% CI, 0.48–1.22;
P=0.27; Fig. 3A) and for
ER−tumors, the HR was 1.30 (95 % CI, 0.66–2.56;
P=0.45).
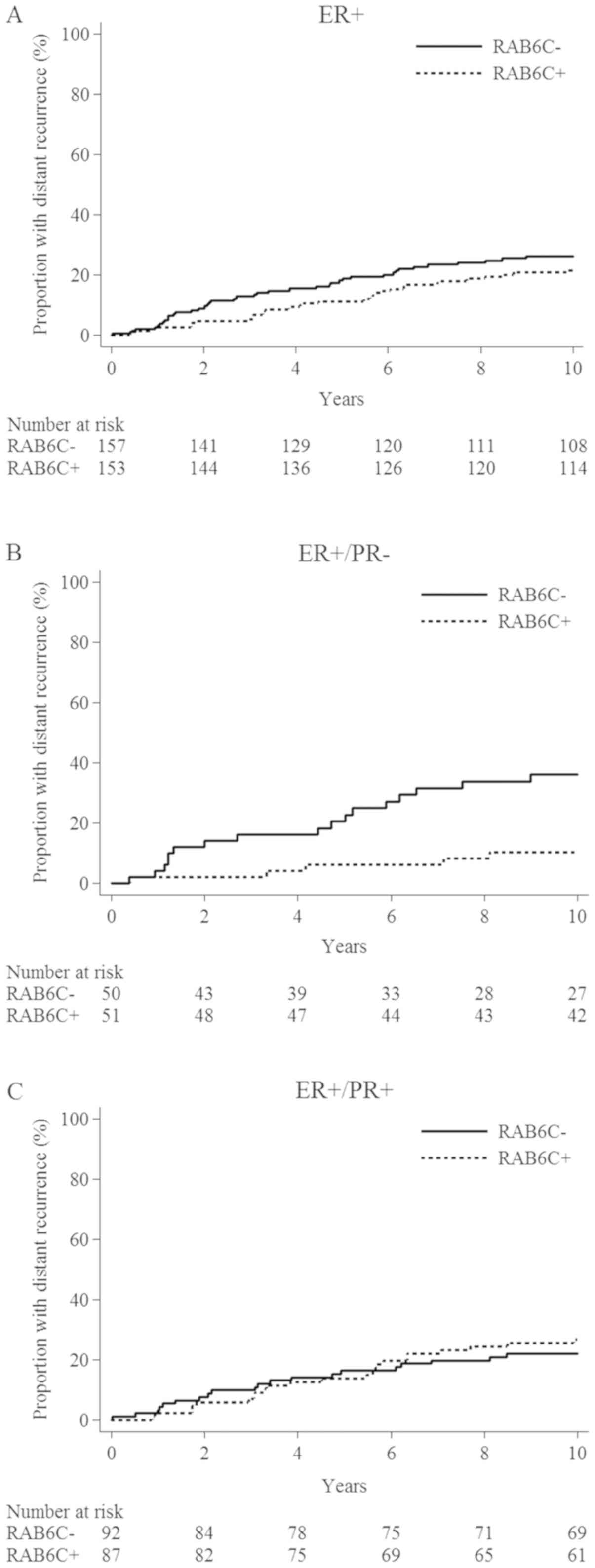 | Figure 3.Cumulative distant recurrence risk in
relation to the protein expression of RAB6C in patients of the
independent cohort. (A) ER+. HR, 0.77 (95% CI,
0.48–1.22; P=0.27). (B) ER+/PR−. HR, 0.24
(95% CI, 0.09–0.66; P=0.005). (C) ER+/PR+.
HR, 1.20 (95% CI, 0.66–2.19; P=0.55). ER, estrogen receptor; HR,
hazard ratio; PR, progesterone receptor; RAB6C, Ras-related protein
Rab-6C. |
For patients with ER+/PR−
tumors, a decreased distant recurrence rate was observed in
RAB6C+ compared with RAB6C− cases (HR, 0.24;
95% CI, 0.09–0.66; P=0.005; Fig.
3B). However, no statistically significant differences were
determined in patients with ER+/PR+ tumors
(HR, 1.20; 95% CI, 0.66–2.19; P=0.55; Fig. 3C). The separate HRs for the
‘low-risk’ and ‘high-risk’ cohorts with
ER+/PR− tumors were 0.22 (95% CI, 0.06–0.79)
and 0.21 (95% CI, 0.041–1.10), respectively. For those with
ER+/PR+ tumors, the HRs were 1.56 (95% CI,
0.72–3.41) and 1.21 (95% CI, 0.44–3.31), respectively (Fig. S2).
When adjusting for age, tumor size, lymph node
status, HER2, grade and PR in the multivariable analysis of distant
recurrence risk in patients with ER+ tumors, RAB6C
retained its prognostic importance, which was stronger in patients
with ER+/PR− tumors as indicated by a
statistically significant interaction between PR and RAB6C
(Table SIV). Stratifying the
results of the multivariable analysis according to PR status
revealed that the distant recurrence rate ratio for high vs. low
RAB6C was markedly reduced among patients with
ER+/PR− tumors (HR, 0.17; 95% CI, 0.05–0.60;
P=0.006), but not in patients with ER+/PR+
tumors (HR, 1.31; 95% CI, 0.69–2.48; P=0.41; Table V).
 | Table V.Distant recurrence rate in the
independent cohort for high RAB6C compared with low RAB6C
stratified by hormone receptor status. |
Table V.
Distant recurrence rate in the
independent cohort for high RAB6C compared with low RAB6C
stratified by hormone receptor status.
|
|
|
| Univariable |
|
Multivariablea |
|
|---|
|
|
|
|
|
|
|
|
|---|
|
| Number of
patients/events | HR (95% CI) |
| HR (95% CI) |
|
|---|
|
|
|
|
|
|
|
|---|
| Hormone receptor
status |
RAB6C+ |
RAB6C− | RAB6C+
vs. RAB6C− | P-value | RAB6C+
vs. RAB6C− | P-value |
|---|
| All | 204/49 | 265/73 | 0.82
(0.57–1.18) | 0.29 |
|
|
| ER+ | 153/32 | 157/40 | 0.77
(0.48–1.22) | 0.27 |
|
|
| ER− | 30/13 | 68/23 | 1.30
(0.66–2.56) | 0.45 |
|
|
|
ER+/PR+ | 87/23 | 92/20 | 1.20
(0.66–2.19) | 0.55 | 1.31
(0.69–2.48) | 0.41 |
|
ER+/PR− | 51/5 | 50/17 | 0.24 (0.09
0.66) | <0.01 | 0.17
(0.05–0.60) | <0.01 |
Discussion
To the best of our knowledge, the current study is
the first to assess RAB6C in a dataset of clinical breast
cancer tissue. The prognostic significance was identified using a
dataset containing gene copy numbers and further investigations
were performed at the protein level in a second independent cohort.
The results indicated that high RAB6C expressions decreased the
distant recurrence risk of patients, a finding that was most
significant in the ER+/PR− subgroup. Many of
these patients receive both endocrine therapy and chemotherapy, but
certain individuals may exhibit a good prognosis even when
receiving reduced systemic treatment. In the cohorts of the present
study, all patients were systemically untreated, which enabled for
the analysis of the natural course associated with RAB6C. The
results of the present study may contribute to the understanding of
biological heterogeneity within ER+/PR−
tumors and the identification of potentially overtreated
patients.
Variations in gene copy number were analyzed in a
dataset of 199 ER+ tumors, after which the 20 most
varied SNPs were selected and tested for prognostic significance.
To counteract the problem of multiple testing, the P-value for each
SNP was assessed at a significance level of α/20=0.0025 according
to the principle of Bonferroni's correction. Furthermore, after
applying the Cox regression model, RAB6C was identified as a
prognostic marker.
The identification of RAB6C as a tumor marker with
prognostic significance in the current study was supported by
previous literature. Bhat et al (22,23) used
next generation sequencing and data from independent published
studies to identify RAB6C as one of nine genes with promoter
methylation. The high sensitivity and specificity of this could
discriminate between normal, premalignant or tumor tissues in
cervical tissues and in head and neck squamous cell carcinoma.
Furthermore, hypermethylation leading to the downregulation of
RAB6C was associated with a poorer survival (22,23).
The function of RAB6C is yet to be fully
elucidated. However, the gene is known to be a member of the RAB6
family, which is highly conserved. By gene duplication and splicing
during evolution, RAB6 has diverged to three different isoforms;
RAB6A, RAB6B and RAB6C. RAB6A has been
subjected to splicing, generating two further variants,
RAB6A and RAB6A′. RAB6C was most likely
generated by the retrotransposition of mRNA from RAB6A′ and
therefore consists of a single exon (24). This retrotransposition theory is also
supported by the fact that RAB6C and RAB6A′ genes
exhibit an identity of 97%. However, the proteins that the genes
encode seem to have different functions. As far as we know, ER and
PR are not target genes of RAB6C, but RAB6C has been demonstrated
to be directly targeted by p53 and several experimental studies
have indicated that high RAB6C expressions may be a favorable
prognostic and predictive factor for cancer (11,13,19). The
expression also seems to be associated with the type of tissue.
Cells with a high metastatic ability possess lower RAB6C levels
than MCF-7 cells, which in turn have lower RAB6C expressions than
normal tissue (13). Furthermore,
RAB6C inhibits cell proliferation, invasion and metastases
(13), and promotes apoptosis
(11). Shan et al (19) revealed that RAB6C may be involved in
chemotherapeutic resistance. Cancer cells expressing RAB6C have
also demonstrated an increased sensitivity to several anti-cancer
drugs, including doxorubicin, paclitaxel, vinblastine and
vincristine (19). Unlike other
proteins of the RAB6 family, which are located in the Golgi
apparatus serving to regulate protein transport, RAB6C is primarily
localized to the centrosome and is involved in cell division
(24). This distinguished function
of RAB6C may contribute to the prognostic impact that was
identified in the current study.
We identified prognostic significance of
RAB6C based on gene copy number with data from a public data
set. Since gene copy number variation is an indicator of genomic
instability, which per se implies poor prognosis, it is important
to consider also the gene expression. Therefore, we also analyzed
RAB6C protein expression in an independent cohort. We did not have
data on both gene copy number and gene expression in either cohort,
which impeded a correlation analysis between gene copy number and
gene expression. Gene expression analysis of RAB6C is complicated
due to the fact that it consists of a single exon and several gene
expression microarrays are not able to distinguish between mRNA
expression of RAB6A and RAB6C (24,25). In
order to distinguish between RAB6A and RAB6C, protein expression
analysis seems to be a more reliable method.
RAB6C is located within a genomic region with
large-scale copy number polymorphism, contributing to genomic
variation between normal humans (26). The RAB6C-like protein is expressed by
a gene located in the same chromosomal region within a distance of
1.38 Mb from RAB6C (24). The
antibody that was used in the current study has been confirmed to
detect RAB6C. However, the proteins are 100% identical according to
NCBI's protein database, which uses the BLAST tool (27) and differ at three amino acids
according to the database of Ensembl (28). Thus, RAB6C and RAB6C-like protein are
very similar and the gene products may possess the same
function.
In the public dataset and the independent cohort
used in the current study, an association was determined between
grade and RAB6C, particularly among ER+/PR−
tumors. In the independent cohort, the current study also analyzed
the association between RAB6C and the three components of grading
(pleomorphism, tubule formation and mitosis). The results revealed
a negative correlation between RAB6C and each of the components,
including mitosis (P<0.001). This seems reasonable, since RAB6C
has been demonstrated to serve a role in DNA replication and it is
known that the depletion of RAB6C generates tetraploid cells with
supernumerary centrosomes (24).
The present study analyzed the prognostic course
associated with RAB6C. Since most ER+ patients receive
systemic therapy and experimental studies have indicated that RAB6C
may be involved in chemotherapeutic resistance, a treatment
predictive value should be investigated in future study.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank data manager Mrs.
Ulla Johansson (Regional Cancer Center of Stockholm Gotland,
Stockholm, Sweden) for updating the database, technician Mrs.
Birgitta Holmlund (Department of Oncology, Linköping University
Hospital, Linköping, Sweden) for technical assistance, and Dr Najme
Wall (Department of Oncology, Linköping University Hospital,
Linköping, Sweden), Dr Hans Olsson (Department of Pathology,
Linköping University Hospital, Linköping, Sweden) and Dr Dennis
Sgroi (Department of Pathology, Massachusetts General Hospital,
Boston, MA, USA) for determining tumor grades.
Funding
The present study was supported by grants obtained
from the Swedish Cancer Society (OS; grant no. 17-0479), The Cancer
Research Foundations of Radiumhemmet (TF), the Cancer Society in
Stockholm (TF) and the King Gustav V Jubilee Clinical Research
Foundation (TF; grant no. 181093), the Onkologiska klinikerna i
Linköpings forskningsfond (HF; grant no. 2016-06-21), ALF grants
Region Östergötland (OS; grant no. LIO-795201) and the County
Council of Östergötland (HF; grant no. LIO-625491). Funding sources
did not have any role in the study design, in the collection,
analysis, interpretation of data, in the writing of the manuscript
or in the decision to submit the manuscript for publication.
Availability of data and materials
The public data set with accession number GSE10099
analyzed during the present study is available in the NCBI GEO
repository (http://www.ncbi.nlm.nih.gov/). The datasets from the
independent cohort used and/or analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
HF, JC and OS conceived and designed the present
study. TF and BN provided the study materials, collected clinical
follow-up data and hormone receptor data from the patients, and
performed the clinical interpretation. TB, JS and HF performed the
experiments and scored RAB6C. HF performed statistical analysis and
drafted the manuscript. HF, JC, OS, TB, JS, BN and TF interpreted
the results, critically revised the manuscript and approved the
final version.
Ethics approval and consent to
participate
The present study was performed in accordance with
the Declaration of Helsinki. For the independent cohort, an ethical
approval for the use of tumor material was provided by the local
Ethical Committee of Karolinska University Hospital (approval no.
KI 97-451 with amendments 030201 and 171027). According to the
approval that was received, patient informed consent was not
required.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CI
|
confidence interval
|
|
ER
|
Estrogen receptor
|
|
HER2
|
human epidermal growth factor receptor
2
|
|
HR
|
hazard ratio
|
|
IHC
|
immunohistochemistry
|
|
PR
|
progesterone receptor
|
|
RAB6C
|
Ras-related protein Rab-6C
|
|
SNP
|
single nucleotide polymorphism
|
|
TMA
|
tissue microarray
|
References
|
1
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gruvberger S, Ringnér M, Chen Y, Panavally
S, Saal LH, Borg A, Fernö M, Peterson C and Meltzer PS: Estrogen
receptor status in breast cancer is associated with remarkably
distinct gene expression patterns. Cancer Res. 61:5979–5984.
2001.PubMed/NCBI
|
|
3
|
Zhang Y, Martens JW, Yu JX, Jiang J,
Sieuwerts AM, Smid M, Klijn JG, Wang Y and Foekens JA: Copy number
alterations that predict metastatic capability of human breast
cancer. Cancer Res. 69:3795–3801. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grann VR, Troxel AB, Zojwalla NJ, Jacobson
JS, Hershman D and Neugut AI: Hormone receptor status and survival
in a population-based cohort of patients with breast carcinoma.
Cancer. 103:2241–2251. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Early Breast Cancer Trialists'
Collaborative Group (EBCTCG), ; Davies C, Godwin J, Gray R, Clarke
M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, et al: Relevance
of breast cancer hormone receptors and other factors to the
efficacy of adjuvant tamoxifen: Patient-level meta-analysis of
randomised trials. Lancet. 378:771–784. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thakkar JP and Mehta DG: A review of an
unfavorable subset of breast cancer: Estrogen receptor positive
progesterone receptor negative. Oncologist. 16:276–285. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Arpino G, Weiss H, Lee AV, Schiff R, De
Placido S, Osborne CK and Elledge RM: Estrogen receptor-positive,
progesterone receptor-negative breast cancer: Association with
growth factor receptor expression and tamoxifen resistance. J Natl
Cancer Inst. 97:1254–1261. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Viale G, Regan MM, Maiorano E,
Mastropasqua MG, Dell'Orto P, Rasmussen BB, Raffoul J, Neven P,
Orosz Z, Braye S, et al: Prognostic and predictive value of
centrally reviewed expression of estrogen and progesterone
receptors in a randomized trial comparing letrozole and tamoxifen
adjuvant therapy for postmenopausal early breast cancer: BIG 1-98.
J Clin Oncol. 25:3846–3852. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Haverty PM, Fridlyand J, Li L, Getz G,
Beroukhim R, Lohr S, Wu TD, Cavet G, Zhang Z and Chant J:
High-resolution genomic and expression analyses of copy number
alterations in breast tumors. Genes Chromosomes Cancer. 47:530–542.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tian K, Wang Y and Xu H: WTH3 is a direct
target of the p53 protein. Br J Cancer. 96:1579–1586. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tian K, Wang Y, Huang Y, Sun B, Li Y and
Xu H: Methylation of WTH3, a possible drug resistant gene, inhibits
p53 regulated expression. BMC Cancer. 8:3272008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gan L, Zuo G, Wang T, Min J, Wang Y, Wang
Y and Lv G: Expression of WTH3 in breast cancer tissue and the
effects on the biological behavior of breast cancer cells. Exp Ther
Med. 10:154–158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM,
Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu
J, et al: Gene-expression profiles to predict distant metastasis of
lymph-node-negative primary breast cancer. Lancet. 365:671–679.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rutqvist LE and Johansson H; Stockholm
Breast Cancer Study Group, : Long-term follow-up of the randomized
Stockholm trial on adjuvant tamoxifen among postmenopausal patients
with early stage breast cancer. Acta Oncol. 46:133–145. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rutqvist LE and Johansson H: Long-term
follow-up of the Stockholm randomized trials of postoperative
radiation therapy versus adjuvant chemotherapy among ‘high risk’
pre- and postmenopausal breast cancer patients. Acta Oncol.
45:517–527. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jerevall PL, Jansson A, Fornander T, Skoog
L, Nordenskjöld B and Stål O: Predictive relevance of HOXB13
protein expression for tamoxifen benefit in breast cancer. Breast
Cancer Res. 12:R532010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wrange O, Nordenskjöld B and Gustafsson
JA: Cytosol estradiol receptor in human mammary carcinoma: An assay
based on isoelectric focusing in polyacrylamide gel. Anal Biochem.
85:461–475. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shan J, Mason JM, Yuan L, Barcia M, Porti
D, Calabro A, Budman D, Vinciguerra V and Xu H: Rab6c, a new member
of the rab gene family, is involved in drug resistance in MCF7/AdrR
cells. Gene. 257:67–75. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shan J, Yuan L, Budman DR and Xu HP: WTH3,
a new member of the Rab6 gene family, and multidrug resistance.
Biochim Biophys Acta. 1589:112–123. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Uhlén M, Björling E, Agaton C, Szigyarto
CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A,
Asplund C, et al: A human protein atlas for normal and cancer
tissues based on antibody proteomics. Mol Cell Proteomics.
4:1920–1932. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bhat S, Kabekkodu SP, Jayaprakash C,
Radhakrishnan R, Ray S and Satyamoorthy K: Gene promoter-associated
CpG island hypermethylation in squamous cell carcinoma of the
tongue. Virchows Arch. 470:445–454. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bhat S, Kabekkodu SP, Varghese VK,
Chakrabarty S, Mallya SP, Rotti H, Pandey D, Kushtagi P and
Satyamoorthy K: Aberrant gene-specific DNA methylation signature
analysis in cervical cancer. Tumour Biol. 39:10104283176945732017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Young J, Menetrey J and Goud B: RAB6C is a
retrogene that encodes a centrosomal protein involved in cell cycle
progression. J Mol Biol. 397:69–88. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Williams TK, Yeo CJ and Brody J: Does this
band make sense? Limits to expression based cancer studies. Cancer
Lett. 271:81–84. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sebat J, Lakshmi B, Troge J, Alexander J,
Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, et al:
Large-scale copy number polymorphism in the human genome. Science.
305:525–528. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Altschul SF, Madden TL, Schäffer AA, Zhang
J, Zhang Z, Miller W and Lipman DJ: Gapped BLAST and PSI-BLAST: A
new generation of protein database search programs. Nucleic Acids
Res. 25:3389–3402. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yates A, Akanni W, Amode MR, Barrell D,
Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil
L, et al: Ensembl 2016. Nucleic Acids Res. 44:D710–D716. 2016.
View Article : Google Scholar : PubMed/NCBI
|