Introduction
The Eph receptor family comprises the largest
receptor tyrosine kinase superfamily, and contains 14 distinct
members, with 9 molecules identified in its ligand ephrin family
(1). According to their sequence
homology and binding specificity, both Ephs and ephrins are
classified as type A and B. Upon engagement of Eph by the cognate
ephrin, the two molecules activate simultaneously and induce
intracellular signal transduction, which initiates a number of
biological processes, including axon guidance, neural crest cell
migration, hindbrain segmentation, somite formation and
vasculogenesis (2,3). Moreover, there is accumulating evidence
that the Eph/ephrin system also serves a pivotal role in the
development and progression of numerous cancer types (4,5).
EPH receptor A2 (EphA2) is the most well
characterized of the Eph receptors, particularly when regarding its
role in tumorigenesis; it has been revealed to be upregulated in a
number of different types of tumor, including prostate, colon and
lung cancer, as well as melanomas (4). Furthermore, overexpression of
EphA2 is able to induce malignant transformation in mammary
epithelial cells (4). The
EphB/ephrinB system is also implicated in tumorigenesis (4). The expression level of EphB2 is
reported to be upregulated in gastrointestinal, liver, ovarian,
lung and renal cancers (4). Although
the majority of studies suggest that Ephs and ephrins serve an
oncogenic role, EphB2 was reported as a tumor suppressor in
prostate and colorectal tumors (6–8). These
findings reflect the complexity of the differential functions of
the Eph/ephrin system, which is capable of exerting
context-dependent agonistic or antagonistic effects. Caspase-8, a
member of the cysteine-aspartic acid protease (caspase) family, is
well characterized as an initiator of death receptor-mediated
apoptosis, and has been implicated in other similar apoptotic
responses (9). Caspase-8 promoter
methylation results in the loss of gene expression, which is
associated with tumor severity in a variety of different tumor
types. The methylation-mediated silencing of key
apoptosis-associated genes serves an important role in the
pathogenesis and development of therapeutic resistance in human
cancer cells (10).
Esophageal cancer represents the sixth most frequent
cause of cancer-associated mortality worldwide (11). Esophageal squamous cell carcinoma
(ESCC) is the most prevalent histological subtype of esophageal
cancer and exhibits high mortality rates and a 5-year overall
survival rate of ≤15% (12,13). The most common pathological subtypes
of esophageal cancer are ESCC and esophageal adenocarcinoma.
Despite the well-characterized pathological progression of ESCC,
the underlying molecular mechanisms are predominantly yet to be
elucidated. Several studies reported that the expression of
EphA2 (and one of its receptors, ephrinA1) were upregulated
in ESCC, and correlated with tumor progression and patient
survival, revealing their predictive potential for the diagnosis
and prognosis of patients with ESCC (14). Previous studies demonstrated that
EPHB4 conferred a survival advantage on tumor cells by
decreasing apoptosis, whereas knockdown of EPHB4 expression
using siRNA induced apoptosis and decreased tumor cell viability
via the activation of caspase-8. However, studies focusing on the
influence that EphB/ephrin-B and caspase-8 exert on ESCC
progression and genesis remain limited. Therefore, the present
study investigated the expression levels of EPHB4, its
cognate ligand ephrin B2 (EFNB2) (1–3) and
caspase-8 in ESCC. In addition, the association between their
relative expression levels and clinical parameters important in the
diagnosis and prognosis of ESCC were also investigated in the
present study. The results from the present study provide
additional understanding, potentially facilitating the development
of diagnostic and therapeutic strategies for the treatment of
ESCC.
Materials and methods
Patients and samples
In the present study, 96 ESCC samples, and their
paired paracancerous esophageal tissues, were obtained from
patients with ESCC treated at the First Affiliated Hospital of
Zhengzhou University (Henan, China), between July 2002 and August
2006, following the provision of written informed consent. The
tumor stage was classified according to the 8th edition of the TNM
classification (15). Cancerous
tissues were surgically resected from patients who had not received
any neo-adjuvant therapy, and the corresponding non-cancerous
‘normal’ tissues, located at least 3 cm away from the tumor site,
were obtained in the same manner. Each specimen was divided into 2
pieces, one of which was fixed in 4% formalin at 4°C overnight,
sectioned and examined using immunohistochemical (IHC) staining,
and the other of which was stored at −80°C. The present study was
approved by the Institutional Review Board of the Institute for
Nutritional Sciences, Chinese Academy of Sciences.
Quantitative (q)PCR
RNA extraction, DNA template synthesis and
amplification reactions were performed as previously described
(16). The primers for the qPCR are
listed as follows: EPHB4 forward, 5′-TCCTTCCTGCGGCTAAAC-3′
and reverse, 5′-CTTTGCAGACGAGGTTGCT-3′; EFNB2 forward,
5′-TCTTTGGAGGGCCTGGATAA-3′ and reverse,
5′-CGTCTGTGCTAGAACCTGGATT-3′; caspase-8 forward,
5′-CTGCAGAGGAACCTGGTACATCC-3′ and reverse,
5′-TCTTACTCCAAGGTGGCCATG-3′; and β-actin forward,
5′-GATCATTGCTCCTCCTGAGC-3′ and reverse, 5′-ACTCCTGCTTGCTGATCCAC-3′.
All primers were designed using PRIMER5 software (version 5.00;
Premier Biosoft International) and purchased from Shanghai Sangong
Pharmaceutical Co., Ltd. Reactions were characterized at the point
during cycling when amplification of the PCR product was first
detected after a fixed number of cycles. Quantification was
performed by measuring the quantitation cycle (Cq) value. The
levels of target genes in each sample were normalized to the
housekeeping gene β-actin via the following formula: Normalized
level (NL)=level(target)/level
(β-actin)=2Cq(target)/2Cq(β−actin)=2Cq(target)−Cq(β−actin)=2∆Cq.
Furthermore, the relative levels (RL) of target genes in cancer
tissues vs. corresponding normal samples were calculated according
to the formula:
RL=NL(cancer)/NL(normal)=2∆Cq(cancer)/2∆Cq(normal)=2[∆Cq(cancer)−∆Cq(normal)]=2∆∆Cq.
As both NL and RL are represented as 2Cq, the present
study used ∆Cq and ∆∆Cq to represent NL and RL, respectively, when
performing statistical analysis.
IHC
The specimens were fixed in 4% formalin at 4°C
overnight. The paraffin-embedded tissues were cut into 5-µm thick
sections, deparaffinized, rehydrated in graded dimethylbenzene and
ethanol solutions, and subjected to antigen retrieval.
Subsequently, the sections were blocked using 5% normal goat serum
(Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature
for 1 h. The tissue sections were then incubated with the following
primary antibodies at 4°C overnight: Rabbit anti-human EPHB4
(1:500; cat. no. sc-365510), rabbit anti-human EFNB2 (1:500,
cat. no. sc-398735) (both Santa Cruz Biotechnology, Inc.) and
rabbit anti-human caspase-8 (1:100; cat. no. 552143; BD Pharmingen;
BD Biosciences). Following primary incubation, the sections were
incubated at 37°C for 1 h with horseradish peroxidase-conjugated
goat anti-rabbit IgG secondary antibody (1:1,000; cat. no. A-10194;
Chemicon International; Thermo Fisher Scientific Inc.). Finally,
all sections were counterstained with hematoxylin at room
temperature for 5–8 min.
Scoring of IHC staining was simultaneously performed
by three independent pathologists. Tumor cells positive and
negative for staining were counted separately under a light
microscope (magnification, ×200). For each slide, 7–10 microscopic
fields with ≥300 cells/microscopic field were randomly selected.
The ratio of positive cells was calculated as the number of
positively stained tumor cells divided by the total tumor cells, in
each high-power field area. The level of protein expression was
quantified by calculating the percentage ratio of positively
stained cells in the esophageal cancer sample compared with that in
the matched paracancerous esophageal tissues. Patients with high
expression of EPHB4, EFNB2 and caspase-8 had protein levels of
≥1.89, ≥1.57 and ≥0.56, respectively; whereas patients with low
expression had protein levels of <1.89, <1.57 and <0.56,
respectively.
Statistical analysis
The χ2 test was used to determine the
association between the expression levels of EPHB4, EFNB2
and caspase-8 in ESCC samples and the clinical characteristics,
respectively. Pearson's correlation analysis was used to estimate
the relative degree. Kaplan-Meier survival curves comparing
patients with high and low expression at the mRNA and protein
levels were plotted and univariate survival analysis was performed
using log-rank test.
Multivariate analyses were performed to estimate the
effects of certain clinicopathological characteristics, and the
expression levels of the two genes, on survival. The data were
analyzed using Student's t-test. P<0.05 were considered to
indicate a statistically significant difference. All statistical
analyses were performed using SPSS version 22 (IBM
Corporation).
Results
Expression of EPHB4, EFNB2 and
caspase-8 genes in ESCC and matched normal esophageal tissues
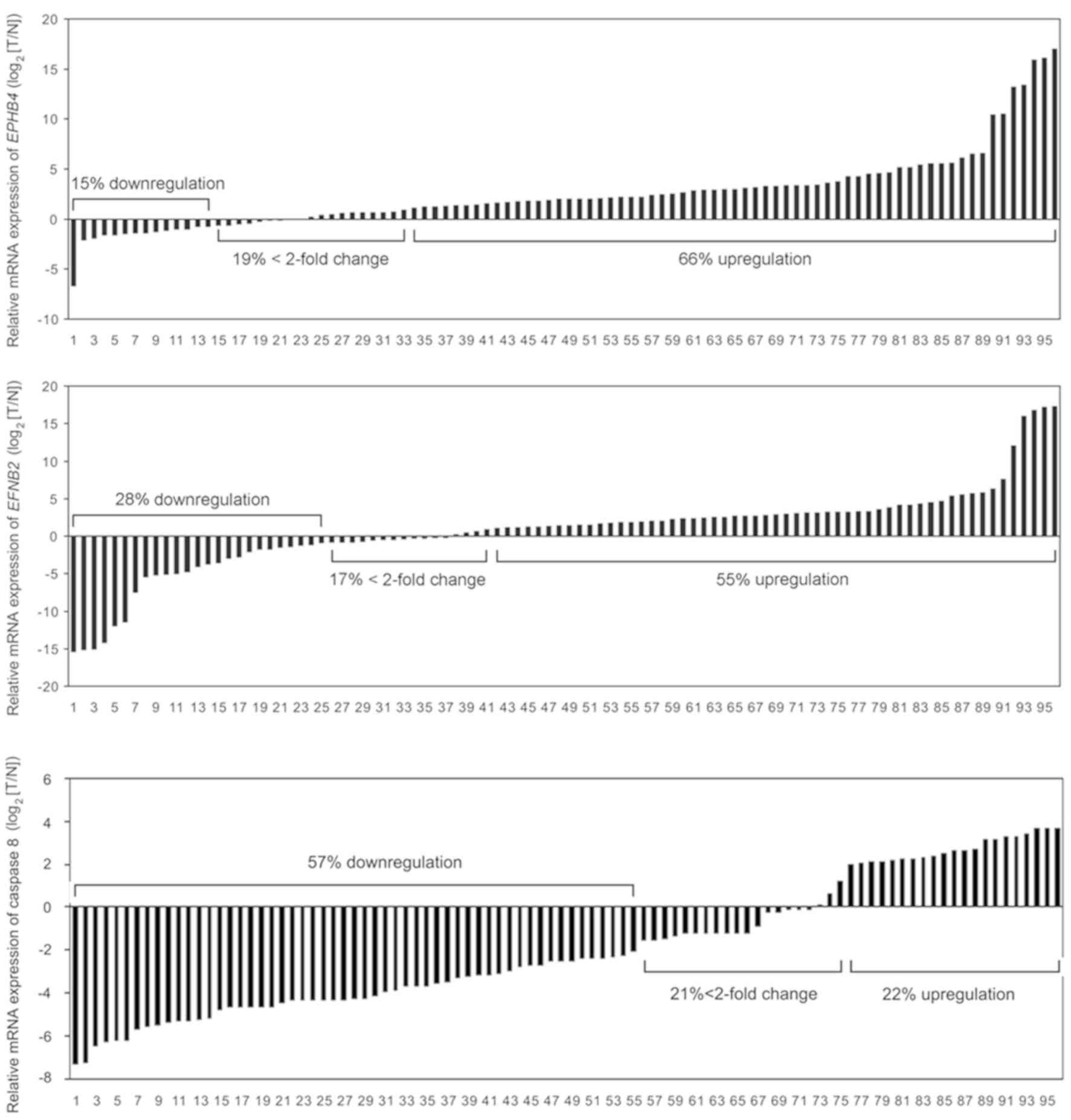
In order to investigate the expression pattern of
EPHB4, EFNB2 and caspase-8 in ESCC, the mRNA levels of the
three genes were quantified in 96 pairs of tumor samples and
matched normal esophageal tissue samples using qPCR. Expression
levels were presented as a ratio between EPHB4, EFNB2 or
caspase-8 and the reference gene β-actin. Upregulation of
EPHB4 and EFNB2 occurred in 63 out of 96 (66%) ESCC
samples, and 53 out of 96 (55%) paired normal esophageal tissues,
respectively. By contrast, downregulation of caspase-8 was observed
in 55 out of 96 (57%) ESCC samples, when compared with that in
normal tissues (Fig. 1). Univariate
analysis revealed that the mRNA level of EPHB4 was
significantly increased in tumor tissues, compared with paired
normal tissues (P=0.001), while the expression level of caspase-8
was significantly lower in the ESCC samples (P=0.001). However,
there was no significant difference in the mRNA level of
EFNB2 between the ESCC and paired normal tissues (P=0.172)
(Table I). Pearson's correlation
analysis demonstrated that the mRNA expression of EPHB4 was
positively correlated with that of EFNB2
(R2=0.620; P<0.001). Notably, EPHB4
(R2=−0.428; P=0.001) and EFNB2
(R2=−0.267, P=0.028) were both negatively
correlated with caspase-8 (Table
II).
 | Table I.Expression of EPHB4, EFNB2 and
caspase-8 genes in esophageal cancer and paired paracancerous
esophageal tissues (n=96 pairs). |
Table I.
Expression of EPHB4, EFNB2 and
caspase-8 genes in esophageal cancer and paired paracancerous
esophageal tissues (n=96 pairs).
| mRNA/protein | n | Cancerous |
Matched-paracancerous | (N-C)/(C/N) | t-test | P-value |
|---|
| EPHB4
mRNA | 96 | 12.89±10.08 | 15.45±10.26 | 2.56±3.92 | 6.411 |
<0.01a |
| EFNB2
mRNA | 96 | 8.05±5.88 | 8.86±5.69 | 0.81±5.77 | 1.375 | 0.172 |
| caspase-8 mRNA | 96 | 7.38±2.47 | 5.46±1.87 | −1.92±2.67 | 7.307 |
<0.001a |
| EPHB4 protein | 96 | 21.35±8.296 | 2.80±0.947 | 8.74±5.65 | −21.603 |
<0.001a |
| EFNB2 protein | 96 | 11.67±2.478 | 1.71±0.597 | 7.83±3.44 | −38.322 |
<0.001a |
| Caspase-8
protein | 96 | 2.51±2.384 | 8.85±7.879 | 0.28±0.15 | −22.761 |
<0.001a |
 | Table II.Pearson's correlation analysis of
mRNA and protein expression of EPHB4, EFNB2 and caspase-8 in
esophageal cancer and matched normal esophageal tissues (n=96
pairs). |
Table II.
Pearson's correlation analysis of
mRNA and protein expression of EPHB4, EFNB2 and caspase-8 in
esophageal cancer and matched normal esophageal tissues (n=96
pairs).
|
| mRNA | Protein |
|---|
|
|
|
|
|---|
| Genes | R-value | P-value | R-value | P-value |
|---|
| EPHB4 and
EFNB2 |
0.620 |
<0.001a | 0.202 | 0.049a |
| EPHB4 and
caspase-8 | −0.428 |
<0.001a | −0.340 | 0.001a |
| EFNB2 and
caspase-8 | −0.267 | 0.028a | −0.198 | 0.041a |
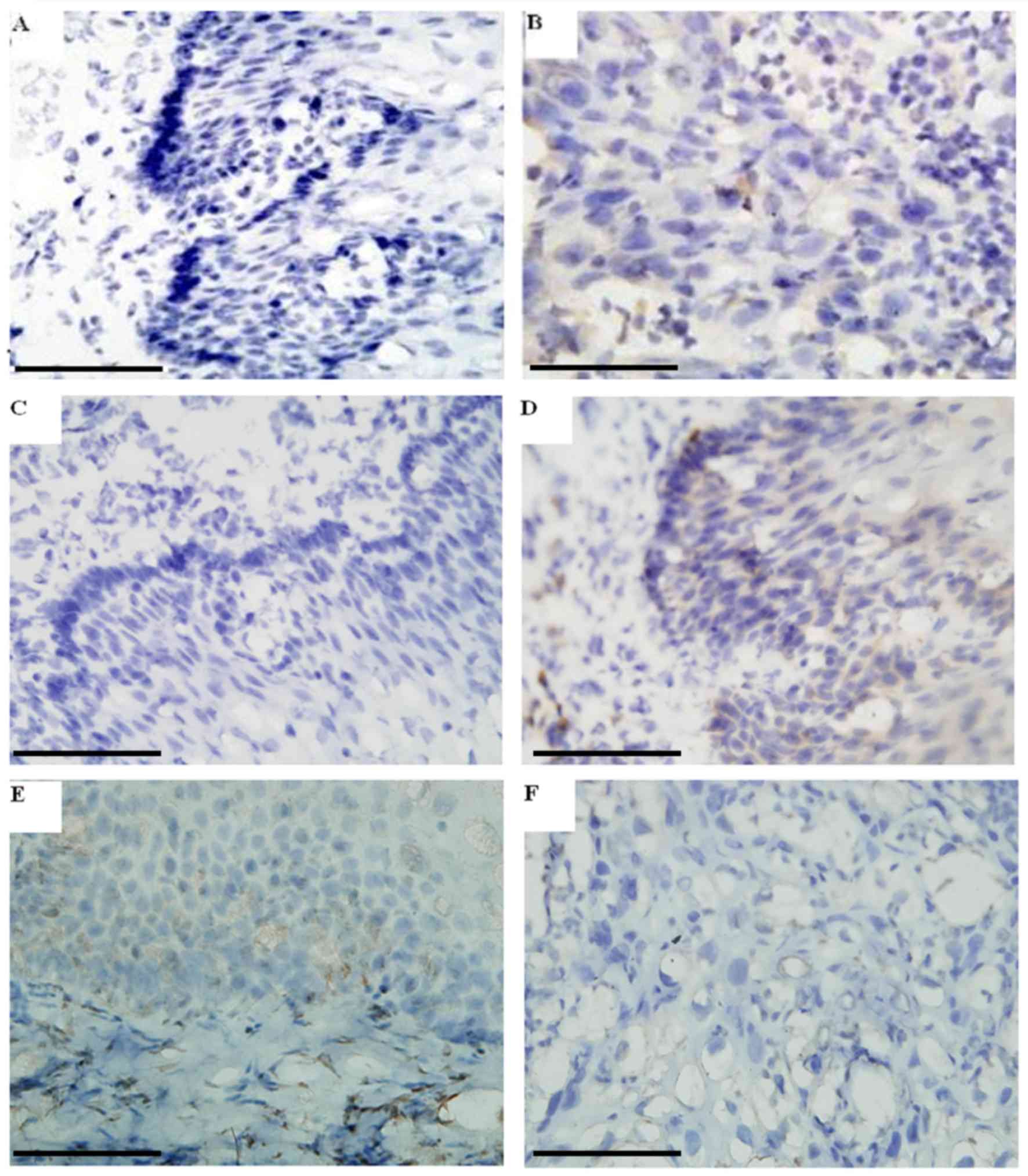
Subsequently, IHC was performed to investigate the
protein expression levels of EPHB4, EFNB2 and caspase-8
proteins in 96 pairs of esophageal tissues. As presented in
Fig. 2A and C, EPHB4 and
EFNB2 proteins were not apparent in the majority of normal
esophageal epithelial cells, while they were highly expressed in
the majority tumor cells in corresponding ESCC tissues (Fig. 2B and D), synonymous with previous
reports (14,17). As presented in Fig. 2E, strong staining of caspase-8 was
observed in the superficial layer of normal esophageal epithelia,
but was almost undetectable in the ESCC samples (Fig. 2F), which is also consistent with
other studies (18,19). The IHC scoring analysis revealed that
the ratio of EPHB4/EFNB2-positive to -negative cells in ESCC
tissues was significantly higher in comparison with that in the
corresponding normal tissue, whereas the ratio of
caspase-8-positive to -negative cells was lower in ESCC tissues
compared with that in their normal counterparts (Table I).
Association between the expression of
EPHB4, EFNB2 and caspase-8, and the clinicopathological features of
patients with ESCC
The univariate analysis revealed a significant
association between the expression of EPHB4 and family
history, metastasis, and tumor size, position and stage. The
expression level of EPHB4 was significantly higher in
patients with a family history of cancer (P<0.001). A
significant association also existed between increased levels of
EPHB4 and metastasis (P=0.001), larger tumors (P=0.001),
ESCC located in the lower segment of the esophagus (P=0.010) and a
higher stage (P=0.043), indicating that the upregulation of
EPHB4 expression was associated with ESCC progression. Sex
and age were not significantly associated with the expression level
of EPHB4 (Table III).
 | Table III.Association between the levels of
EPHB4/EFNB2 and caspase-8 mRNA expression and the clinical
and pathological features of individuals with esophageal cancer
(n=96 pairs). |
Table III.
Association between the levels of
EPHB4/EFNB2 and caspase-8 mRNA expression and the clinical
and pathological features of individuals with esophageal cancer
(n=96 pairs).
|
| EPHB4 |
|
| EFNB2 |
|
| Caspase-8 |
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|---|
| Factors | High, n | Low, n | χ2 | P-value | High, n | Low, n | χ2 | P-value | High, n | Low, n | χ2 | P-value |
|---|
| Sex |
|
| 0.233 | 0.629 |
|
| 1.149 | 0.284 |
|
| 0.526 | 0.506 |
|
Male | 45 | 22 |
|
| 36 | 31 |
|
| 26 | 41 |
|
|
|
Female | 18 | 11 |
|
| 19 | 10 |
|
| 15 | 14 |
|
|
| Age, years |
|
| 2.433 | 0.303 |
|
| 2.001 | 0.368 |
|
| 1.733 | 0.494 |
|
≤35 | 2 | 0 |
|
| 2 | 0 |
|
| 1 | 1 |
|
|
|
35–50 | 12 | 10 |
|
| 11 | 11 |
|
| 12 | 10 |
|
|
|
≥50 | 49 | 23 |
|
| 42 | 30 |
|
| 28 | 44 |
|
|
| Metastasis |
|
| 11.039 | 0.001a |
|
| 0.970 | 0.325 |
|
| 41.552 |
<0.001a |
|
Yes | 45 | 12 |
|
| 20 | 19 |
|
| 9 | 48 |
|
|
| No | 18 | 21 |
|
| 35 | 22 |
|
| 32 | 7 |
|
|
| Family history |
|
| 53.940 |
<0.001a |
|
| 15.668 |
<0.001a |
|
| 7.327 | 0.012a |
|
Yes | 53 | 2 |
|
| 41 | 14 |
|
| 17 | 38 |
|
|
| No | 10 | 31 |
|
| 14 | 27 |
|
| 24 | 17 |
|
|
| Tumor size,
cm3 |
|
| 14.415 | 0.001a |
|
| 5.060 | 0.080 |
|
| 32.714 |
<0.001a |
|
≤100 | 14 | 20 |
|
| 15 | 19 |
|
| 27 | 7 |
|
|
|
100–200 | 32 | 10 |
|
| 25 | 17 |
|
| 13 | 29 |
|
|
|
>200 | 17 | 3 |
|
| 15 | 5 |
|
| 1 | 19 |
|
|
| TNM stage |
|
| 6.294 | 0.043a |
|
| 1.942 | 0.379 |
|
| 16.816 |
<0.001a |
| I | 14 | 14 |
|
| 13 | 15 |
|
| 20 | 8 |
|
|
| II | 21 | 12 |
|
| 20 | 13 |
|
| 14 | 19 |
|
|
|
III | 28 | 7 |
|
| 22 | 13 |
|
| 7 | 28 |
|
|
| Tumor position |
|
| 9.167 | 0.010a |
|
| 6.075 | 0.048a |
|
| 7.887 | 0.019a |
|
Upper | 12 | 16 |
|
| 11 | 17 |
|
| 18 | 10 |
|
|
|
Middle | 37 | 13 |
|
| 34 | 16 |
|
| 18 | 32 |
|
|
|
Lower | 14 | 4 |
|
| 10 | 8 |
|
| 5 | 13 |
|
|
Statistical analysis also demonstrated that the
expression level of EFNB2 was significantly associated with
several clinical features, including tumor position and family
history. The mRNA level of EFNB2 was significantly higher in
the patients with a family history of cancer (P<0.001). ESCCs
located in the lower segment of the esophagus exhibited higher
EFNB2 expression than those in the upper segment (P=0.048).
However, no associations were observed between the expression level
of EFNB2 and sex, age, metastasis or tumor size and stage
(Table III).
The expression level of caspase-8 was significantly
downregulated in patients with family history (P=0.012).
Downregulated expression levels of caspase-8 were significantly
associated with metastasis (P<0.000), increased tumor size
(P<0.000), ESCC at the lower segment of the esophagus (P=0.019)
and a higher stage (P<0.000), indicating that low caspase-8
expression is associated with the progression of ESCC (Table III). However, there was no
significant association observed between caspase-8 expression and
sex or age.
IHC scoring analysis revealed that the ratio of
EPHB4-positive to -negative cells in tissue samples was
higher in patients with a family history of cancer (P=0.002). There
was also a significant association between a higher
positive-staining ratio and metastasis (P=0.005), larger tumors
(P<0.001) and higher tumor stages (P=0.004). The increased ratio
of EFNB2-positive to -negative cells, as well as a decreased
ratio of caspase-8 was identified in patients with metastasis or
greater tumors, respectively (EFNB2, P=0.004 and P=0.018,
respectively; and caspase-8, P=0.000 and P=0.000, respectively)
(Table IV). Taken together, the
associations between protein levels of EPHB4, EFNB2 or
caspase-8 and certain clinicopathological features of patients with
ESCC (according to IHC), were consistent with the results
concerning the mRNA levels.
 | Table IV.Associations between EPHB4/Ephrinb2
and caspase-8 protein expression and the clinical and pathological
features of individuals with esophageal cancer (n=96 pairs). |
Table IV.
Associations between EPHB4/Ephrinb2
and caspase-8 protein expression and the clinical and pathological
features of individuals with esophageal cancer (n=96 pairs).
|
|
| EPHB4 | EFNB2 | Caspase-8 |
|---|
|
|
|
|
|
|
|---|
| Factors | n | C/N | t/F | P-value | C/N | t/F | P-value | C/N | t/F | P-value |
|---|
| Sex |
|
| −1.005 | 0.318 |
| 2.589 | 0.011a |
| 1.592 | 0.115 |
|
Male | 67 | 8.36±5.95 |
|
| 8.41±3.67 |
|
| 0.27±0.54 |
|
|
|
Female | 29 | 9.62±4.86 |
|
| 6.48±2.41 |
|
| 0.30±0.35 |
|
|
| Age, years |
|
| 2.044 | 0.135 |
| 2.044 | 0.135 |
| 0.832 | 0.438 |
|
≤35 | 2 | 10.33±0.94 |
|
| 10.33±0.94 |
|
| 0.28±0.54 |
|
|
|
35–50 | 22 | 6.65±3.43 |
|
| 6.65±3.43 |
|
| 0.27±0.40 |
|
|
|
≥50 | 72 | 9.34±6.11 |
|
| 9.34±6.11 |
|
| 0.28±0.44 |
|
|
| Metastasis |
|
| −2.862 | 0.005a |
| −2.987 | 0.004a |
| 5.072 |
<0.001a |
|
Yes | 57 |
10.06±6.05a |
|
| 8.66±3.38 |
|
| 0.21±0.31 |
|
|
| No | 39 | 6.82±4.41 |
|
| 6.61±3.20 |
|
| 0.38±0.47 |
|
|
| Family history |
|
| −3.181 | 0.002a |
| −0.136 | 0.892 |
| 1.703 | 0.092 |
|
Yes | 55 | 10.26±6.03 |
|
| 7.78±3.40 |
|
| 0.29±0.43 |
|
|
| No | 41 | 6.72±4.40 |
|
| 7.88±3.55 |
|
| 0.28±0.33 |
|
|
| Tumor size |
|
| 13.129 |
<0.001a |
| 4.174 | 0.018a |
| 16.534 |
<0.001a |
| I | 34 | 5.71±2.60 |
|
| 6.50±2.59 |
|
| 0.36±0.41 |
|
|
| II | 42 | 9.23±4.33 |
|
| 8.47±3.23 |
|
| 0.25±0.31 |
|
|
|
III | 20 | 12.89±8.49 |
|
| 8.73±4.50 |
|
| 0.21±0.23 |
|
|
| I vs.
II |
|
|
| 0.003a |
|
| 0.012a |
|
|
<0.001a |
| I vs.
III |
|
|
|
<0.001a |
|
| 0.020a |
|
|
<0.001a |
| II vs.
III |
|
|
| 0.009 |
|
| 0.777 |
|
| 0.108 |
| Tumor stage |
|
| 5.882 | 0.004a |
| 0.820 | 0.443 |
| 4.083 | 0.020a |
| I | 28 | 5.91±2.50 |
|
| 7.73±3.13 |
|
| 0.32±0.22 |
|
|
| II | 33 | 9.33±4.56 |
|
| 7.32±3.33 |
|
| 0.28±0.26 |
|
|
|
III | 35 | 10.46±5.65 |
|
| 8.38±3.79 |
|
| 0.25±0.49 |
|
|
| I vs.
II |
|
|
| 0.015a |
|
| 0.642 |
|
| 0.308 |
| I vs.
III |
|
|
| 0.001a |
|
| 0.460 |
|
| 0.006a |
| II vs.
III |
|
|
| 0.385 |
|
| 0.208 |
|
| 0.070 |
| Tumor position |
|
| 2.986 | 0.055 |
| 0.278 | 0.758 |
| 1.337 | 0.268 |
|
Upper | 28 | 6.65±3.12 |
| 0.017a | 7.44±3.23 |
|
| 0.28±0.43 |
|
|
|
Middle | 50 | 9.83±6.66 |
| 0.169 | 7.92±3.59 |
|
| 0.27±0.25 |
|
|
|
Lower | 18 | 8.97±4.90 |
| 0.570 | 8.17±3.49 |
|
| 0.31±0.31 |
|
|
| Upper
vs. middle |
|
|
|
|
|
| 0.560 |
|
| 0.114 |
| Upper
vs. lower |
|
|
|
|
|
| 0.490 |
|
| 0.272 |
| Middle
vs. lower |
|
|
|
|
|
| 0.797 |
|
| 0.877 |
Expression of EphB4, EFNB2 or
caspase-8 and clinical outcomes of ESCC
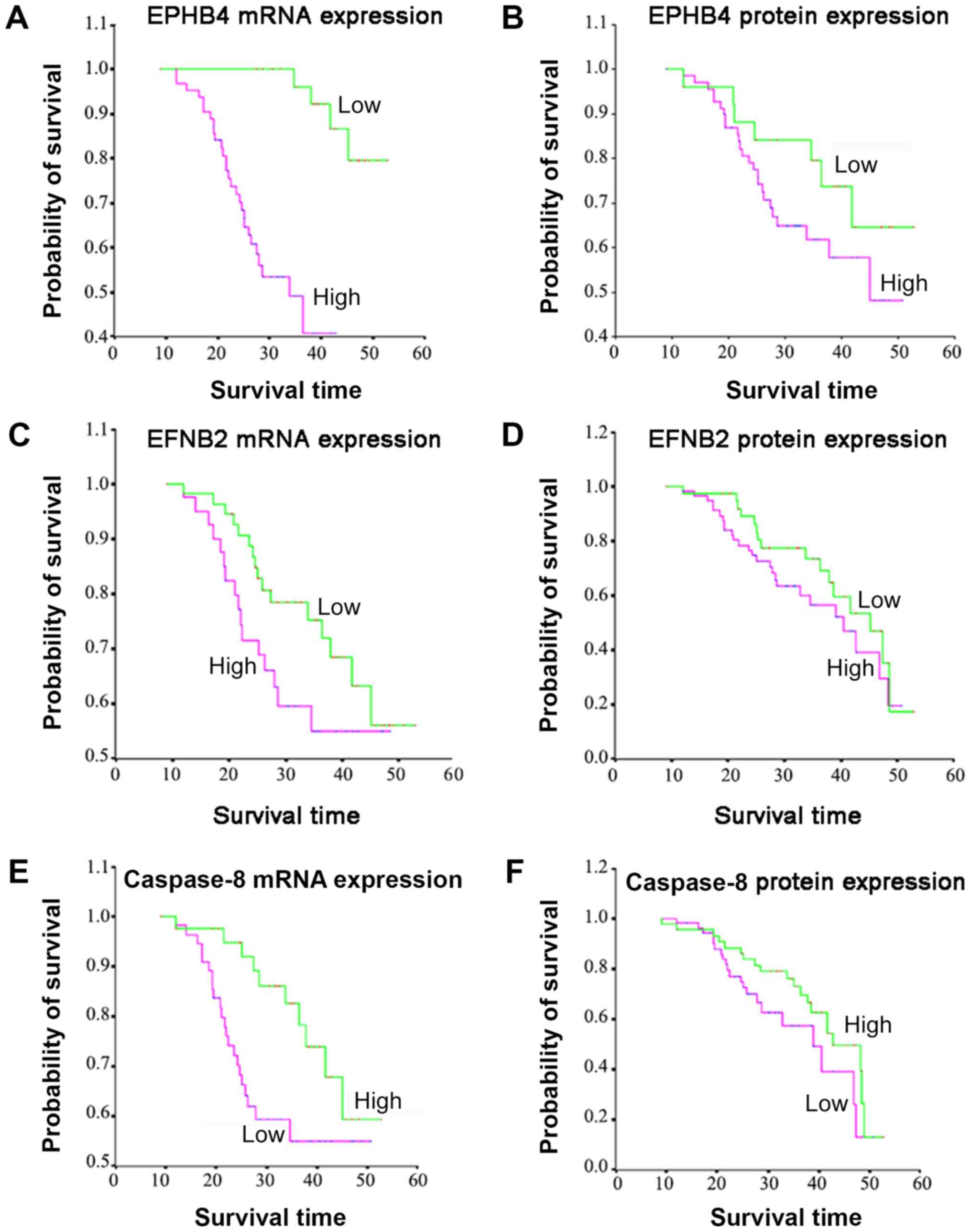
The univariate survival analysis demonstrated that
patient age, family history and tumor metastasis were all
significantly associated with survival time. The mRNA expression
levels of EPHB4 and EFNB2 but not caspase-8, was
associated with survival time and the protein expression levels of
EPHB4 and caspase-8, but not EFNB2, was associated
with survival time (Table V).
Kaplan-Meier curves indicated that patients with higher mRNA
(P<0.0001) and protein (P<0.0001) expression levels of
EPHB4 exhibited a significantly shortened median survival
time, compared with patients with lower expression levels (Fig. 3A and B; Table V). Similarly, patients with higher
mRNA level of EFNB2 expression (P=0.041; Fig. 3C), or patients with lower protein
level expression of caspase-8 expression (P=0.045; Fig. 3F) also exhibited a significantly
shortened median survival time (Table
V). In addition, as presented in Table V, patients with a family history of
cancer exhibited a significantly decreased survival time compared
with those without a family history of cancer (P<0.001).
Furthermore, older patients also had a shortened survival time
compared with those patients that were younger (P<0.001), and
patients with metastatic tumors exhibited a markedly decreased
survival time compared with those without tumor metastasis
(P<0.001) (Table V). However,
there were no significant associations observed between survival
and sex, tumor size, stage or position. The multivariate analysis
results revealed that the mRNA expression level of EPHB4 and
EFNB2, the protein expression level of EPHB4 and
caspase-8, metastasis and family history were all significant
independent risk factors for ESCC, with hazard ratios of 5.290,
3.146, 1.394, 2.784, 1.885 and 1.786, respectively (Table VI).
 | Table V.Univariate survival analysis of the
association between expression levels of EPHB4, EFNB2 and
caspase-8 and certain clinicopathological characteristics in
patients with esophageal cancer. |
Table V.
Univariate survival analysis of the
association between expression levels of EPHB4, EFNB2 and
caspase-8 and certain clinicopathological characteristics in
patients with esophageal cancer.
| Factors | Cases (n=96) | Events, n | Median survival,
months | SE | Log-rank | P-value |
|---|
| Sex |
|
|
|
| 1.22 | 0.259 |
|
Male | 67 | 47 | 35.585 | 1.179 |
|
|
|
Female | 29 | 22 | 37.177 | 1.765 |
|
|
| Age, years |
|
|
|
| 47.37 |
<0.001a |
|
<30 | 2 | 2 | 37.661 | 0.100 |
|
|
|
30–50 | 22 | 16 | 36.040 | 2.179 |
|
|
|
>50 | 72 | 51 | 20.500 | 1.077 |
|
|
| Metastasis |
|
|
|
| 25.30 |
<0.001a |
|
Yes | 57 | 36 | 28.868 | 1.624 |
|
|
| No | 39 | 33 | 40.579 | 1.079 |
|
|
| Family history |
|
|
|
| 15.95 |
<0.001a |
|
Yes | 55 | 31 | 31.126 | 1.152 |
|
|
| No | 41 | 38 | 40.150 | 1.125 |
|
|
| Tumor size,
cm3 |
|
|
|
| 2.13 | 0.334 |
|
≤100 | 34 | 29 | 38.291 | 1.325 |
|
|
|
100–200 | 42 | 30 | 34.593 | 1.609 |
|
|
|
>200 | 20 | 10 | 33.607 | 2.517 |
|
|
| TNM stage |
|
|
|
| 0.44 | 0.809 |
| I | 28 | 23 | 37.233 | 1.662 |
|
|
| II | 33 | 21 | 35.072 | 1.660 |
|
|
|
III | 35 | 25 | 34.835 | 1.761 |
|
|
| Tumor position |
|
|
|
| 3.48 | 0.175 |
|
Upper | 28 | 24 | 37.937 | 1.779 |
|
|
|
Middle | 50 | 34 | 35.072 | 1.424 |
|
|
|
Lower | 18 | 11 | 34.835 | 1.740 |
|
|
| EPHB4
(mRNA) |
|
|
|
| 20.77 |
<0.001a |
|
Low | 33 | 33 | 41.400 | 1.154 |
|
|
|
High | 63 | 36 | 31.358 | 0.960 |
|
|
| EFNB2
(mRNA) |
|
|
|
| 3.03 | 0.041a |
|
Low | 41 | 38 | 37.863 | 1.391 |
|
|
|
High | 55 | 31 | 33.898 | 1.245 |
|
|
| Caspase-8
(mRNA) |
|
|
|
| 0.532 | 0.466 |
|
Low | 37 | 33 | 48.500 | 3.911 |
|
|
|
High | 59 | 36 | 52.815 | 3.307 |
|
|
| EPHB4
(protein) |
|
|
|
| 7.420 | 0.006a |
|
Low | 48 | 36 | 39.525 | 1.274 |
|
|
|
High | 48 | 33 | 30.168 | 1.342 |
|
|
| EFNB2
(protein) |
|
|
|
| 2.715 |
0.095 |
|
Low | 46 | 37 | 38.573 | 0.641 |
|
|
|
High | 50 | 32 | 32.417 | 2.169 |
|
|
| Caspase-8
(protein) |
|
|
|
| 4.016 |
0.045a |
|
Low | 36 | 26 | 34.898 | 1.245 |
|
|
|
High | 60 | 43 | 39.863 | 1.391 |
|
|
 | Table VI.Multivariate Cox proportional hazards
regression analysis (n=96 pairs). |
Table VI.
Multivariate Cox proportional hazards
regression analysis (n=96 pairs).
| Variables | Hazard ratio | 95% CI | P-value |
|---|
| Sex, male vs.
female | 1.253 | 0.533–2.946 | 0.606 |
| Age, years |
|
|
|
| 35–50
vs. ≤35 | 0.741 | 0.341–1.641 | 0.451 |
| ≥50 vs.
≤35 | 0.762 | 0.355–1.693 | 0.493 |
| Metastasis, yes vs.
no | 1.885 | 1.545–2.517 | 0.037a |
| Family history, yes
vs. no | 1.786 | 1.217–2.389 | 0.026a |
| Tumor size,
cm3 |
|
|
|
| 100–200
vs. ≤100 | 1.472 | 0.668–2.115 | 0.107 |
| >200
vs. ≤100 | 1.662 | 0.715–2.262 | 0.227 |
| Tumor stage |
|
|
|
| II vs.
I | 1.001 | 0.441–1.379 | 0.110 |
| III vs.
I | 1.009 | 0.449–1.382 | 0.172 |
| Tumor position |
|
|
|
| Middle
vs. upper | 0.915 | 0.473–1.771 | 0.752 |
| Lower
vs. upper | 0.936 | 0.484–1.817 | 0.912 |
| High vs. low
expression |
|
|
|
|
EPHB4 (mRNA) | 5.290 | 3.723–7.706 | 0.012a |
|
EFNB2 (mRNA) | 3.146 | 2.070–5.248 | 0.037a |
|
Caspase-8 (mRNA) | 0.936 | 0.323–2.713 | 0.903 |
| EPHB4
(protein) | 1.394 | 1.011–1.968 | 0.035a |
| EFNB2
(protein) | 1.350 | 0.596–3.058 | 0.472 |
|
Caspase-8 (protein) | 2.784 | 1.888–5.727 | 0.031a |
Discussion
A number of studies have reported that EPHB4
and/or EFNB2 expression is upregulated in multiple
malignancies, including gastric (20), colon (21), uterine endometrial (22,23),
breast (24), cervical (25) and ovarian cancer (26), melanoma (27), esophageal squamous cell carcinoma
(14,16) and squamous cell carcinoma of the head
and neck (28), which suggests that
EPHB4 and EFNB2 may serve an oncogenic role in these
tumor types. In the present study, it was observed that the
expression of either EPHB4 or EPNB2 was increased in
ESCC samples compared with that in corresponding normal esophageal
tissues. It has been previously demonstrated that the upregulation
of EPHB4 or EPNB2 is associated with metastasis and decreased
survival in patients with ESCC (14,16);
however, the present study revealed that it is also associated with
tumor size and position, and family history, as well as confirming
its association with decreased survival. Therefore, EPHB4
and EFNB2 may also serve oncogenic roles in the development
and progression of ESCC.
The present study revealed that EPHB4
expression exhibited a positive correlation with EFNB2
expression, at both the mRNA and protein level; furthermore, IHC
demonstrated that both molecules were expressed in the majority of
ESCC cells. Considering they are cognate receptors and ligands, the
aforementioned results suggested their potential ligation and the
activation of downstream pathways in ESCC. Previous studies
demonstrated that the activation of EPHB4 and/or
EFNB2 triggered ‘forward’ and ‘reverse’ bidirectional
signaling (2,3), which may stimulate angiogenesis in
vivo (25,29–32), and
stimulated the growth of primary and metastatic tumor cells
(33,34). The EPHB4 ‘forward’ signaling
was able to promote the proliferation and migration of endothelial
cells via the Pl-3 kinase pathway, which increased the formation of
new cancer vasculature (35). The
EFNB2 ‘reverse’ signaling, upon activation by EPHB4,
not only induced an angiogenic response in cultured endothelial
cells, but also promoted angiogenesis in breast cancer xenografts
in vivo (35). In addition,
EPHB4 and EFNB2 were also revealed to promote
angiogenesis-independent tumor formation, in which the
EFNB2-dependent EPHB4 ‘forward’ signaling enhanced
the migration and invasion of melanoma cells (36), via the activation of RhoA GTPase. The
present study determined that EPHB4 expression was
associated with tumor size, metastasis and stage, indicating that
EPHB4 may influence ESCC cell proliferation and migration.
EPHB4 has been reported to promote the proliferation and
migration of tumor cells in a variety of different cancer types
(22,23,28,36–42),
which supports the results of the present study. In addition, the
present study demonstrated that the upregulation of EPHB4
and EFNB2 was associated with poor outcome, and there have
been similar reports in squamous cell carcinoma of the head and
neck (28), as well as in
endometrial (22) and ovarian cancer
(26,43).
Resistance to apoptosis is required for tumor
growth, and is a hallmark of cancer cells (44). Apoptosis resistance contributes to
tumorigenesis, and results in the failure of cytotoxic therapies
and a poor prognosis in patients, suggesting that targeting
apoptotic pathways may represent a promising therapeutic approach
for anticancer treatment. Accumulating evidence has demonstrated
that apoptosis resistance, caused by downregulation of proapoptotic
signaling molecules (such as caspase-8), frequently occurs in
tumors of various origins. The present study demonstrated that the
mRNA and protein level of caspase-8 was significantly downregulated
in ESCC tissues compared with that in paracancerous tissues,
indicating that this molecule may influence escape from endogenous
growth control in the development and progression of ESCCs, which
was similar to the findings previously reported (19). However, the present study also
revealed that the expression of caspase-8 was associated with
certain clinicopathological characteristics, including metastasis,
tumor size, position and stage, and patient prognosis, in contrast
to certain previously reported results (18). In conclusion, the downregulation of
caspase-8 expression in ESCC suggested that it may serve as a
useful predictor of prognosis in this type of cancer. Furthermore,
the present study analyzed the associations between EPHB4, EFNB2
and caspase-8 in ESCC. The results revealed that, in ESCC tissues,
the expression levels of EPHB4 and EFNB2 were negatively correlated
with caspase-8 at both the mRNA and protein levels, which, to the
best of our knowledge, has not been yet reported elsewhere.
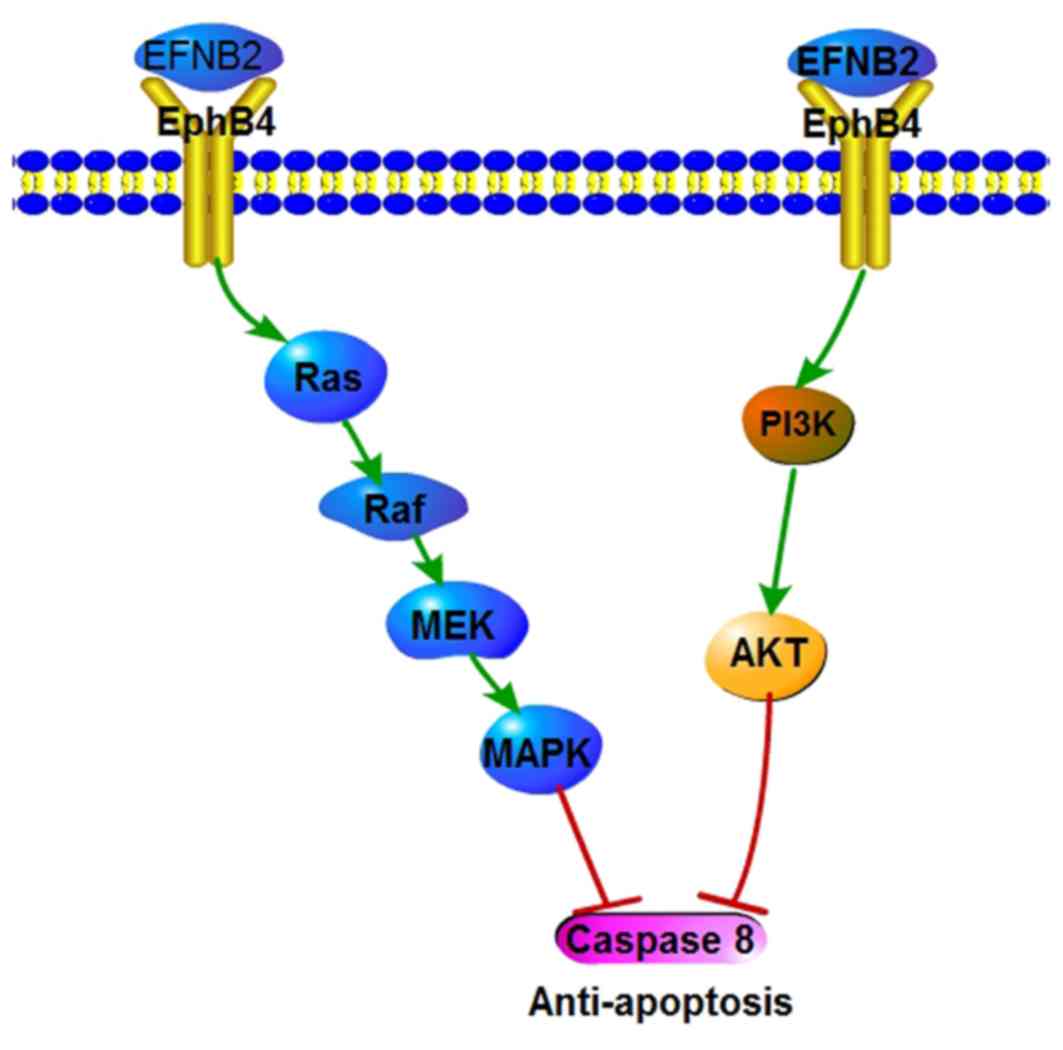
The present study indicates that the upregulation of
EPHB4 and EFNB2 expression in tumor cells promotes growth (via the
inhibition of apoptotic pathways), which may be facilitated by a
decrease in caspase-8 expression, resulting from regulation of the
downstream effectors of EPHB4/EFNB2. A diagram representing the
underlying molecular mechanism concerning the role of EPHB4, EFNB2
and caspase-8 in ESCC cells is exhibited in Fig. 4. The negative association between
caspase-8 activation and EPHB4 expression has been previously
reported in ovarian carcinoma (26),
and is consistent with the results of the present study. The
Ras/MAPK/ERK and Akt signaling pathways, downstream of EPHB4, could
confer anti-apoptotic characteristics. However, the molecular
mechanism underlying the negative correlation between EPHB4/EFNB2
and caspase-8 expression requires further investigation. Overall,
the upregulation of EPHB4 and EFNB2 in tumor cells may disrupt
caspase-8-mediated apoptosis and confer a survival advantage in
tumor cells.
In summary, the present study reported that both
EPHB4 and EFNB2 were upregulated, while caspase-8 was
downregulated, in ESCC tissues compared with that in matched normal
tissues. Expression levels were closely associated with a number of
clinicopathological features, as well as patient survival. The
current findings indicate the importance of the three molecules
studied with regard to the genesis and progression of ESCC.
Consequently, the expression levels of EPHB4, EFNB2 and
caspase-8 may serve as biological signatures and useful prognostic
indicators in ESCC, as well as potentially representing novel
therapeutic targets in this type of cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Key
R&D Program of China (grant no. 2018YFC1603002 and
2018YFC1604404); the ‘Personalized Medicines-Molecular
Signature-based Drug Discovery and Development’, Strategic Priority
Research Program of the Chinese Academy of Sciences (grant no.
XDA12010316); the National Natural Science Foundation of China
(grant nos. 31520103907, 81730083) to Dong Xie; the National
Natural Science Foundation of China (grant nos. 31771538,
81972757); the Youth Innovation Promotion Association of the
Chinese Academy of Sciences Fund and the Sanofi-SIBS 2018 Young
Faculty Award; and Postdoctoral Science Foundation of China (grant
no. 2017M622677).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XM, DX and JL conceived and designed the
experiments. QN, BZ and PC performed the experiments. QN wrote the
manuscript. QN collected and analyzed the data. PC assisted with
revising the manuscript. All authors read and approved the final
manuscript.
Ethical approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
institutional and/or national research committee and with the 1964
Helsinki declaration and its later amendments or comparable ethical
standards. The present study was approved by the Institutional
Review Board of the Institute for Nutritional Sciences, Chinese
Academy of Sciences (project number 30930023). Written informed
consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Unified nomenclature for Eph family
receptors and their ligands, the ephrins. Eph Nomenclature
Committee. Cell. 90:403–404. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wilkinson DG: Multiple roles of EPH
receptors and ephrins in neural development. Nat Rev Neurosci.
2:155–164. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Palmer A and Klein R: Multiple roles of
ephrins in morphogenesis, neuronal networking, and brain function.
Genes Dev. 17:1429–1450. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heroult M, Schaffner F and Augustin HG:
Eph receptor and ephrin ligand-mediated interactions during
angiogenesis and tumor progression. Exp Cell Res. 312:642–650.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kd gsbrun M and Eichmann A: A role for
axon guidance receptors and ligands in blood vessel development and
tumor angiogenesis. Cytokine Growth Factor Rev. 16:535–548. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Batlle E, Bacani J, Begthel H, Jonkheer S,
Gregorieff A, van de Born M, Malats N, Sancho E, Boon E, Pawson T,
et al: EphB receptor activity suppresses colorectal cancer
progression. Naturec. 435:1126–1130. 2005. View Article : Google Scholar
|
|
7
|
Huusko P, Ponciano-Jackson D, Wolf M,
Kiefer JA, Azorsa DO, Tuzmen S, Weaver D, Robbins C, Moses T,
Allinen M, et al: Nonsense-mediated decay microarray analysis
identifies mutations of EPHB2 in human prostate cancer. Nat Genet.
36:979–983. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okumura F, Joo-Okumura A, Obara K,
Petersen A, Nishikimi A, Fukui Y, Nakatsukasa K and Kamura T:
Ubiquitin ligase SPSB4 diminishes cell repulsive responses mediated
by EphB2. Mol Biol Cell. 8:3532–3541. 2017. View Article : Google Scholar
|
|
9
|
Teitz T, Lahti JM and Kidd VJ: Aggressive
childhood neuroblastomas do not express caspase-8: An important
component of programmed cell death. J Mol Med (Berlin, Germany).
79:428–436. 2001. View Article : Google Scholar
|
|
10
|
Teng Y, Dong YC, Liu Z, Zou Y, Xie H, Zhao
Y, Su J, Cao F, Jin H and Ren H: DNA methylation-mediated caspase-8
downregulation is associated with anti-apoptotic activity and human
malignant glioma grade. Int J Mol Med. 39:725–733. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y: Epidemiology of esophageal
cancer. World J Gastroenterol. 19:5598–5606. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Domper Arnal MJ, Ferrandez Arenas A and
Lanas Arbeloa A: Esophageal cancer: Risk factors, screening and
endoscopic treatment in Western and Eastern countries. World J
Gastroenterol. 21:7933–7943. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tachibana M, Tonomoto Y, Hyakudomi R,
Hyakudomi M, Hattori S, Ueda S, Kinugasa S and Yoshimura H:
Expression and prognostic significance of EFNB2 and EPHB4 genes in
patients with oesophageal squamous cell carcinoma. Dig Liver Dis.
39:725–732. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rice TW, Ishwaran H, Ferguson MK,
Blackstone EH and Goldstraw P: Cancer of the esophagus and
esophagogastric junction: An eighth edition staging primer. J
Thorac Oncol. 12:36–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Liu DP, Chen PP, Koeffler HP, Tong
XJ and Xie D: Involvement of IFN regulatory factor (IRF)-1 and
IRF-2 in the formation and progression of human esophageal cancers.
Cancer Res. 67:2535–2543. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hasina R, Mollberg N, Kawada I, Mutreja K,
Kanade G, Yala S, Surati M, Liu R, Li X, Zhou Y, et al: Critical
role for the receptor tyrosine kinase EPHB4 in esophageal cancers.
Cancer Res. 73:184–194. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takikita M, Hu N, Shou JZ, Wang QH, Giffen
C, Taylor PR and Hewitt SM: Biomarkers of apoptosis and survival in
esophageal squamous cell carcinoma. BMC Cancer. 9:3102009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xue LY, Hu N, Song YM, Zou SM, Shou JZ,
Qian LX, Ren LQ, Lin DM, Tong T, He ZG, et al: Tissue microarray
analysis reveals a tight correlation between protein expression
pattern and progression of esophageal squamous cell carcinoma. BMC
Cancer. 6:2962006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yin J, Cui Y, Li L, Ji J and Jiang WG:
Overexpression of EPHB4 is associated with poor survival of
patients with gastric cancer. Anticancer Res. 37:4489–4497.
2017.PubMed/NCBI
|
|
21
|
Liu W, Ahmad SA, Jung YD, Reinmuth N, Fan
F, Bucana CD and Ellis LM: Coexpression of ephrin-Bs and their
receptors in colon carcinoma. Cancer. 94:934–939. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Alam S, Fujimoto J, Jahan I, Sato E and
Tamaya T: Overexpression of ephrinB2 and EPHB4 in tumor advancement
of uterine endometrial cancers. Ann Oncol. 18:485–490. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takai N, Miyazaki T, Fujisawa K, Nasu K
and Miyakawa I: Expression of receptor tyrosine kinase EPHB4 and
its ligand ephrin-B2 is associated with malignant potential in
endometrial cancer. Oncol Rep. 8:567–573. 2001.PubMed/NCBI
|
|
24
|
Li X, Song C, Huang G, Sun S, Qiao J, Zhao
J, Zhao Z and Li M: The coexpression of EPHB4 and EphrinB2 is
associated with poor prognosis in HER2-positive breast cancer.
OncoTargets Ther. 10:1735–1742. 2017. View Article : Google Scholar
|
|
25
|
Zhang S, Jiang T and Liang M: Expression
of Eph B4 and Ephrin B2 in cervical cancer tissues and
angiogenesis. Int J Gynaecol Obstet. 96:46–47. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kumar SR, Masood R, Spannuth WA, Singh J,
Scehnet J, Kleiber G, Jennings N, Deavers M, Krasnoperov V, Dubeau
L, et al: The receptor tyrosine kinase EphB4 is overexpressed in
ovarian cancer, provides survival signals and predicts poor
outcome. Br J Cancer. 96:1083–1091. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Neuber C, Belter B, Meister S, Hofheinz F,
Bergmann R, Pietzsch HJ and Pietzsch J: Overexpression of receptor
tyrosine kinase EPHB4 triggers tumor growth and hypoxia in A375
melanoma xenografts: Insights from multitracer small animal imaging
experiments. Molecules. 23(pii): E4442018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Masood R, Kumar SR, Sinha UK, Crowe DL,
Krasnoperov V, Reddy RK, Zozulya S, Singh J, Xia G, Broek D, et al:
EPHB4 provides survival advantage to squamous cell carcinoma of the
head and neck. Int J Cancer. 119:1236–1248. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Erber R, Eichelsbacher U, Powajbo V, Korn
T, Djonov V, Lin J, Hammes HP, Grobholz R, Ullrich A and Vajkoczy
P: EphB4 controls blood vascular morphogenesis during postnatal
angiogenesis. EMBO J. 25:628–641. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kertesz N, Krasnoperov V, Reddy R,
Leshanski L, Kumar SR, Zozulya S and Gill PS: The soluble
extracellular domain of EPHB4 (sEphB4) antagonizes EPHB4-EphrinB2
interaction, modulates angiogenesis, and inhibits tumor growth.
Blood. 107:2330–2338. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He S, Ding Y, Zhou J, Krasnoperov V,
Zozulya S, Kumar SR, Ryan SJ, Gill PS and Hinton DR: Soluble EPHB4
regulates choroidal endothelial cell function and inhibits
laser-induced choroidal neovascularization. Invest Ophthalmol Vis
Sci. 46:4772–4779. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Noren NK, Lu M, Freeman AL, Koolpe M and
Pasquale EB: Interplay between EPHB4 on tumor cells and vascular
ephrin-B2 regulates tumor growth. Proc Natl Acad Sci USA.
101:5583–5588. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Folkman J: Angiogenesis in cancer,
vascular, rheumatoid and other disease. Nat Med. 1:27–31. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bouck N: Angiogenesis: A mechanism by
which oncogenes and tumor suppressor genes regulate tumorigenesis.
Cancer Treat Res. 63:359–371. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Steinle JJ, Meininger CJ, Forough R, Wu G,
Wu MH and Granger HJ: Eph B4 receptor signaling mediates
endothelial cell migration and proliferation via the
phosphatidylinositol 3-kinase pathway. J Biol Chem.
277:43830–43835. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang NY, Pasquale EB, Owen LB and Ethell
IM: The EPHB4 receptor-tyrosine kinase promotes the migration of
melanoma cells through Rho-mediated actin cytoskeleton
reorganization. J Biol Chem. 281:32574–32586. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Meyer S, Hafner C, Guba M, Flegel S,
Geissler EK, Becker B, Koehl GE, Orso E, Landthaler M and Vogt T:
Ephrin-B2 overexpression enhances integrin-mediated ECM-attachment
and migration of B16 melanoma cells. Int J Oncol. 27:1197–1206.
2005.PubMed/NCBI
|
|
38
|
Xia G, Kumar SR, Masood R, Koss M,
Templeman C, Quinn D, Zhu S, Reddy R, Krasnoperov V and Gill PS:
Up-regulation of EPHB4 in mesothelioma and its biological
significance. Clin Cancer Res. 11:4305–4315. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xia G, Kumar SR, Masood R, Zhu S, Reddy R,
Krasnoperov V, Quinn DI, Henshall SM, Sutherland RL, Pinski JK, et
al: EPHB4 expression and biological significance in prostate
cancer. Cancer Res. 65:4623–4632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xia G, Kumar SR, Stein JP, Singh J,
Krasnoperov V, Zhu S, Hassanieh L, Smith DL, Buscarini M, Broek D,
et al: EPHB4 receptor tyrosine kinase is expressed in bladder
cancer and provides signals for cell survival. Oncogene.
25:769–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lian H, Jia X, Shi N, Xie S, Wang J, Wang
W, Ma F, Liu H, Wang A, Cheng X and Liu C: Notch signaling promotes
serrated neoplasia pathway in colorectal cancer through epigenetic
modification of EPHB2 and EPHB4. Cancer Manag Res. 10:6129–6141.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lv J, Xia Q, Wang J, Shen Q, Zhang J and
Zhou X: EPHB4 promotes the proliferation, invasion, and
angiogenesis of human colorectal cancer. Exp Mol Pathol.
100:402–408. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu Q, Suo Z, Kristensen GB, Baekelandt M
and Nesland JM: The prognostic impact of EphB2/B4 expression on
patients with advanced ovarian carcinoma. Gynecol Oncol. 102:15–21.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|