Introduction
The occurrence and development of malignant tumors
is a complicated process, and numerous oncogenes and tumor
suppressor genes are involved. Overexpressed in lung cancer 1
(OLC1) is a relatively novel candidate oncogene, which was
originally discovered by Yuan et al (1) using suppression subtractive
hybridization. It was identified that OLC1 was more highly
expressed in squamous cell lung tumors compared with normal
bronchial epithelial cells, and that it promoted tumor formation in
nude mice. Similarly, it was also demonstrated that OLC1 was up
regulated in human esophageal carcinoma and contributed to the
proliferation of esophageal cancer cells (2).
The OLC1 gene was first designated as increased
sodium tolerance 1 (IST1), which has also been identified and
studied in non-lethal yeast mutants (3,4). It
encodes a protein that participates in the disassembly of endosomal
sorting complexes (5,6). It was reported that the human IST1 gene,
now known as OLC1, may serve a significant function in cytokinesis
in mitosis, as demonstrated in HeLa cells (7). As it is highly conserved and vital for
organisms ranging from yeast to humans, studying the mechanisms
that control OLC1 expression may provide a greater insight into its
physiopathological function in human tumor genesis and
progression.
A preliminary investigation indicated that OLC1
maybe degraded through the ubiquitin proteasome pathway and that
cigarette smoke condensate (CSC) may affect this degradation
(8). Through a bioinformatics
analysis of the OLC1 protein, destruction (D)-box motif (amino acid
sequence, RXXLXXXXN) was identified at amino acids 12–20. This
motif is essential for the recognition and subsequent degradation
of the protein by the anaphase-promoting complex/cyclosome (APC/c)
(9,10). As the destruction of APC targets is
regulated by two activators, cell-division cycle protein 20 (Cdc20)
and cadherin 1 (Cdh1) (11–13), these activators are also predicted to
govern the degradation of OLC1. Previous studies regarding this
topic predominantly focused on the effects of CSC on OLC1 stability
(8), whereas the present study
explores the in-depth mechanism by which OLC1 is degraded through
the ubiquitin proteasome pathway.
In the present study, it was revealed that OLC1
interacted with the APC/c through the APC2 subunit, assisted by two
coactivator proteins, Cdh1 and Cdc20. The up regulation of Cdh1 and
Cdc20 accelerated OLC1 degradation, whereas the down regulation of
Cdh1/Cdc20 resulted in OLC1 protein accumulation. In addition, the
D-box sequence was characterized, as this motif was a critical
determinant of OLC1 degradation. Mutations of the D-box motif
enhanced OLC1 protein stability and induced an increase in cell
growth and colony formation.
Materials and methods
Cell lines, cell culture and
synchronization procedures
A stable OLC1-overexpressing cell line
KYSE150/GFP-OLC1 and its null control cell line KYSE150/GFP were
constructed as previously described (2). H1299 human lung carcinoma cells were
purchased from the American Type Culture Collection (Manassas, VA,
USA). KYSE150/GFP, KYSE150/GFP-OLC1 and H1299 cells were cultured
in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
HyClone; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100
U/ml penicillin, 100 µg/ml streptomycin and 400 µg/ml G418 in a
humidified atmosphere with 5% CO2 at 37°C.
To synchronize the cells at the G0 phase,
the cells were incubated in RPMI-1640 medium without serum for 72
h. The cells were transferred into fresh RPMI-1640 medium with
serum for subsequent incubation. Then cells were collected every 2
h.
For G2/M phase arrest, cells were
incubated with 400 ng/ml nocodazole (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 16 h. Following nocodazole treatment, cells
were transferred into completeRPMI-1640 medium as described, and
collected every 2 h.
Reverse transcription polymerase chain
reaction (RT-PCR)
Total RNA was extracted from treated KYSE150/GFP
cells with TRIzol reagent (Thermo Fisher Scientific, Inc.), and RNA
(6 µg) was reverse-transcribed according to the manufacturer's
protocol (Super Script™ First-Strand Synthesis System for RT-PCR
kit; Thermo Fisher Scientific, Inc., Waltham, MA, USA). For PCR
amplification, 1 µl cDNA solution was used as a template. The
primer sequences used for the 435-bp OLC1 product were as follows:
Forward, 5′-ACAGTGGGAGAGAGCACGTT-3′ and reverse,
5′-GCACCTTGTCCTTTCTCTGC-3′. The sequences for the 299-bp GAPDH
product were as follows: Forward, 5′-GCTGAGAACGGGAAGCTTGT-3′ and
reverse, 5′-GCCAGGGGTGCTAAGCAG-3′. The thermo cycling conditions
for OLC1 are: 94°C for 5 min, 94°C for 30 sec, 58°C for 30 sec,
72°C for 1 min, 30 cycles, and 70°C for 7 min. The thermo cycling
conditions for GAPDH are: 94°C for 4 min, 94°C for 30 sec, 58°C for
30 sec, 72°C for 30 sec for 20 cycles, and 70°C for 4 min.
Protein stability experiments
In order to identify the half-life of OLC1, the
protein synthesis inhibitor cycloheximide (CHX; Sigma-Aldrich and
Merck KGaA, Darmstadt, Germany, 100 µg/ml) was added to the cell
culture, and cells were collected at a range of time intervals. In
order to ensure the effect of the proteasome on OLC1 degradation,
MG132, a proteasome inhibitor, was added to the cell culture. The
cells were incubated with CHX alone or with CHX and 20 µM MG132 for
a range of durations (4, 8, 12 and 16 h), or with CHX and a range
of MG132 concentrations (5, 10 and 20 µM). For each experiment,
dimethyl sulfoxide (DMSO) was used as a control.
Immunoprecipitation and western blot
analyses
Cells were harvested by washing twice with PBS and
scraping away the cells, which were lysed in a lysis buffer on ice
for 40 min. The composition of the lysis buffer was as follows:
PBS, 2 µg/ml aprotinin, 2 µg/ml leupeptin, 1% NonidetP-40 and 100
µg/ml phenylmethylsulfonyl fluoride. The lysates were harvested via
centrifugation at 4°C at 16,000 × g for 20 min, and the
supernatants were extracted to collect complete protein
samples.
For immunoprecipitation, 500 µg total cellular
protein lysate was incubated with ~2 µg of the indicated
antibodies, including OLC1 rabbit anti-human polyclonal antibody
(ready for use), which was prepared and purified using the purified
recombinant glutathione-S-transferase-OLC1 by Wuhan Sanying
Biotechnology, Wuhan, China (1) and
actin mouse anti-human monoclonal antibody (ready for use; cat.
no., sc-8432; Santa Cruz Biotechnologies, Santa Cruz, CA, USA) at
4°C for 6 h and then incubated with 20 µl A/G-agarose beads (Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. Next,
immune complexes was harvested via centrifugation at 4°C at 1,000 ×
g for 5 min and washed 7–8 times with 1% Nonidet P-40. The
immunocomplexed proteins were then subjected to a western blot
analysis.
For the western blot analysis, 80–100 µg cellular
protein was resolved via 10% SDS-PAGE and transferred to
Immobilon-P polyvinylidene fluoride membranes (Millipore, Bedford,
MA, USA). Then the membranes were incubated in PBS containing 5%
nonfat milk at room temperature for 30 min to block nonspecific
binding. Next the membranes were incubated at 37°C for 2 h with the
following antibodies: OLC1 (dilution, 1:100; Wuhan Sanying
Biotechnology, Wuhan, China), APC2 (dilution, 1:500; cat. no.,
sc-517022), cyclin A (dilution, 1:500; cat. no., sc-271682),
cyclinD1 (dilution, 1:500; cat. no., sc-246), cyclin E (dilution,
1:500; cat. no., sc-247), actin (dilution, 1:1,000; cat. no.,
sc-8432), GFP (dilution, 1:500; cat. no., sc-9996) (all from Santa
Cruz Biotechnology, Inc., Dallas, TX, USA), Cdc20 (dilution,
1:2,000; cat. no., NB100-59828; Novus Biologicals, LLC, Littleton,
CO, USA,) ubiquitin (dilution, 1:1,000; cat. no., U5379; Sigma
Aldrich; Merck KGaA) and Cdh1 (dilution, 1:1,000; cat. no., C7855;
Sigma Aldrich; Merck KGaA) and cyclin B1 (dilution, 1:250; cat.
no., 554178; BD Biosciences, Franklin Lakes, NJ, USA). The
membranes were washed in PBS with 0.1% Tween 20 five times and then
incubated at room temperature for 1 h with a goat anti-mouse (cat.
no., sc-2005) or goat anti-rabbit (cat. no., sc-2004) horseradish
peroxidase-conjugated secondary antibody (the two antibodies were
diluted at 1:1,000 and were purchased from Santa Cruz
Biotechnology, Inc.). Next the membranes were washed five times,
and antibody reactivity was visualized using a FUJIFILM LAS-4000
machine. Images were edited using Multi-Gauge Fujifilm (version 3;
Fujifilm Life Science, Tokyo, Japan) and Photoshop CS software
(Adobe Systems, Inc., San Jose, CA, USA).
Cell protein ubiquitination assay
Cultured cells were treated with 20 µM MG132 or
DMSO. Then cells were collected andlysed on ice for 30 min with
thelysis buffer. Then the lysates were centrifuged at 16,000 × g
for 20 min at 4°C. In each immunoprecipitation reaction, as
described previously, 500 µg total cellular protein lysate was
incubated with 10 µl of the indicated antibody. Then the immune
complexes were pulled down and analyzed with western blotting.
Using an anti-ubiquitin antibody, the polyubiquitinated OLC1
protein was probed.
Plasmid mutation and cell
transfection
pEGFP-N1 and pEGFP-N1-OLC1 plasmids were supplied by
Professor Shujun Cheng of the Chinese Academy of Medical Sciences
and Peking Union Medical College (Beijing, China). pCMV-Myc-Cdh1
and pCMV-Myc-Cdc20 plasmids were provided by Professor James Hsieh
of Washington University (Seattle, WA, USA) and pCMV-3 plasmids
were obtained from the State Key Laboratory of Molecular Oncology
(Beijing, China).
In order to identify the specific sequences that
acted as potential degradation signals for OLC1, a generated
D-boxsite-directed mutation construct, pEGFP-N1-mut-OLC1, was
produced in conjunction with Shanghai Gene Chem Co., Ltd.
(Shanghai, China). For cell transfection, H1299 cells were seeded
onto 35-mm plates 1 day prior to transfection to allow cells to
reach 95% confluence at the time of transfection. To prepare each
plate, 10 µl Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) and 4 µg plasmid DNA were added to 500 µl
RPMI-1640 medium without antibiotics. The solutions were mixed
lightly, left to stand for 20 min at room temperature, diluted with
3 ml RPMI-1640 medium without antibiotics, and added to the plates
to stand for 6 h at 37°C. Next, 4 ml RPMI-1640 medium from each
plate was exchanged for an equal volume of RPMI-1640 medium
supplemented with 10% FBS, and plates were incubated for an
additional 48 h. Cells were then harvested.
Small interfering RNA (siRNA)
construction and cell transfection
siRNA for Cdh1 and Cdc20 were constructed by Jikai
Biotechnology, Inc. The Cdh1 siRNA sequence was
5′-UGAGAAGUCUCCCAGUCAGdTdT-3′ and the Cdc20 siRNA sequence was
5′-AAACCTGGCGGTGACCGCTAT-3′.
For cell transfection, H1299 cells were seeded onto
35-mm plates 1 day prior to transfection to allow the cells to
reach 70% confluence at the time of transfection. To prepare each
plate, 4 µg siRNA and 10 µl Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) were added to 500 µl
RPMI-1640 medium without antibiotics. The solutions were mixed
gently, sitting for 20 min at room temperature, diluted with 3 ml
RPMI-1640 medium without antibiotics and added to the plates to sit
for 6 h at 37°C. Next, 4 ml RPMI-1640 medium from each plate was
exchanged for an equal volume of medium with 10% FBS, and plates
were incubated for 48 h. Cells were then harvested.
Cell proliferation assay and colony
formation
Cells in the exponential growth phase were seeded at
a density of 3,000 cells per well in 12-well plates, in triplicate.
Cells were then counted every 24 h for 5 days to produce a growth
curve. All experiments were repeated three times.
At 24 h after transfection, cells transfected with
each specific plasmid were seeded in RPMI-1640 with 10% FBS and 400
µg/ml G418 at a density of 1,000 cells per well in six-well plates
in triplicate. After 14 days, the cells were washed with PBS, fixed
in cold methanol and stained with 0.5% crystal violet for 10 min at
room temperature. Colonies with more than 50 cells were counted.
All experiments were repeated three times.
Statistical analysis
SPSS version 11.5.0 (SPSS, Inc., Chicago, IL, USA)
was used to perform all statistical analyses. A Student's t-test
was used for comparison between two groups. The differences between
multiple groups were analyzed using a one-way analysis of variance
and Fisher's least significant difference test as a post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
OLC1 expression is cell
cycle-dependent
To explore whether the expression of OLC1 is
associated with cell cycle progression, two cell cycle
synchronization approaches were adopted in order to detect the OLC1
protein expression in KYSE150/GFP cells. By means of serum
starvation, cells were synchronized at the G0 phase.
Cells were arrested at the mitotic phase with the microtubule
inhibitor nocodazole. Cells were subsequently collected every 2 h,
and the expression of OLC1 protein was analyzed with western blot
assays. The expression of cyclin B1, D1, A and E were analyzed to
assess cell cycle progression. As presented in Fig. 1A, following serum starvation, OLC1
protein was highly expressed during most of the
G0/G1 phase, remained at relatively low
levels at the S phase and demonstrated a slightly elevated level in
the G2/M phase. Similar results were obtained in the
experiments performed using no codazole to synchronize cells at the
mitotic phase (Fig. 1B). In an
additional experiment, the mRNA expression of OLC1 was examined,
which demonstrated relatively little change throughout the whole
cell cycle (Fig. 1C). In conclusion,
these observations indicate that OLC1 protein expression is cell
cycle-dependent and that it is primarily regulated via
post-translational, and not transcriptional, modification.
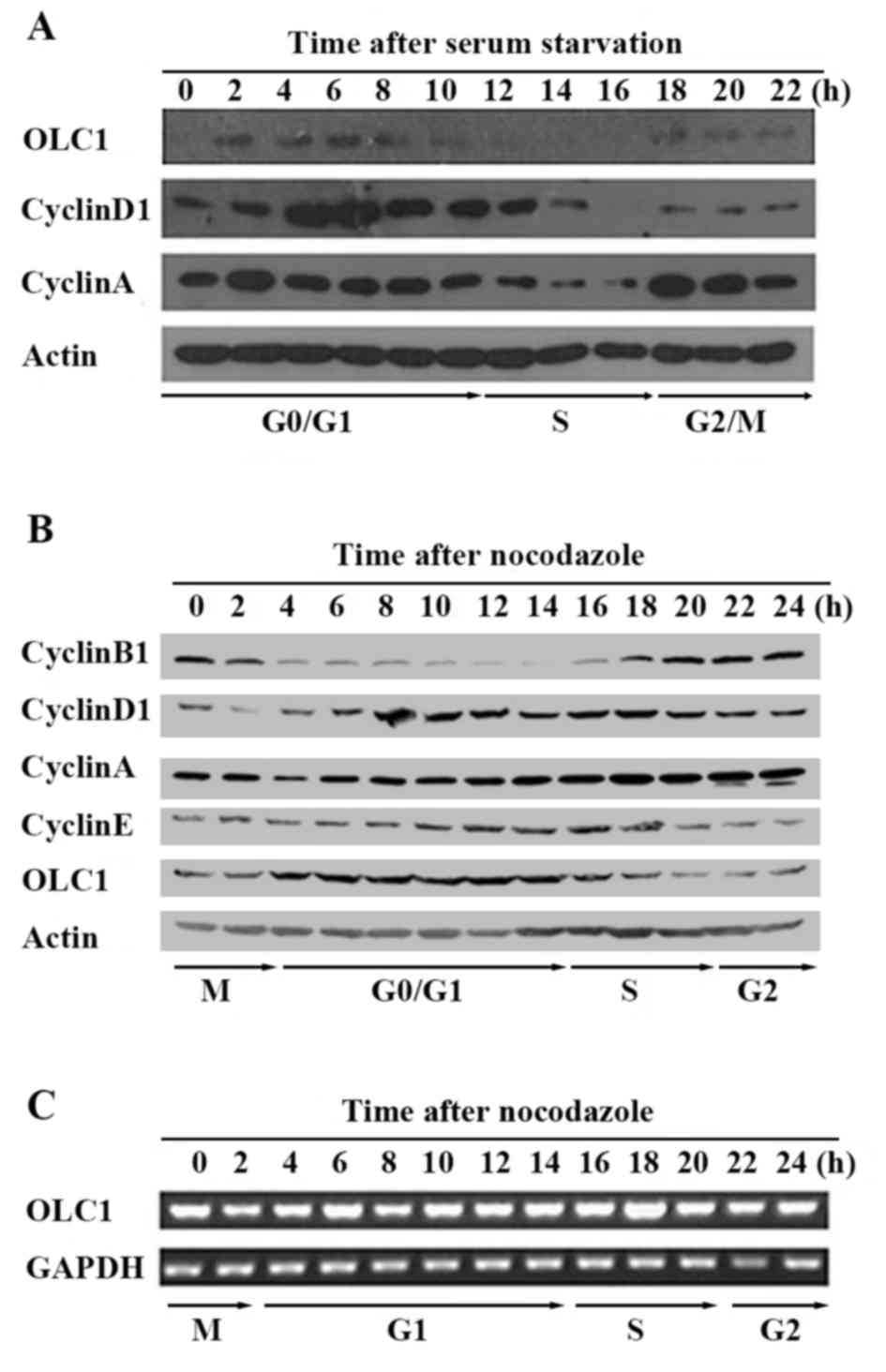 | Figure 1.Expression of OLC1 throughout the cell
cycle. (A) Using serum starvation, KYSE150/GFP cells were
synchronized at the G0 phase. Following release, cells
were collected every 2 for 24 h. The protein expression of OLC1,
cyclin D1, cyclin A and actin was analyzed using western blot
analysis. (B) Using 0.4 µg/ml nocodazole, KYSE150/GFP cells were
synchronized at the mitotic phase. Following release, cells were
collected every 2 for 24 h. The protein expression of OLC1, cyclin
B1, cyclin D1, cyclin A, cyclin E and actin was analyzed using
western blot analysis. (C) Using reverse transcription polymerase
chain reaction, the mRNA expression of OLC1 and GAPDH were
detected. OLC1, overexpressed in lung cancer 1. |
OLC1 protein is short-lived and its
stability is regulated by the ubiquitin proteasome pathway
Considering that the OLC1 protein expression was
determined to be cell cycle-dependent, similar to the cyclins, the
half-life of OLC1 protein was not expected to be long. Following
treatment with the protein synthesis inhibitor CHX for a range of
times, the half-life of OLC1 was analyzed. OLC1 protein expression
decreased to ~50% at 8 h of CHX treatment compared with the
expression observed at 0 h and in the untreated control cells
(Fig. 2A), suggesting that OLC1
protein has a rapid turnover rate. Given that 80–90% of
intracellular proteins are degraded by the ubiquitin proteasome
pathway, it was predicted that OLC1 protein stability may be
regulated though this mechanism. Therefore, the selective
proteasome inhibitor MG132 was used to treat KYSE150/GFP and
KYSE150/GFP-OLC1 cells with or without CHX, which was followed by
an analysis of endogenous and exogenous OLC1 protein expression.
The addition of different concentrations of MG132 resulted in an
increase in OLC1 expression compared with cells treated only with
CHX. Similarly, with increasing MG132 treatment times, OLC1 protein
expression levels were increased accordingly (Fig. 2B and C). This suggests that OLC1
protein degradation is regulated by the ubiquitin proteasome
pathway. Subsequently, immune precipitation as says with an
anti-OLC1 antibody was performed to further confirm this. Ubiquitin
was detected in the OLC1 immuno complex, and as expected, the
amount present was increased following the addition of MG132
(Fig. 2D).
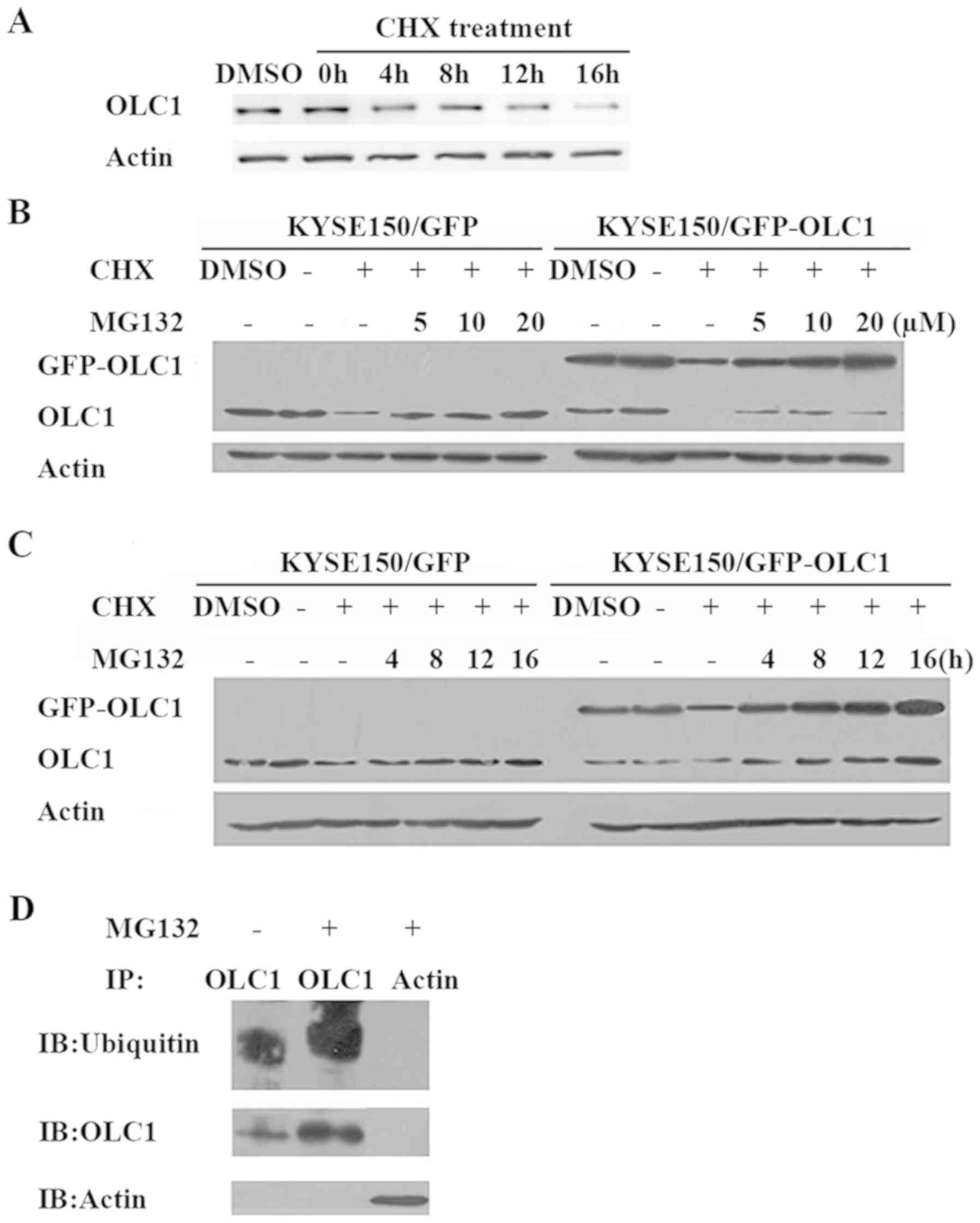 | Figure 2.OLC1 protein expression with CHX and
MG132 treatment. (A) Following treatment with 100 µg/ml CHX,
KYSE150/GFP cells were collected for western blot analysis of OLC1
protein expression at the indicated times. KYSE150/GFP and
KYSE150/GFP-OLC1 cells were co-treated with CHX (100 µg/ml) and
MG132 at (B) different concentrations (5, 10 and 20 µM MG132) or
(C) for different lengths of time (4, 8, 12 and 16 h) with 20 µM
MG132, with the DMSO, negative control and MG132-negative groups
collected for analysis after 16 h of treatment. Results are
representative of three independent experiments. (D) With (+) or
without (−) treatment with 20 µM MG132, KYSE150/GFP-OLC1 cells were
incubated for 16 h, collected and lysed for IP with an anti-OLC1
antibody. Ubiquitins were detected in the immunocomplex, and the
presence of OLC1 was verified. IP with an anti-actin antibody was
used as a negative control. OLC1, overexpressed in lung cancer 1;
CHX, cycloheximide; IP, immunoprecipitation. |
OLC1 protein is degraded by APC/c
The OLC1 protein sequence was analyzed and a D-box
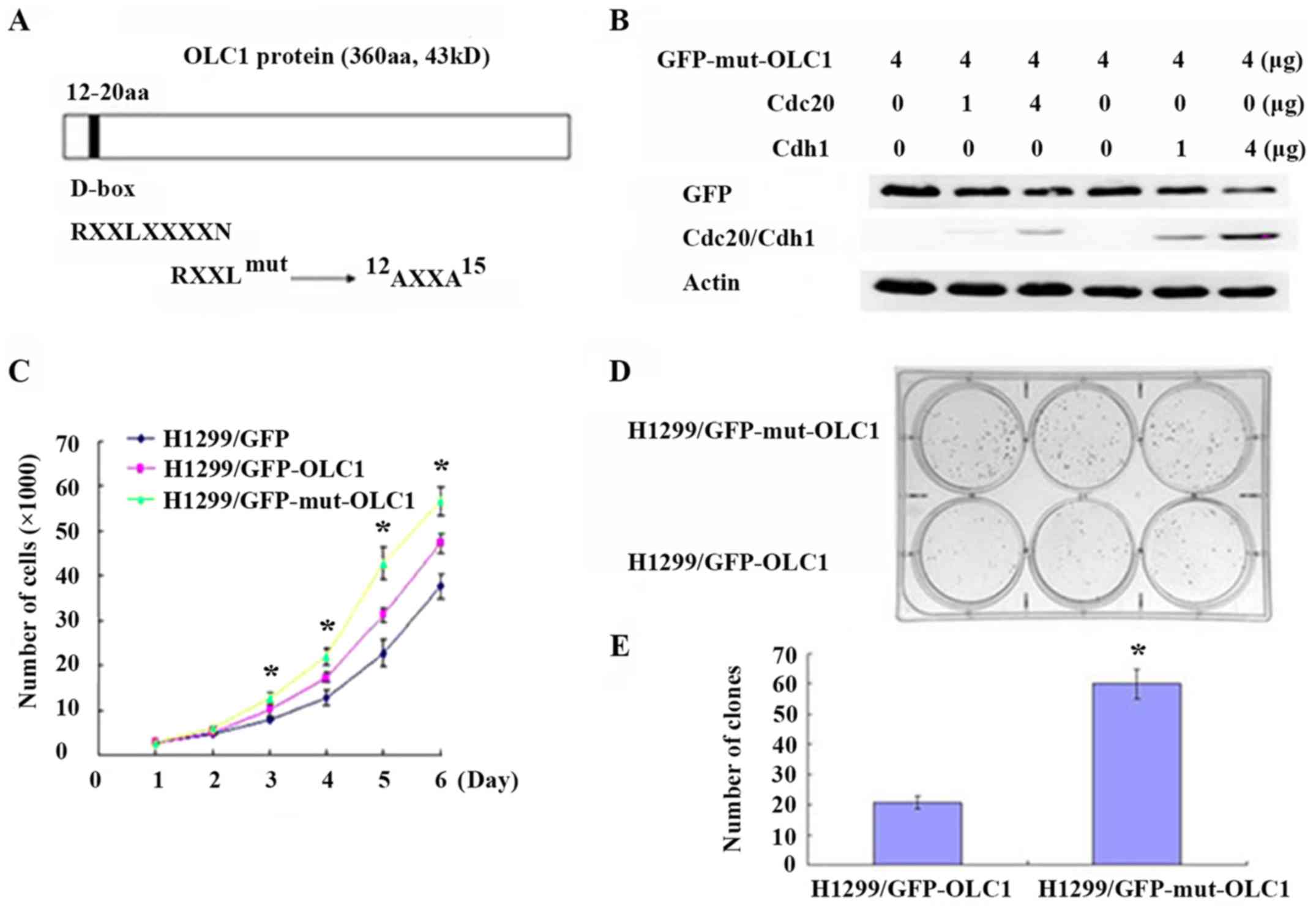
motif was identified (amino acids 12–20; Fig. 3A). In the process of ubiquitination
and degradation of proteins, D-box motifs are often the recognition
sites for the E3 ligase of the APC/c (7,8). Thus, it
was hypothesized that the E3 ligase of APC/c may facilitate OLC1
ubiquitination. In the APC/c, APC2 is the most important subunit,
as it acts as the connector with the target protein. Therefore, the
interaction between OLC1 with APC2 was examined. The results
demonstrated that OLC1 directly interacts with APC/c through its
APC2 component (Fig. 3B), which
indicates that OLC1 protein ubiquitination and degradation are
mediated by the APC/c pathway.
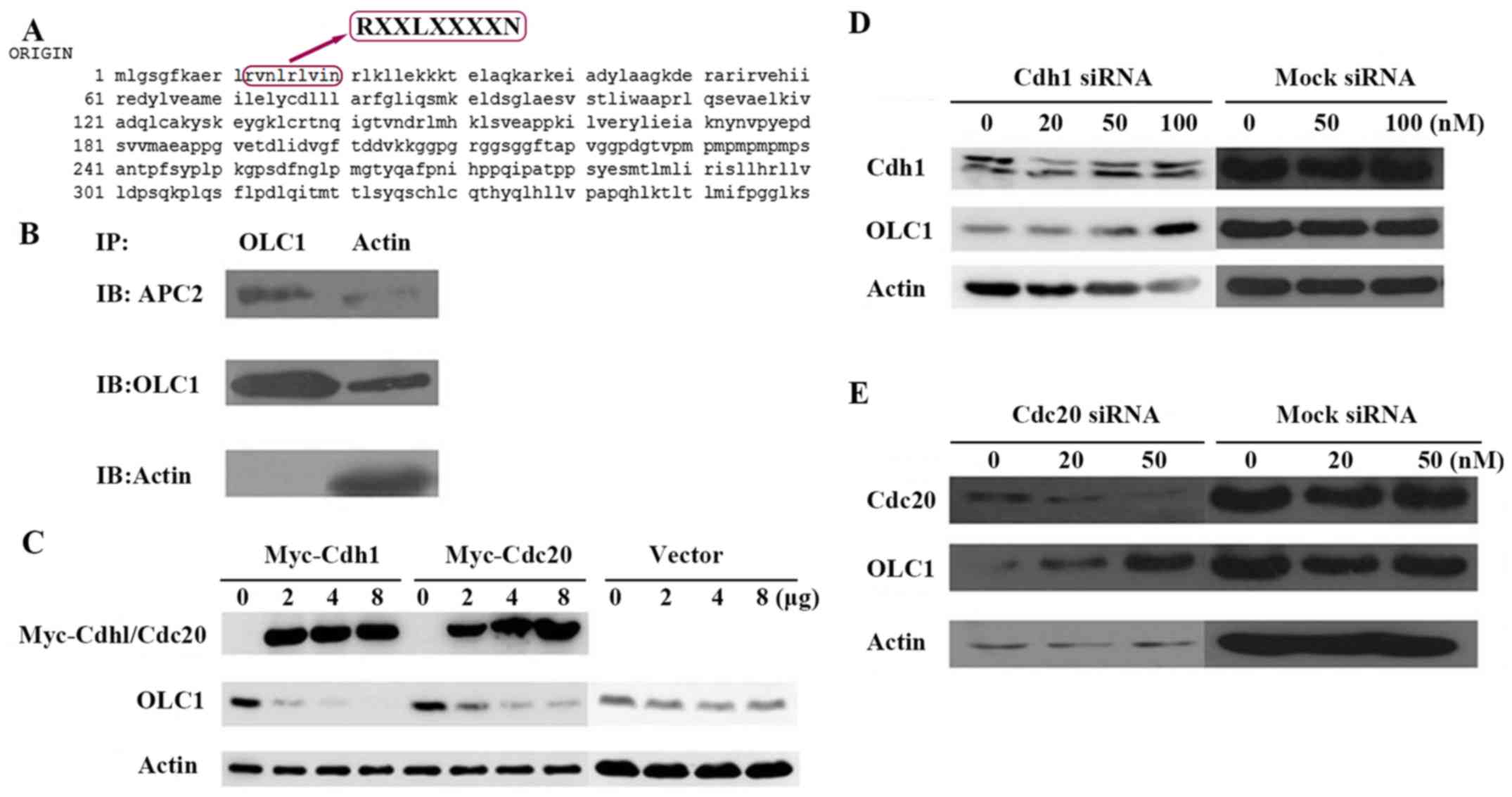 | Figure 3.OLC1 protein is degraded by the APC/c.
(A) Analysis of the OLC1 protein sequence indicated that it
contains a destruction box at the site of amino acids 12–20, which
may be recognized by APC/c. (B) To investigate whether OLC1 may
bind directly to the components of the APC/c, KYSE150/GFP-OLC1
cells were collected, lysed and subjected to IP. The presence of
actin, OLC1 and APC2 in the immunocomplex were verified. (C)
Different concentrations (2, 4 and 8 µg) of Myc-Cdh1/Cdc20
expression or mock vectors were transiently transfected into H1299
cells for 48 h. Then OLC1, Myc-Cdh1 and Myc-Cdc20 protein
expression were evaluated. Actin was included as a loading control.
Different concentrations (20, 50 and 100 nM) of (D) Cdh1 or (E)
Cdc20 or mock siRNA were transiently transfected into H1299 cells
for 48 h. Then OLC1, Cdh1 and Cdc20 protein expressions were
evaluated. Actin was included as a loading control. OLC1,
overexpressed in lung cancer 1; APC/c, anaphase-promoting cyclosome
complex; IP, immunoprecipitation; Cdh1, cadherin 1; Cdc20,
cell-division cycle protein 20; siRNA, small interfering RNA. |
The activation of APC/c also requires the
association with proteins containing tryptophan aspartate. Cdc20
(termed Fizzy in Drosophila) and Cdh1 (Fizzy-associated in
Drosophila) have been identified as two of these activators,
and have been demonstrated to activate APC/c E3 ligase and
stimulate substrate degradation. These two proteins may directly
interact with the APC/c and serve important functions as limiting,
substrate-specific activators of APC-dependent proteolysis
(11,12). To confirm whether Cdh1 or Cdc20 were
required for OLC1 protein degradation, Cdh1 and Cdc20 expression
was transiently up regulated individually in H1299 cells; it was
revealed that the increase of each protein resulted in a reduction
in OLC1 protein expression (Fig. 3C).
On the contrary, the down regulation of the endogenous Cdh1 or
Cdc20 via siRNAs resulted in the increased expression of OLC1
(Fig. 3D and E). From the above
results, it was posited that Cdh1 and Cdc20 are involved in the
process of OLC1 degradation.
Mutated D-box domains interfere with
OLC1 protein degradation and accelerate cell growth and colony
formation
In the present study, the analysis of the OLC1
protein sequence revealed that it contained a D-box (amino acids
12–20). Generally, substrates containing a D-box or KEN-box domain
are recognized by the APC/c E3 ligase, assisted by Cdc20 and Cdh1
(11,12). Therefore, it was hypothesized that the
D-box domain mediated the ubiquitination-dependent destruction of
OLC1 proteins. The conserved D-box motif was point-mutated, as
presented in Fig. 4A, and it was
assessed whether this mutation had an effect on OLC1 protein
stability. H1299 cells were co-transfected with D-box-mutated
GFP-OLC1 (GFP-mut-OLC1) and different concentrations of Cdh1 or
Cdc20 expression vectors. As expected, the increased expression of
Cdh1 and Cdc20 did not promote the degradation of the mutated OLC1,
suggesting that the identified D-box motif was the critical
sequence required for OLC1 ubiquitination and the subsequent
degradation (Fig. 4B).
Previous studies have demonstrated that OLC1 is an
oncogenic protein, as the induced overexpression of OLC1 results in
anchorage-independent growth in vitro and malignant
transformation in vivo. Additionally, OLC1 has been
identified as overexpressed in multiple types of malignant tumor,
including in lung cancer and esophageal squamous carcinoma
(1,2).
Thus, additional experiments were performed to confirm if the cells
with mutated OLC1 may exhibit more malignant characteristics.
First, a cell growth assay was performed; the growth curve revealed
that H1299 cells expressing OLC1 with a mutated D-box grew
significantly faster compared with those expressing wild-type OLC1
(Fig. 4C). This indicated that the
D-box mutated OLC1 exhibited a greater capacity to facilitate cell
growth. Colony formation assays were also conducted; the cells
expressing mutant OLC1 developed a significantly higher number of
colonies compared with the control cells (Fig. 4D and E). These results indicated that
the mutated D-box motif was not recognized by APC/c E3 ligase,
affecting the subsequent degradation of the OLC1 protein. Overall,
non-degradable OLC1 presents a greater oncogenic capacity compared
with wild-type OLC1.
Discussion
The OLC1 gene is located in chromosome 16q22.2. When
it was first identified in non-lethal yeast mutants in 1999, it was
named the IST1 gene (14). In yeast,
IST1 participates in the multi vesicular body (MVB) sorting
pathway, in which certain membrane proteins are sorted into the
lumen of the vacuole for their eventual degradation (15,16). The
gene product of IST1/OLC1 in different organisms is highly
conserved, including between yeast and humans. In humans, a
previous study identified that OLC1 was essential for cytokinesis,
another membrane scission event that is topologically similar to
MVB formation, and that the depletion of OLC1 resulted in the
accumulation of multinucleated cells (7). Cytokinesis is the last stage of the cell
cycle, where a cell divides into two daughter cells, passing the
same amount of genetic material to each daughter cell. Therefore,
abnormal cytokinesis will result in the uneven distribution of the
chromosomes in the cell, inducing cell genome instability and
potentially leading to the development of a tumor (17,18).
A previous study regarding OLC1 studied it from
another angle; the OLC1 gene was identified as a potential
oncogene, that was highly expressed in lung cancer, in 2008
(1). It was identified that the over
expression of OLC1 was associated with smoking history in patients
with lung cancer, and that this overexpression induced tumor
formation in athymic mice, whereas the knockdown of OLC1 increased
apoptosis and decreased colony formation. It was also revealed that
cigarette smoke may increase OLC1 protein expression at the
cellular level (8). Previous studies
have revealed that the OLC1 protein is highly expressed in numerous
malignant tumor types, including esophageal squamous cancer,
colorectal cancer, breast cancer and ovarian cancer. Furthermore,
high expression levels of OLC1 are associated with a poor prognosis
in a number of these cancer types (2,19–21).
However, until the present study, few studies
focused on the molecular regulatory mechanism controlling OLC1
expression. Given the critical function of OLC1 from yeast to
humans, studying how OLC1 is regulated may provide further insight
into human tumor genesis. In the present study, it was demonstrated
that OLC1 protein may be cell-cycle-dependent with a short
half-life, and that it may be degraded through the APC/c-mediated
proteasome pathway.
Two approaches for cell cycle synchronization were
conducted in order to study the expression pattern of OLC1 during
the entire cell cycle. Given that the expression of numerous other
cyclins fluctuates in different phases of the cell cycle, cyclins
A, B1, D1 and E were examined to determine the dynamic changes in
OLC1 expression in the cell cycle. The OLC1 protein exhibited
higher expression levels during the G0/G1
phase compared with the other phases, whereas the mRNA expression
levels of OLC1 demonstrated little change across the entire cell
cycle. In conclusion, the expression of OLC1 is regulated through
post-transcriptional mechanisms during the cell cycle.
Mammalian cells contain two distinct major
proteolytic pathways. One important non-lysosomal mechanism for the
degradation of intracellular proteins is the ubiquitin-proteasome
pathway (22,23). Ubiquitin and ubiquitin ligase
post-translationally modify the abundance of target proteins,
including oncogenes or tumor suppressor genes, and thereby alter
their effect. For example, the Akt signaling pathway has many
important biological functions, while its deregulation is
associated to the development of numerous types of malignant tumors
in humans. Although previous studies have primarily focused on Akt
phosphorylation, other post-translational modifications to Akt,
including ubiquitination, have been demonstrated to serve an
important function in Akt activation. A previous study revealed
that a cancer-associated Akt mutation within the Akt PH domain
(E17K) was identified in a range of human cancer types, including
colon and breast cancer. Akt E17K mutants displayed enhanced Akt
ubiquitination, contributing to Akt hyperactivation and
constitutive Akt membrane recruitment, suggesting a potential role
for Akt ubiquitination in cancer (24). MDM2 is an E3 ubiquitin ligase with
strong clinical relevance due to its ability to regulate the tumor
suppressor p53. By recruiting an E2 ubiquitin-conjugating enzyme,
MDM2 facilitates the export of p53 from the nucleus to the
cytoplasm, and targets p53 for ubiquitin-dependent proteasome
degradation (25).
Similar to the processes revealed for other
oncogenes and tumor suppressor genes, ubiquitin-mediated
proteolysis was determined as an important regulator of OLC1
protein expression. In the present study, it was demonstrated that
the expression of OLC1 was elevated following treatment with MG132,
a proteasome inhibitor. OLC1 protein degradation decreased, and
increased ubiquitin was ligated to OLC1 proteins following MG132
treatment. All these results indicate that OLC1 degradation must be
regulated by the ubiquitin-proteasome pathway.
A previous study has demonstrated that the
interactions between OLC1 and other proteins are involved in a
range of biological processes, including MVB biogenesis,
cytokinesis and enveloped virus budding (16). In mammalian cells, the complete
function of OLC1 is required for efficient abscission during
cytokinesis (5). Additionally, in the
process of cytokinesis, the E3 ligase APC/c mediates the
ubiquitin-dependent proteolysis of cell-cycle-regulating proteins
(26,27). Thus, we hypothesized that APC/c may
function as an E3 ligase for OLC1 ubiquitination. In a further
study, it was revealed that there was a conserved D-box motif of
RVNLRLVINR within the OLC1 protein sequence, which is a conserved
and well-recognized site for E3 ligase APC/c in a number of
ubiquitinated substrates (28).
Additionally, co activators containing tryptophan aspartate are
also required to assist the activation of APC/c. Two of these
proteins have been identified as Cdc20 and Cdh1 (13,29).
Usually, Cdc20 targets D-box-containing substrates, whereas Cdh1
may interact with either the D-box or KEN box of proteins (11,30).
Consistent with this, in the present study, immune precipitation as
says revealed that OLC1 may directly bind to APC/c through the
subunit APC2, and either Cdc20 or Cdh1 up regulation induced
decreased OLC1 expression. Likewise, cells with Cdc20 and Cdh1 down
regulated via siRNA exhibited OLC1 protein stabilization.
Furthermore, mutations to the OLC1 D-box significantly reduced OLC1
degradation. Collectively, it was confirmed that the APC/c mediated
ubiquitin-proteasome pathway regulated the degradation of OLC1.
Using constructed wild-type and D-box-mutated OLC1
plasmids, functional experiments were performed. Growth curve and
colony formation assays demonstrated that the overexpression of the
non-degradable OLC1 protein not only facilitated cell growth, but
also enhanced the clone-forming capability of the cells. These
findings reveal that the destruction of the degradation domain
results in the abnormal accumulation of the OLC1 mutant, which
could not be degraded through the APC/c-mediated
ubiquitin-proteasome pathway. These malignant cell phenotypes
support the idea that OLC1 exhibits on cogenic properties.
In conclusion, these findings have the potential to
make important contributions in clarifying the mechanism of the
APC/c-mediated destruction of the candidate oncogene OLC1.
Acknowledgements
The authors would like to thank Dr Shimada of Kyoto
University (Kyoto, Japan) for providing the KYSE150 cells and
Professor Shujun Cheng of the Chinese Academy of Medical Sciences
and Peking Union Medical College, Cancer Institute and Cancer
Hospital (Beijing, China), for providing the pEGFP-N1-OLC1 and
pEGFP-N1 plasmids. pCMV-Myc-Cdh1 and pCMV-Myc-Cdc20 plasmids were
generously provided by Professor James Hsieh of Washington
University (Seattle, WA, USA).
Funding
This research was supported by Grants from the
National High Technology Research and Development Program of China
(grant no., 2006AA02A403).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author contributions
QMZ and TT obtained funding and participated in the
study coordination. ZXJ and WC were responsible for study design.
ZXJ, WC and NY performed the experimental procedures. LJD and MF
performed data analysis, and ZXJ, WC and TT were responsible for
editing and review of the manuscript. TT was also responsible for
study design.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yuan J, Ma J, Zheng H, Shi T, Sun W, Zhang
Q, Lin D, Zhang K, He J, Mao Y, et al: Overexpression of OLC1,
cigarette smoke, and human lung tumorigenesis. J Natl Cancer Inst.
100:1592–1605. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li X, Suo J, Shao S, Xue L, Chen W, Dong
L, Shi J, Fu M, Lu N, Zhan Q and Tong T: Overexpression of OLC1
promotes tumorigenesis of human esophageal squamous cell carcinoma.
PLoS One. 9:e909582014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dimaano C, Jones CB, Hanono A, Curtiss M
and Babst M: Ist1 regulates Vps4 localization and assembly. Mol
Biol Cell. 19:465–474. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rue SM, Mattei S, Saksena S and Emr SD:
Novel Ist1-Did2 complex functions at a late step in multivesicular
body sorting. Mol Biol Cell. 19:475–484. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Agromayor M, Carlton JG, Phelan JP,
Matthews DR, Carlin LM, Ameer-Beg S, Bowers K and Martin-Serrano J:
Essential role of hIST1 in cytokinesis. Mol Biol Cell.
20:1374–1387. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bajorek M, Schubert HL, McCullough J,
Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill
CP and Sundquist WI: Structural basis for ESCRT-III protein
autoinhibition. Nat Struct Mol Biol. 16:754–762. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bajorek M, Morita E, Skalicky JJ, Morham
SG, Babst M and Sundquist WI: Biochemical analyses of human IST1
and its function in cytokinesis. Mol Biol Cell. 20:1360–1373. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Xiao T, Cheng S, Tong T and Gao
Y: Cigarette smoke suppresses the ubiquitin-dependent degradation
of OLC1. Biochem Biophys Res Commun. 407:753–757. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Castro A, Bernis C, Vigneron S, Labbe JC
and Lorca T: The anaphase-promoting complex: A key factor in the
regulation of cell cycle. Oncogene. 24:314–325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Manchado E, Eguren M and Malumbres M: The
anaphase-promoting complex/cyclosome (APC/C): Cell-cycle-dependent
and -independent functions. Biochem Soc Trans. 38:65–71. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
da Fonseca PC, Kong EH, Zhang Z, Schreiber
A, Williams MA, Morris EP and Barford D: Structures of APC/C(Cdh1)
with substrates identify Cdh1 and Apc10 as the D-box co-receptor.
Nature. 470:274–278. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Robbins JA and Cross FR: Regulated
degradation of the APC coactivator Cdc20. Cell Div. 5:232010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang S, Chang L, Alfieri C, Zhang Z, Yang
J, Maslen S, Skehel M and Barford D: Molecular mechanism of APC/C
activation by mitotic phosphorylation. Nature. 533:260–264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Entian KD, Schuster T, Hegemann JH, Becher
D, Feldmann H, Güldener U, Götz R, Hansen M, Hollenberg CP, Jansen
G, et al: Functional analysis of 150 deletion mutants in
Saccharomyces cerevisiae by a systematic approach. Mol Gen Genet.
262:683–702. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Allison R, Lumb JH, Fassier C, Connell JW,
Ten Martin D, Seaman MN, Hazan J and Reid E: An ESCRT-spastin
interaction promotes fission of recycling tubules from the
endosome. J Cell Biol. 202:527–543. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo EZ and Xu Z: Distinct mechanisms of
recognizing endosomal sorting complex required for transport III
(ESCRT-III) protein IST1 by different microtubule interacting and
trafficking (MIT) domains. J Biol Chem. 290:8396–8408. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
D'Avino PP, Giansanti MG and Petronczki M:
Cytokinesis in animal cells. Cold Spring Harb Perspect Biol.
7:a0158342015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tormos AM, Taléns-Visconti R and Sastre J:
Regulation of cytokinesis and its clinical significance. Crit Rev
Clin Lab Sci. 52:159–167. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jia C, Li X, Sun H and Sui L:
Over-expression of the overexpressed in lung cancer 1 is associated
with poor prognosis in epithelial ovarian cancer. J Surg Oncol.
107:847–852. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu J, Song H, Yao L, Liu Y, Zhang Y, Zhao
H, Ji H and Wang Y: Over-expression of the overexpressed in lung
cancer-1 is associated with poor prognosis in colorectal cancer.
Anticancer Res. 34:367–372. 2014.PubMed/NCBI
|
|
21
|
Ou-Yang QH, Duan ZX, Jin Z and Lei JX:
OLC1 is overexpressed in breast cancer and its expression
correlates with poor patient survival. Tumour Biol. 35:8823–8827.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chondrogianni N, Petropoulos I, Grimm S,
Georgila K, Catalgol B, Friguet B, Grune T and Gonos ES: Protein
damage, repair and proteolysis. Mol Aspects Med. 35:1–71. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jain CK, Arora S, Khanna A, Gupta M,
Wadhwa G and Sharma SK: The ubiquitin-proteasome pathway an
emerging anticancer strategy for therapeutics: A patent analysis.
Recent Pat Anticancer Drug Discov. 10:201–213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang WL, Wu CY, Wu J and Lin HK:
Regulation of Akt signaling activation by ubiquitination. Cell
Cycle. 9:487–497. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chao CC: Mechanisms of p53 degradation.
Clin Chim Acta. 438:139–147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lindon C: Control of mitotic exit and
cytokinesis by the APC/C. Biochem Soc Trans. 36:405–410. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chang L and Barford D: Insights into the
anaphase-promoting complex: A molecular machine that regulates
mitosis. Curr Opin Struct Biol. 29:1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barford D: Structure, function and
mechanism of the anaphase promoting complex (APC/C). Q Rev Biophys.
44:153–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Van Voorhis VA and Morgan DO: Activation
of the APC/C ubiquitin ligase by enhanced E2 efficiency. Curr Biol.
24:1556–1562. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cho HJ, Lee EH, Han SH, Jeong JH, Kwon J
and Kim H: Degradation of human RAP80 is cell cycle regulated by
Cdc20 and Cdh1 ubiquitin ligases. Mol Cancer Res. 10:615–625. 2012.
View Article : Google Scholar : PubMed/NCBI
|