Introduction
Liver cancer is one of the most common malignant
tumors with high morbidity and mortality rates, with 854,000 new
cases and 810,000 mortality cases per year (1). Liver cancer is the fifth most common
cancer and the second leading cause of cancer-associated mortality
worldwide in 2018 (2). Multiple
treatment options are available for patients with liver cancer,
including surgical resection, transarterial chemoembolization,
radiotherapy and sorafenib (3).
However, the prognosis of patients with liver cancer after surgery
remains poor due to the postoperative recurrence and early blood
vessel invasion (4,5). Liver cancer has been demonstrated to be
associated with mutations of numerous genes, including catenin beta
1, tumor protein p53 and axin 1 (6).
Novel molecular and cellular targets, including cancer stem wells,
were identified, allowing the development of a novel therapy for
patients with advanced liver cancer, resulting in favorable
curative effects and significantly prolonging the patients'
survival time (7). Since preliminary
progress has been made in the molecular-targeted therapy of liver
cancer, the present study will determine potential novel biomarker
for the early diagnosis of liver cancer and the prognosis of
patients.
Budding uninhibited by benzimidazoles 1 (BUB1) is a
mitotic checkpoint serine/threonine kinase that serves a central
role in aligning chromosomes and establishing the mitotic spindle
checkpoint (8). In addition, BUB1
also serves an important role in the accurate partitioning of
chromosomes during the cleavage of daughter cells from mother cells
(9,10). BUB1 contains three primary regions: A
conserved N-terminal region containing the kinetochore localization
domain; an intermediate, non-conserved region that acts as a
scaffold for the recruitment of proteins; and a C-terminal region
that contains a catalytic serine/threonine kinase domain (11). The function of BUB1 as oncogene or
tumor suppressor gene has been observed in various types of cancer,
including breast cancer, pancreatic ductal adenocarcinoma, prostate
and gastric cancer (12–15). Several studies have demonstrated the
unfavorable prognostic role of BUB1 in liver cancer based on
bioinformatics analysis (16–18).
However, the molecular biological function of BUB1 in liver cancer
still remains unclear.
In the present study, the importance of BUB1 in the
progression of liver cancer was investigated. Initially, reverse
transcription-quantitative (RT-q)PCR and immunohistochemistry were
used to determine the expression of BUB1 in liver cancer and
adjacent normal tissues. In addition, the significance of BUB1 in
tumor cell proliferation was demonstrated in vitro and the
molecular mechanism underlying BUB1 function in liver cancer growth
was evaluated.
Materials and methods
Liver cancer tissue samples
A total of 24 pairs of primary liver cancer tissues
and their corresponding adjacent normal tissues were obtained from
patients who underwent hepatectomy between February 2002 and July
2012 at the Lianshui County People's Hospital (Huaian, China). The
median age of patients was 54 years, and there were 18 men and 6
women. The inclusion criteria were as follows: i) Patients
clinically diagnosed with liver cancer following surgery; ii) R0
resection of all patients based on histologic examinations, and
iii) paired normal tissue was adjacent to tumor tissue with
distance <2 cm. The exclusion criteria were as follows: i)
Patients with distant metastasis and ii) patients who had received
radiotherapy or chemotherapy before surgery. The present study was
approved by the Institutional Review Board of The Institute for
Lianshui County People's Hospital. Written informed consent was
obtained from all patients prior to enrollment. The 24 paired
samples were subjected to RNA extraction for RT-qPCR.
Microarray data
The relative mRNA expression levels of BUB1 in liver
tumor tissues and their corresponding adjacent normal tissues was
obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Survival curves
of patients with liver cancer stratified according to the median
expression levels of BUB1 were also obtained from TGCA.
Immunohistochemistry
Clinical liver cancer tissues and paired
non-cancerous tissues were fixed in formalin at room temperature
for 24 h, embedded in paraffin and cut into 5-µm consecutive
sections. Following deparaffinization and antigen recovery in a
sodium citrate solution (pH 6.0) for 20 min at 98°C, the sections
were washed thrice with 0.01 mol/l PBS for 5 min each time, blocked
for 1 h in 0.01 mol/l PBS containing 0.3% Triton X-100 (Santa Cruz
Biotechnology, Inc.) and 5% BSA (Gibco; Thermo Fisher Scientific,
Inc.), and incubated with an anti-BUB1 antibody (cat. no. ab195268;
1:200; Abcam) overnight at 4°C. Following washing with 0.01 mol/l
PBS, the sections were incubated with 0.01 mol/l PBS containing a
horseradish peroxidase-conjugated anti-rabbit immunoglobulin G
antibody (cat. no. ab6759; 1:500; Abcam) for 2 h at room
temperature, followed by development with 0.003%
H2O2 and 0.03% 3,3′-diaminobenzidine in 0.05
mol/l Tris-HCl (pH 7.6). The categories and percentages of
immunohistochemical stained cells were assessed in five independent
high-power microscopic fields for each tissue sample using a light
microscope (magnification, ×400).
RNA extraction and RT-qPCR
The specimens were snap-frozen in liquid nitrogen
and stored at −80°C use. The total RNA of tumor tissues and
adjacent noncancerous tissues from the 24 patients was isolated
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and reverse-transcribed to cDNA using the
PrimeScript RT reagent kit (Takara Bio, Inc.) according to the
manufacturer's instructions. SYBR® Premix Ex Taq (Takara
Bio, Inc.) was used for qPCR. The thermocycling conditions used for
the PCR were as follows: 95°C for 1 min; 40 cycles of 95°C for 12
sec and 58.5°C for 40 sec. The primers were as follows: BUB1
forward, 5′-TGGGAAAGATACATAAGTGGGT-3′ and reverse,
5′-AGGGGATGACAGGGTTCCAAT-3′; GAPDH forward,
5′-ATGACCCCTTCATTGACCTCA-3′ and reverse,
5′-GAGATGATCACCCTTTTGGCT-3′. GAPDH was used as the internal
control. The relative mRNA expression level of BUB1 in each sample
was calculated using the comparative expression level
2−∆∆Cq method (19).
Cell culture
Liver cancer cell lines YY-8103, MHCC97-L, HepG2,
and Huh7 were purchased from The Cell Bank of The Type Culture
Collection of Chinese Academy of Sciences, and all cell lines were
authenticated by STR profiling. Cells were maintained in a
humidified atmosphere containing 5% CO2 at 37°C in DMEM
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 100
U/ml penicillin, 100 mg/ml streptomycin and 10% FBS (both
Invitrogen; Thermo Fisher Scientific, Inc.).
Cell transfection
The full-length cDNA encoding human BUB1 was
obtained from human whole blood by RT-PCR. The human BUB1 gene
primer pair was designed using Primer version 5 (PREMIER Biosoft).
BUB1 cDNA was cloned into a p23-3×flag-GFP vector (Takara Bio,
Inc.) according to the manufacturer's instructions. Lentiviral
supernatants were produced using the Lenti-X HTX packaging system
(Clontech Laboratories, Inc.) and used for transduction of YY-8103
and MHCC97-L cell lines. For negative controls, cell lines were
transduced with supernatants from empty vector cells. The
fluorescence and infection efficiency were determined using an
inverted fluorescence microscope by GFP sorting (magnification,
×200; IX-71; Olympus Corporation). Over-expressed BUB1 with a Flag
tag was detected in cell lines with an anti-Flag (cat. no. 8164;
1:1,000; Cell Signaling Technology, Inc.).
shRNA plasmids for BUB1, which were designed against
the BUB1 gene and constructed in Phblv-u6-puro vectors, were
purchased from Shanghai GenePharma Co., Ltd. A non-target scrambled
oligonucleotide served as the negative control (shcontrol; Shanghai
GenePharma Co., Ltd.). All plasmids were verified by sequencing. To
generate stable BUB1-silenced cell lines, HepG2 and Huh7 cells were
cultured in 6-well plates until they reached 40% confluence. The
medium was then replaced with 1 ml fresh FBS-free culture medium
supplemented with 40 µl viral supernatant (1×108 UT/ml)
and 6 µg/ml polybrene (Han Heng Biotechnology Co., Ltd.) for 24 h.
Cells were cultured and screened in medium containing 2.5 µg/ml
puromycin (Han Heng Biotechnology Co., Ltd.). Individual
puromycin-resistant colonies were isolated during drug screening.
The knockdown efficiency was verified by western blotting. The
shRNA sequences used in the present study were as follows: shBUB1,
forward
5′-CCGGGAATTTCAATTGGGTTCTAAGCTCGAGCTTAGAACCCAATTGAAATTCTTTTTG-3′,
reserve
5′-AATTCAAAAAGAATTTCAATTGGGTTCTAAGCTCGAGCTTAGAACCCAATTGAAATTC-3′;
and shcontrol, forward
5′-CCGGCAAACTTTGTATGCCCGCTTTCTCGAGAAAGCGGGCATACAAAGTTTGTTTTTG-3′
and reserve
5′-AATTCAAAAACAAACTTTGTATGCCCGCTTTCTCGAGAAAGCGGGCATACAAAGTTTG-3′.
Western blot analysis
Cells were lysed in RIPA Lysis Buffer and
phenylmethylsulfonyl fluoride (PMSF) (Thermo Fisher Scientific,
Inc.) and the lysates were centrifuged at 10,000 × g for 15 min at
4°C according to the manufacturer's protocols. Protein
concentration was determined using Bradford reagent (Sigma-Aldrich;
Merck KGaA). Proteins (15 µg) were separated by 10% SDS-PAGE and
transferred onto nitrocellulose membranes (EMD Millipore).
Membranes were blocked with 5% fat-free milk for 1 h at room
temperature and incubated with primary antibodies against BUB1
antibody (cat. no. ab195268; 1:1,000; Abcam), anti-SMAD2 (cat. no.
5339; 1:1,000; Cell Signaling Technology, Inc.), phospho- (p) SMAD2
(cat. no. 3104; 1:1,000; Cell Signaling Technology, Inc.), PCNA
(cat. no. 13110; 1:1,000 dilution; Cell Signaling Technology,
Inc.), Ki67 (cat. no. 2586; 1:1,000; Cell Signaling Technology,
Inc.), Flag (cat. no. 8164; 1:1,000; Cell Signaling Technology,
Inc.) and GAPDH (cat. no. 5174; 1:1,000; Cell Signaling Technology,
Inc.) at 4°C overnight. Membranes were then incubated with
anti-rabbit horseradish peroxidase-conjugated secondary antibody
(cat. no. 7074; 1:5,000; Cell Signaling Technology, Inc.) for 2 h
at room temperature. The immunoreactive protein bands were
visualized using an enhanced chemiluminescence kit (Pierce; Thermo
Fisher Scientific, Inc.) and a Gel Dox XR system (Bio-Rad
Laboratories, Inc.).
Crystal violet assay
A total of 1×103 cells/well were seeded into 6-well
plates and the cells were cultured in medium with 10% FBS. The
medium was changed every three days. After two weeks, the medium
was removed and cells were fixed with 20% methanol at room
temperature for 10 min and stained with 0.5% crystal violet
(Sigma-Aldrich; MerckKGaA). Subsequently, cells were washed with
PBS and images were captured using digital camera. Then, 1 ml
glacial acetic acid was added to the cells, and the optical density
(OD) was detected at 600 nm using a microplate reader.
MTT assay
A total of 1×103 cells/well were seeded into 96-well
plates, and cell viability was detected using MTT. After 1, 2, 3,
4, 5, 6, or 7 days of incubation, 20 µl 5 mg/ml MTT was added to
each well and incubated at 37°C for a further 4 h. Subsequently,
the medium was aspirated and the wells washed with PBS and drained
for ~2 h. Any remaining solution was carefully aspirated and 200 µl
DMSO was added to dissolve the formazan crystal with gentle
agitation. The optical density was measured at 490 nm using a
microplate reader.
Statistical analysis
Statistical evaluations were performed using
GraphPad Prism 5 (GraphPad Software Inc.), and the data are
presented as mean ± standard deviation unless otherwise stated.
Cell proliferation rates were compared using an unpaired Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
BUB1 expression is upregulated in
liver cancer tissues
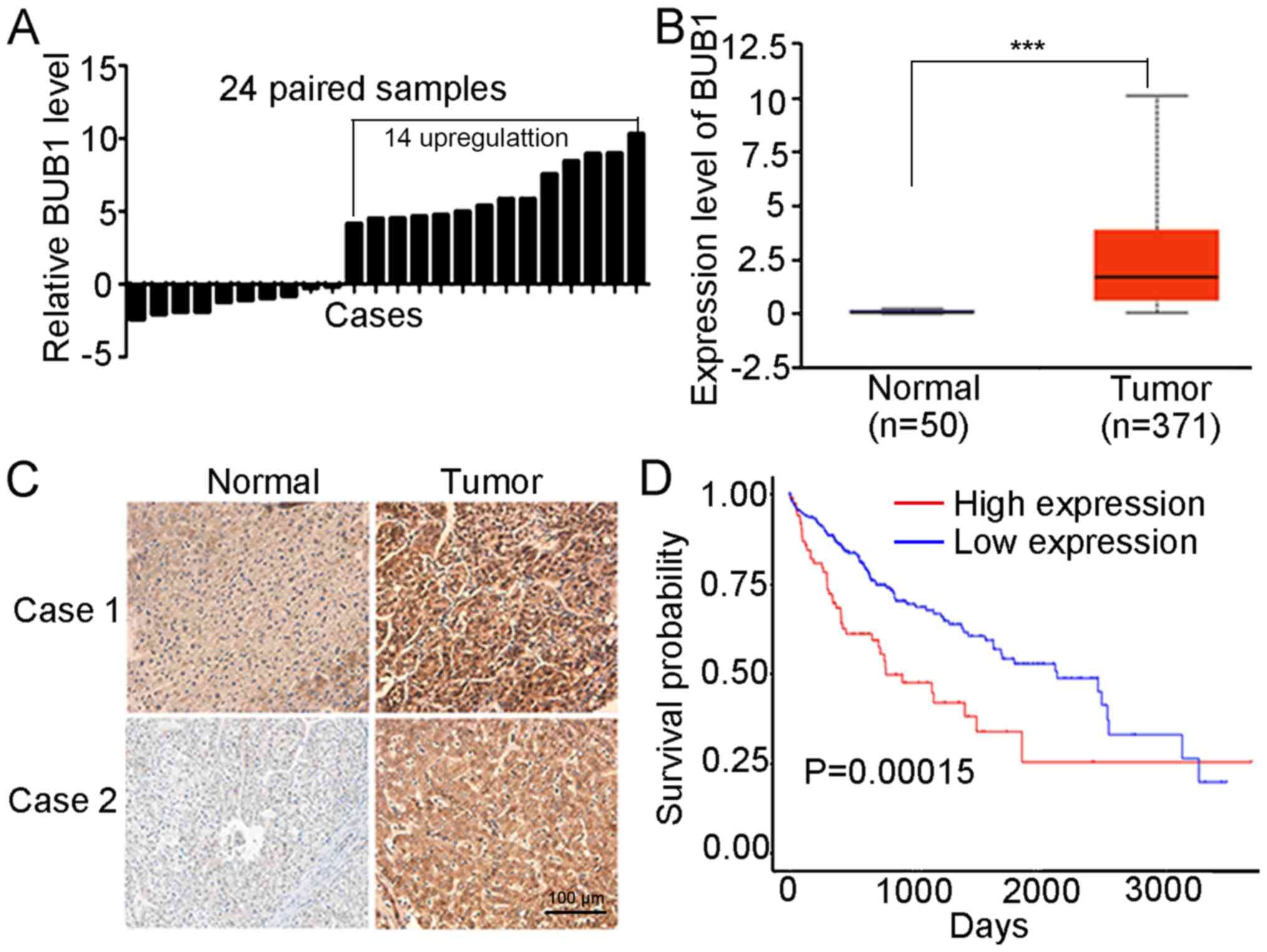
The mRNA expression levels of BUB1 in 24 pairs of
liver cancer tissues and the corresponding normal tissue were
examined. The results of the RT-qPCR demonstrated that mRNA
expression of BUB1 was significantly higher in 14 of the tumor
samples compared with the matched normal tissues (Fig. 1A). Based on the obtained data from
TCGA, BUB1 expression was significantly higher in 371 tumor tissues
compared with 50 normal tissues (P<0.001; Fig. 1B). Immunohistochemistry analysis
revealed that BUB1 was primarily expressed in the cytoplasm, and
the tumor tissues exhibited increased staining intensity compared
with the paired normal tissues in five patients (Figs. 1C and S1), consistent with the results of RT-q
PCR. In addition, the overall survival rates were significantly
higher in the BUB1-high group compared with the BUB1-low group (P
<0.001; Fig. 1D). These results
suggested that BUB1 was upregulated in liver cancer tissues.
Overexpression and knockdown of BUB1
in liver cancer cell lines
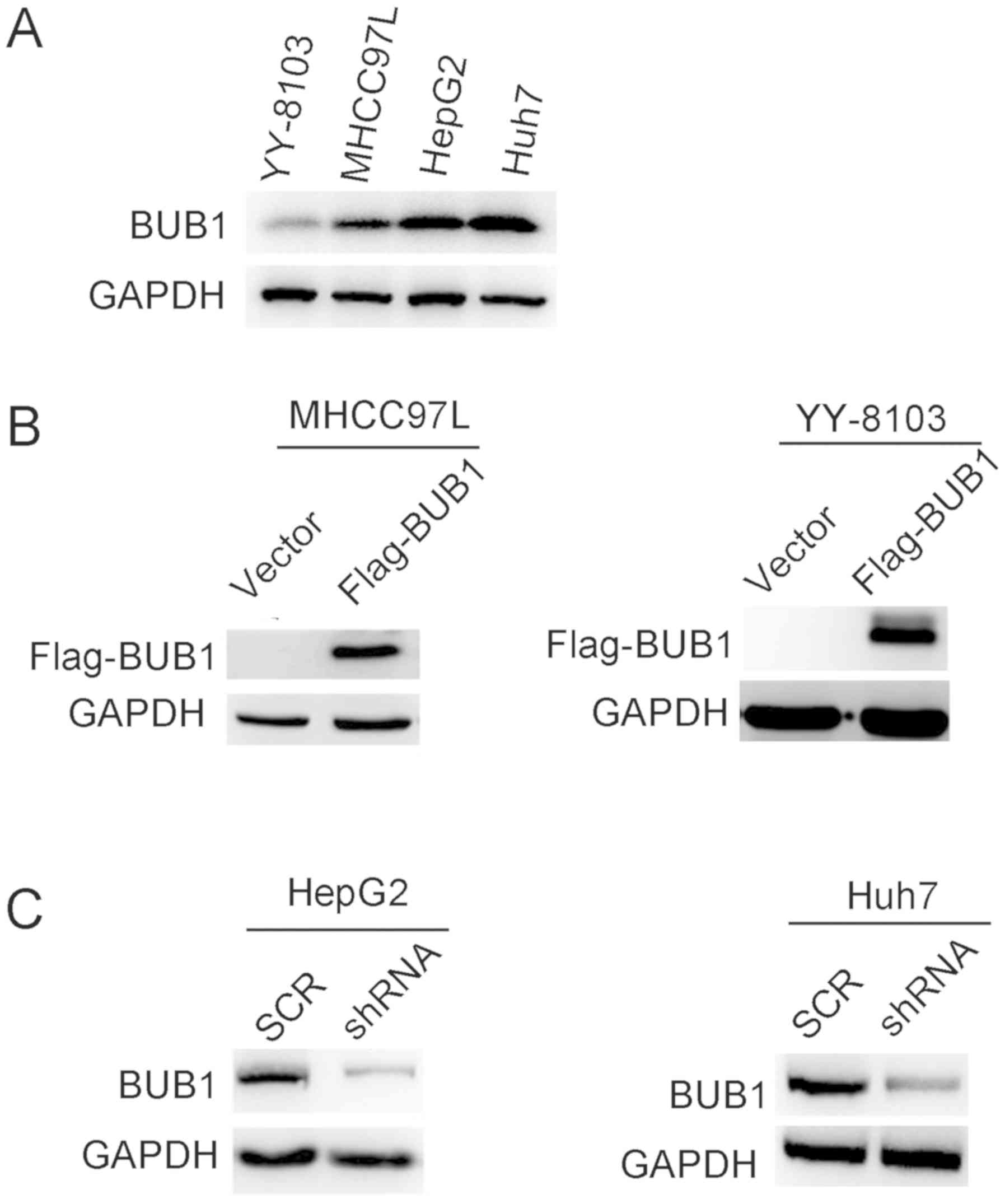
Based on the clinical data, it was hypothesized that
BUB1 may promote the proliferation of liver cancer cells. To
determine the effects of BUB1, the expression levels were
determined in several liver cancer cell lines. The results
demonstrated that BUB1 protein expression levels were higher in
HepG2 and Huh7 cell lines compared with YY-8103 and MHCC97-L cells
(Fig. 2A). To determine the effects
of BUB1 in liver cancer cells, the MHCC97-L and YY-8103 cells were
transfected with plasmids containing either an empty p23 vector or
BUB1 overexpression vectors (Flag-BUB1), whereas shRNA targeting
BUB1 was transfected into HepG2 and Huh7 cells. Western blotting
results revealed that the establishment of overexpression and
knockdown of BUB1 in liver cancer cell lines was successful
(Fig. 2B and C).
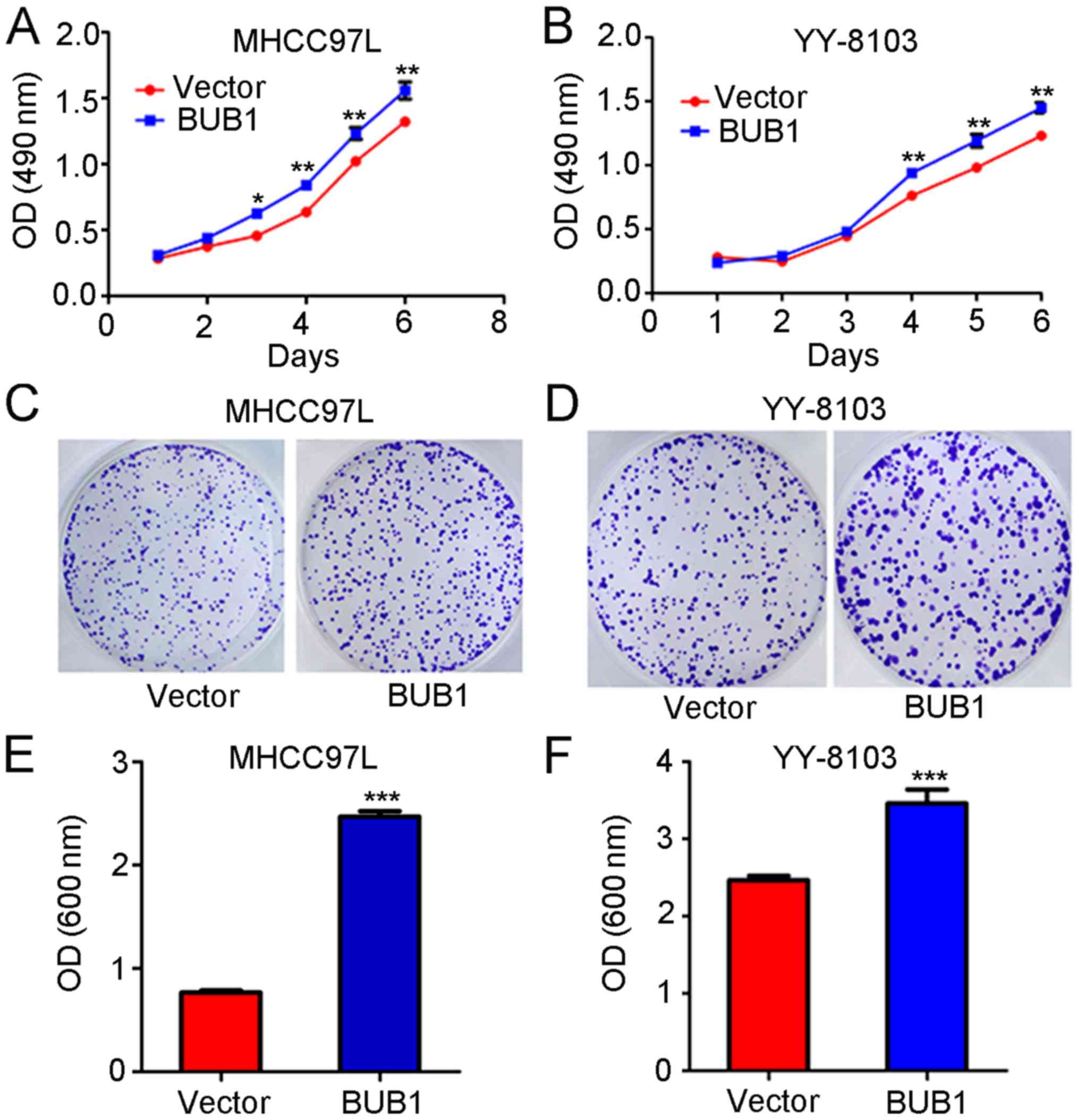
BUB1 overexpression promotes the
proliferation of the liver cancer cells
MTT assay revealed that the absorbance values of the
MHCC97-L cells 3, 4, 5 and 6 days after transfection with the BUB1
overexpression vector were significantly higher compared with
untreated cells (P<0.01; Fig.
3A). Similarly, the absorbance values of YY-8103 cells at 4, 5
and 6 days after transfection with the BUB1 overexpression vector
were significantly increased compared with the untreated cells
(P<0.01; Fig. 3B). The results of
the crystal violet assay demonstrated that the absorbance values of
MHCC97-L and YY-8103 cells following transfection with the BUB1
overexpression vector were significantly higher compared with
untreated cells (all P<0.01; Fig.
3C-F).
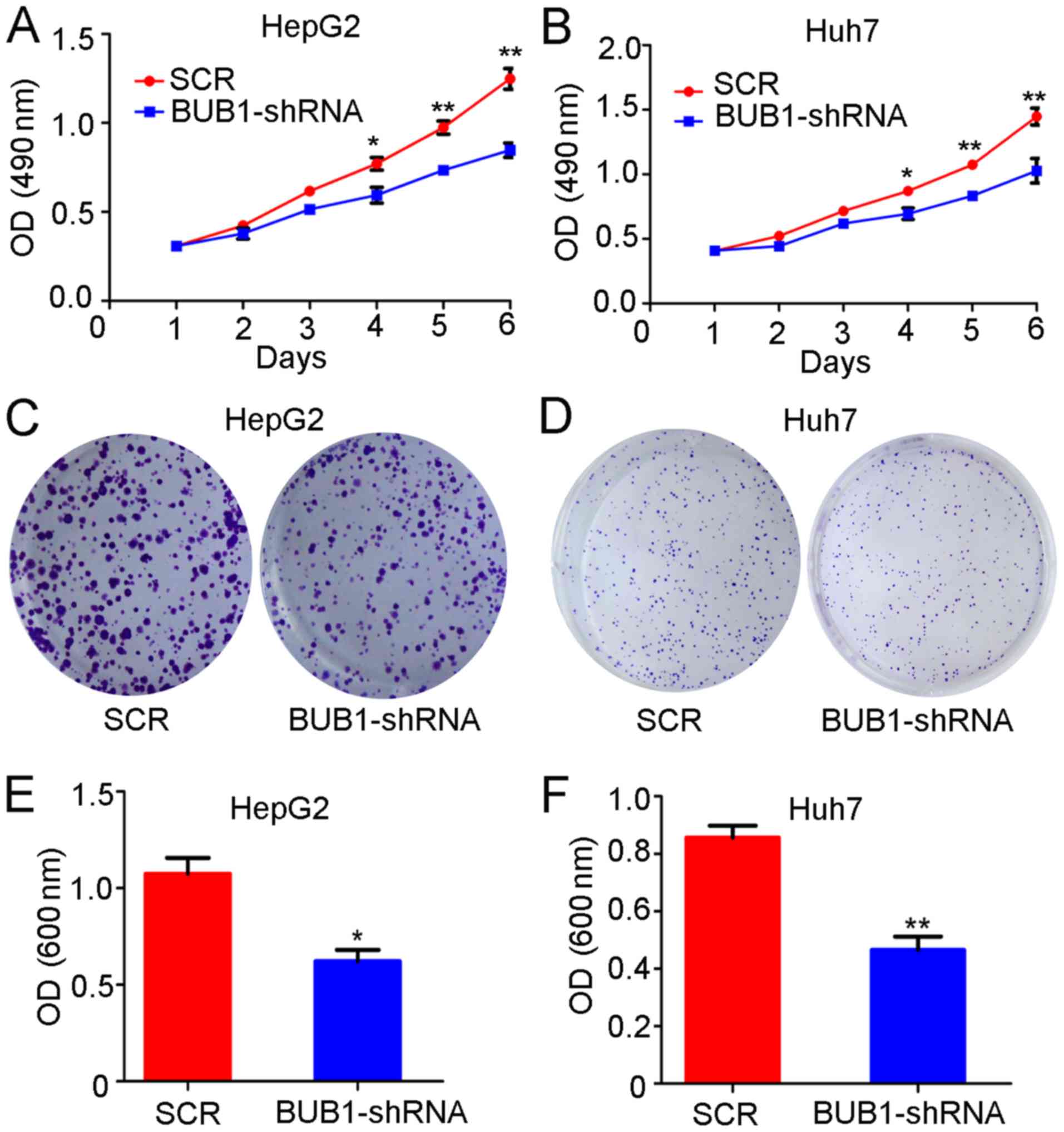
BUB1 knockdown inhibits the
proliferation of liver cancer cells
Similar to the overexpression experiments, the
proliferation of the control and BUB1-shRNA cell lines was tested.
The results of the MTT assay demonstrated that the absorbance
values of HepG2 cells 3, 4, 5 and 6 days after BUB1 knockdown were
significantly lower compared with those of untreated cells
(P<0.01; Fig. 4A). Similarly, the
absorbance values of Huh-7 cells 4, 5 and 6 days after BUB1
knockdown were also significantly lower compared with untreated
cells (P<0.01; Fig. 4B). In
addition, the crystal violet assay demonstrated that the absorbance
of HepG2 (P<0.05; Fig. 4C and E)
and Huh-7 cells (P<0.01; Fig. 4D and
F) after BUB1 knockdown was significantly lower compared with
untreated cells.
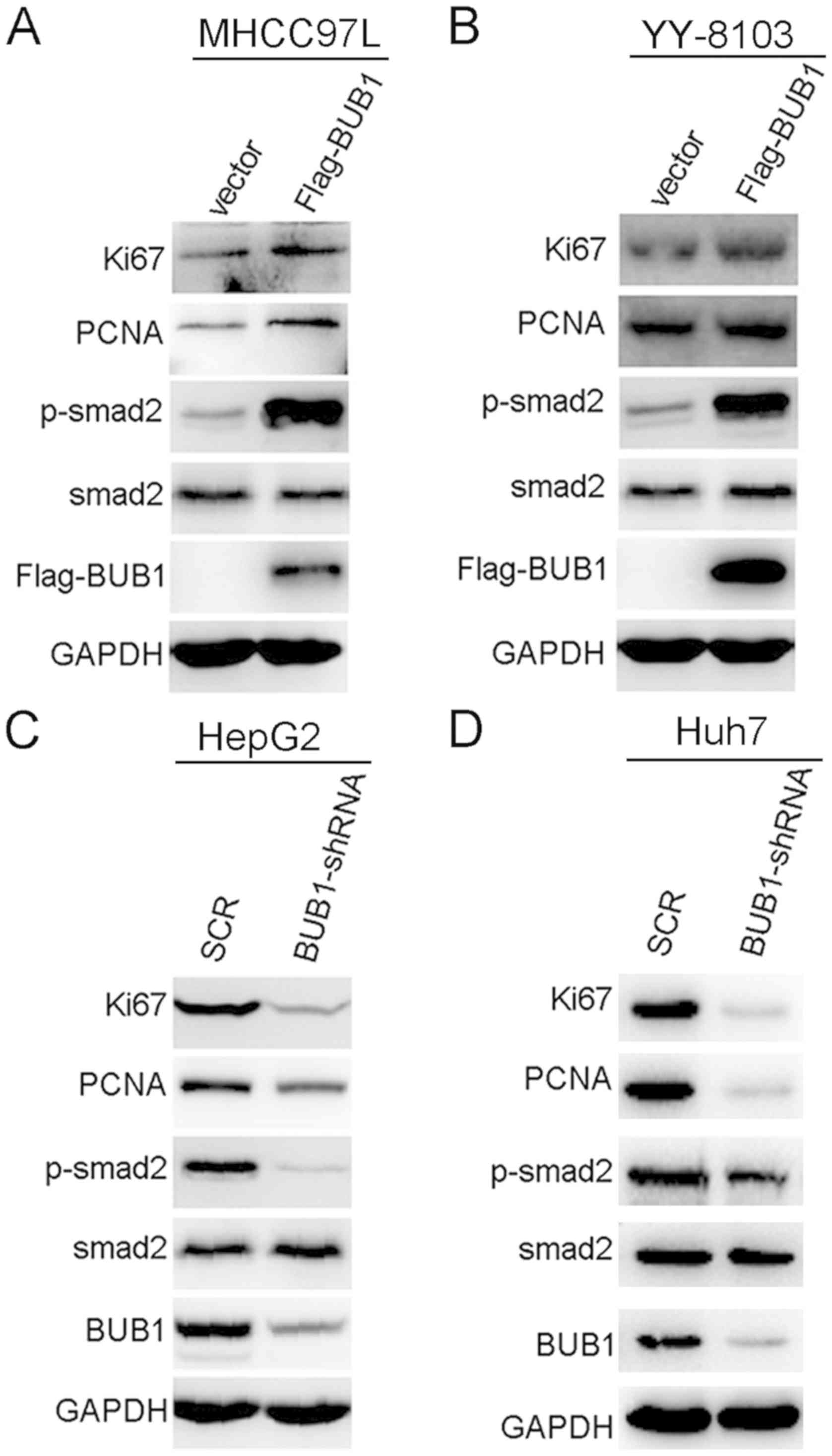
BUB1 activates the phosphorylation of
SMAD2 in liver cancer cells
To explore the molecular mechanism by which BUB1
affected liver cancer, p-SMAD2 and total SMAD2 protein expression
levels in BUB1-overexpressing and knockdown cell lines were
measured. Expression levels of p-SMAD2 were increased when BUB1 was
overexpressed (Fig. 5A and B). The
reverse was observed in HepG2 and Huh-7 cells; knockdown of BUB1
decreased the expression levels of p-SMAD2 (Fig. 5C and D). In addition, BUB1
overexpression notably increased the expression levels of cell
proliferation markers Ki67 and PCNA, whereas BUB1 knockdown
decreased the expression levels of Ki67 and PCNA (Fig. 5C and D). As demonstrated in Fig. S2, further knockdown of BUB1 in the
BUB1-overexpressing MHCC97-L and YY-8103 cells decreased the
expression levels of p-SMAD2, Ki67 and PCNA.
Discussion
Aberrant expression and mutations in BUB1 are
associated with aneuploidy and several types of cancer, including
breast cancer and pancreatic ductal adenocarcinoma (20). To date, several studies have
demonstrated that BUB1 is significantly upregulated in various
types of cancer, such as breast cancer, pancreatic ductal
adenocarcinoma, prostate and gastric cancer (12–14), and
is associated with unfavorable outcomes. However, BUB1 has been
reported to serve differing roles in different types of cancer. In
endometrial carcinoma (21),
low-grade breast cancer (22), and
gastric adenocarcinoma (15), high
expression levels of BUB1 were associated with a good prognosis,
whereas in invasive breast cancer (23) and ovarian cancer (24), high expression of BUB1 was associated
with an unfavorable prognosis.
The results of the present study demonstrated that
BUB1 mRNA and protein expression levels were significantly
increased in liver cancer tissues compared with normal tissues. In
addition, western blotting confirmed successful overexpression and
knockdown of BUB1 in liver cancer cell lines. It was observed that
cell proliferation was significantly increased following BUB1
overexpression, whereas knockdown of BUB1 inhibited liver cancer
cell proliferation. Expression levels of p-SMAD2 were significantly
increased when BUB1 was overexpressed, whereas knockdown of BUB1
decreased the expression levels of p-SMAD2. The present study
demonstrated that BUB1 may promote liver cancer cell proliferation
by activating the phosphorylation of SMAD2. It has been reported
that constitutive activation of the TGF-β/SMAD signaling pathway
serves a crucial role in the development and progression of liver
cancer (25). TGF-β exerts its
effect on gene expression via interaction with SMAD protein
transcription factors, including SMAD2 and SMAD3, followed by
R-SMAD and a common mediator SMAD heterodimer formation (26,27).
Overactivation of TGF-β signaling serves a complicated role in the
development and progression of a range of diseases, including
Parkinson's disease and cardiovascular diseases, which is
associated with increased growth and invasion at later stages of
tumor progression (28,29). To the best of our knowledge, the
present study is the first to demonstrate the involvement of BUB1
in the proliferation of liver cancer.
In summary, the present study demonstrated that BUB1
increased the proliferation of liver cancer cells. These results
provided an improved understanding of the mechanisms for the role
of BUB1 in tumor development and may highlight BUB1 as a potential
target in the diagnosis and/or treatment of liver cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Huaian
Science and Technology Bureau (grant no. HAB201847).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
LZ and YP performed the experiments. PH designed the
study. XC analyzed the data. LZ, YP and PH wrote the manuscript.
All authors read and approved the final manuscript
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of The Institute for Lianshui County People's Hospital
(Huaian, China). Written informed consent was obtained from all
patients prior to enrollment in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Galle PR, Forner A, Llovet JM, Mazzaferro
V, Piscaglia F, Raoul JL, Schirmacher P and Vilgrain V; European
Association for the Study of the Liver, : EASL Clinical Practice
Guidelines: Management of hepatocellular carcinoma. J Hepatol.
69:182–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lu XJ, Shi Y, Chen JL and Ma S:
Krüppel-like factors in hepatocellular carcinoma. Tumour Biol.
36:533–541. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu WB, Rao A, Vu V, Xu L, Rao JY and Wu
JX: Management of centrally located hepatocellular carcinoma:
Update 2016. World J Hepatol. 9:627–634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Portolani N, Coniglio A, Ghidoni S,
Giovanelli M, Benetti A, Tiberio GA and Giulini SM: Early and late
recurrence after liver resection for hepatocellular carcinoma:
Prognostic and therapeutic implications. Ann Surg. 243:229–235.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu J and Gao DZ: Distinction immune genes
of hepatitis-induced heptatocellular carcinoma. Bioinformatics.
28:3191–3194. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marquardt JU, Galle PR and Teufel A:
Molecular diagnosis and therapy of hepatocellular carcinoma (HCC):
An emerging field for advanced technologies. J Hepatol. 56:267–275.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bolanos-Garcia VM and Blundell TL: BUB1
and BUBR1: Multifaceted kinases of the cell cycle. Trends Biochem
Sci. 36:141–150. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Johnson VL, Scott MI, Holt SV, Hussein D
and Taylor SS: Bub1 is required for kinetochore localization of
BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell
Sci. 117:1577–1589. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu H and Tang Z: Bub1 multitasking in
mitosis. Cell Cycle. 4:262–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ricke RM, Jeganathan KB and van Deursen
JM: Bub1 overexpression induces aneuploidy and tumor formation
through Aurora B kinase hyperactivation. J Cell Biol.
193:1049–1064. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu X, Chen G, Cai ZD, Wang C, Liu ZZ, Lin
ZY, Wu YD, Liang YX, Han ZD, Liu JC, et al: Overexpression of BUB1B
contributes to progression of prostate cancer and predicts poor
outcome in patients with prostate cancer. Onco Targets Ther.
9:2211–2220. 2016.PubMed/NCBI
|
|
13
|
Piao J, Zhu L, Sun J, Li N, Dong B, Yang Y
and Chen L: High expression of CDK1 and BUB1 predicts poor
prognosis of pancreatic ductal adenocarcinoma. Gene. 701:15–22.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Han JY, Han YK, Park GY, Kim SD and Lee
CG: Bub1 is required for maintaining cancer stem cells in breast
cancer cell lines. Sci Rep. 5:159932015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stahl D, Braun M, Gentles AJ, Lingohr P,
Walter A, Kristiansen G and Gütgemann I: Low BUB1 expression is an
adverse prognostic marker in gastric adenocarcinoma. Oncotarget.
8:76329–76339. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen QF, Xia JG, Li W, Shen LJ, Huang T
and Wu P: Examining the key genes and pathways in hepatocellular
carcinoma development from hepatitis B virus-positive cirrhosis.
Mol Med Rep. 18:4940–4950. 2018.PubMed/NCBI
|
|
17
|
Zhang L, Huang Y, Ling J, Zhuo W, Yu Z,
Shao M, Luo Y and Zhu Y: Screening and function analysis of hub
genes and pathways in hepatocellular carcinoma via bioinformatics
approaches. Cancer Biomark. 22:511–521. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wen DY, Lin P, Pang YY, Chen G, He Y, Dang
YW and Yang H: Expression of the long intergenic non-protein coding
RNA 665 (LINC00665) gene and the cell cycle in hepatocellular
carcinoma using the Cancer Genome Atlas, the Gene Expression
Omnibus, and Quantitative Real-Time Polymerase Chain Reaction. Med
Sci Monit. 24:2786–2808. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Liu DP, Chen PP, Koeffler HP, Tong
XJ and Xie D: Involvement of IFN regulatory factor (IRF)-1 and
IRF-2 in the formation and progression of human esophageal cancers.
Cancer Res. 67:2535–2543. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Williams GL, Roberts TM and Gjoerup OV:
Bub1: Escapades in a cellular world. Cell Cycle. 6:1699–1704. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li L, Xu DB, Zhao XL and Hao TY:
Combination analysis of Bub1 and Mad2 expression in endometrial
cancer: Act as a prognostic factor in endometrial cancer. Arch
Gynecol Obstet. 288:155–165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mukherjee A, Joseph C, Craze M,
Chrysanthou E and Ellis IO: The role of BUB and CDC proteins in
low-grade breast cancers. Lancet. 385 (Suppl 1):S722015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takagi K, Miki Y, Shibahara Y, Nakamura Y,
Ebata A, Watanabe M, Ishida T, Sasano H and Suzuki T: BUB1
immunolocalization in breast carcinoma: Its nuclear localization as
a potent prognostic factor of the patients. Horm Cancer. 4:92–102.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ocaña A, Pérez-Peña J, Alcaraz-Sanabria A,
Sánchez-Corrales V, Nieto-Jiménez C, Templeton AJ, Seruga B,
Pandiella A and Amir E: In silico analyses identify gene-sets,
associated with clinical outcome in ovarian cancer: Role of mitotic
kinases. Oncotarget. 7:22865–22872. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dituri F, Mancarella S, Cigliano A, Chieti
A and Giannelli G: TGF-β as multifaceted orchestrator in HCC
progression: signaling, EMT, immune microenvironment, and novel
therapeutic perspectives. Semin Liver Dis. 39:53–69. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-beta family signalling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wrana JL, Attisano L, Wieser R, Ventura F
and Massagué J: Mechanism of activation of the TGF-beta receptor.
Nature. 370:341–347. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi Y and Massagué J: Mechanisms of
TGF-beta signaling from cell membrane to the nucleus. Cell.
113:685–700. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Massagué J: TGFbeta in Cancer. Cell.
134:215–230. 2008. View Article : Google Scholar : PubMed/NCBI
|