Introduction
Colorectal cancer (CRC) is one of the most common
malignancies globally at present. It was estimated that >1.8
million new CRC cases and 881,000 CRC-associated mortality cases
occurred in 2018, accounting for ~1/10 cancer cases. CRC ranks
therefore third in incidence and second in mortality (1). Patients with CRC commonly present with
a survival rate <5 years due to the early development of
metastasis (2). Although numerous
treatments, including surgery, radiotherapy, chemotherapy and
targeted therapy, can be used in CRC, the recurrence remains high
(54.5%) (3), the mortality of CRC
accounts for 9.5% of various cancers (1) and the prognosis of patients is poor
(4). At present, certain biomarkers
have been associated with CRC occurrence and progression and
prognosis of patients with CRC, for example carcinoembryonic
antigen, CA19-9 and CA72-4 (5), were
used to detect whether the patient had recurrence and progression
(6–8). In addition, microsatellite instability
and mutations in the p53 or KRAS genes have been used
as prognostic factors (9,10). However, the reliability of these
aforementioned biomarkers remains controversial. It is therefore
crucial vital to identify novel diagnostic and prognostic
biomarkers and therapeutic targets for CRC.
ATP binding cassette subfamily D member 3 (ABCD3),
also known as ZWS2, ABC43, CBAS5, PMP70 and PXMP1, is a member of
the superfamily of ATP-binding cassette (ABC) transporters
associated with the peroxisomal import of fatty acids and/or fatty
acyl-CoAs in the organelle (11).
ABC transporters serve crucial roles in the establishment of
chemoresistance by regulating the flow of anticancer agents into
the cancer cells (12). Inhibition
of fatty acid oxidation has been reported to induce apoptosis in
colorectal cancer cells (13).
Furthermore, the use of gene co-expression network analysis in CRC
demonstrated that ABCD3 is involved in the regulation of ABC
transporters, transmembrane transport, fatty acid β-oxidation and
ATP synthesis following nutrient catabolism (14). Seborova et al (15) demonstrated that ABCD3 downregulation
is associated with a better sensitivity to chemotherapy and time to
progression in patients with ovarian cancer. In addition, Reams
et al (16) reported that
high expression of ABCD3 mRNA is associated with the Gleason Score
in Caucasian American men with prostate tumors. Elsnerova et
al (17) demonstrated that ABCD3
mRNA expression was higher in high-grade serous ovarian carcinoma
compared with other subtypes of epithelial ovarian cancer, and may
therefore be considered as a progression biomarker for ovarian
carcinoma. Although ABCD3 was demonstrated to be less expressed in
colorectal cancer tissues compared with normal tissues (18), the diagnostic and prognostic
abilities of ABCD3 mRNA expression in CRC have rarely been
reported.
The current study demonstrated that decreased ABCD3
mRNA expression was associated with poor survival in patients with
CRC, and that ABCD3 had a good diagnostic value with high
sensitivity and specificity in patients with CRC, according to
analysis of datasets from The Cancer Genome Atlas (TCGA) and Gene
Expression Omnibus (GEO). To identify the biological pathways in
which ABCD3 may be involved in patients with CRC, Gene Set
Enrichment Analysis (GSEA) was used. The results demonstrated that
the ABCD3 high-expression phenotype was differentially enriched in
five biological pathways in CRC, including apoptosis, cell cycle,
renal cell carcinoma, thyroid cancer and CRC.
Materials and methods
Data collection and processing
The Level 3 HTSeq-FPKM files, comprising 612
Transcriptome Profiling RNA-Seqs of 544 cases, were collected from
a TCGA dataset (portal.gdc.cancer.gov/) that included information on
452 and 96 patients with colon and rectal cancer, respectively, on
March 2019. Clinicopathological data was available for 548 patients
but only 544 of these patients also had RNA-Seq data. The 612
transcripts included 568 tumor samples (some patients provided
multiple tumor samples) and 44 normal samples. Patients had not
received neoadjuvant treatment before tumor excision. The survival
time (follow-up ≥30 days) and survival status information were
available for 506 patients with cancer. The clinicopathological
characteristics, including sex, age, clinical stage, T stage, M
stage and lymph node status, were available for 448 samples
obtained from 548 cases and the 100 samples with missing clinical
information were removed. The ABCD3 mRNA expression was collected
for colorectal adenocarcinoma tissues and adjacent normal tissues.
Since the association between ABCD3 mRNA expression and the
clinicopathological characteristics of patients was independent of
the follow-up days, the RNA transcript data for 448 cancer samples
were used for further association analysis (100/548 were excluded
from this analysis due to incomplete clinical data). Meanwhile,
only 506 patients had a survival time ≥30 days and were used for
survival analysis. Furthermore, comparison of ABCD3 expression
between tumor and normal samples from the GEO database was
performed. The four gene expression datasets (series_matrix)
GSE21510 (19), GSE25071 (20), GSE41258 (21) and GSE39582 (22) associated with CRC were downloaded
from GEO and included 19, 46, 186 and 566 tumor samples,
respectively and 25, 4, 54 and 19 normal samples, respectively. In
addition, GSE39582 dataset included the complete clinical
information, including survival time, survival status, sex, age,
clinical stage, T stage, M stage, and lymph node status; however,
the three other datasets didn't have complete clinical information.
Furthermore, the protein expression of ABCD3 was determined by
using the Human Protein Atlas (http://www.proteinatlas.org/).
GSEA of ABCD3 in CRC
GSEA is a computing tool used to identify classes of
genes or proteins that are over-represented in a large set of genes
or proteins, and which may be associated with certain disease
phenotypes (gsea-msigdb.org/gsea/) (23). In the present study, all genes
associated with ABCD3 expression were sequenced by this method in
TCGA dataset using gsea-3.0.jar. Each analysis ran 1,000 genome
sequences. ABCD3 mRNA was treated as a phenotypic marker and
samples were divided into high and low expression groups based on
the median expression value. The signaling pathways enriched in
each phenotype were based on nominal (NOM) P-value <0.05 and
false discovery rate (FDR)<0.25.
Statistical analysis
All statistical analyses were performed using R
language (version 3.5.1) (mirrors.tuna.tsinghua.edu.cn/CRAN/). The comparison
between ABCD3 mRNA expression in paired CRC and normal tissues from
the TCGA and GEO databases was performed using Wilcoxon rank sum
tests, some patients in the TCGA database has both tumor samples
and normal samples, which was analyzed by Wilcoxon signed-rank
test. The diagnostic value of ABCD3 mRNA expression was evaluated
by the receiver operating characteristic (ROC) curve using pROC
package (24). The association
between clinicopathological characteristics and ABCD3 mRNA
expression levels was determined using Kruskal-Wallis test,
Bonferroni's test (when >2 groups were compared) or Wilcoxon
rank sum test (when 2 non-parametric groups were compared),
logistic regression and χ2 test. The association between
clinicopathological characteristics, including sex, age, clinical
stage, T stage, M stage and lymph node status and ABCD3 mRNA
expression and patients' overall survival (OS), was evaluated using
univariate Cox regression analysis and Kaplan-Meier method with the
Survival package in R and P-values were calculated using log-rank
test. Multivariate Cox regression analysis was used to identify
whether ABCD3 mRNA expression could be considered as an independent
prognostic factor in CRC. All P<0.05 was considered to indicate
a statistically significant difference.
Results
Clinicopathological characteristics of
patients
The clinical data of the 548 patients collected from
the TCGA database, including sex, age, ethnicity, clinical stage, T
stage, distant metastasis, and lymph node status and cancer type
are presented in Table I.
 | Table I.Clinicopathological characteristics
of patients with colorectal cancer from The Cancer Genome
Atlas. |
Table I.
Clinicopathological characteristics
of patients with colorectal cancer from The Cancer Genome
Atlas.
| Variables | Number | Percentage |
|---|
| Sex |
|
|
|
Male | 292 | 53.28 |
|
Female | 256 | 46.72 |
| Age, years |
|
|
|
Range | 31-90 |
|
|
Median | 67.5 |
|
| Ethnicity |
|
|
|
American Indian | 1 | 0.18 |
|
Asian | 12 | 2.19 |
|
Black | 59 | 10.77 |
|
White | 252 | 45.99 |
|
Unidentified | 224 | 40.88 |
| Stage |
|
|
| I | 96 | 17.52 |
| II | 210 | 38.32 |
|
III | 149 | 27.19 |
| IV | 78 | 14.23 |
|
Unidentified | 15 | 2.74 |
| T stage |
|
|
|
T1+Tis | 16 | 2.92 |
| T2 | 96 | 17.52 |
| T3 | 373 | 68.07 |
| T4 | 63 | 11.50 |
| Lymph node
status |
|
|
| N0 | 323 | 58.94 |
| N1 | 130 | 23.72 |
| N2 | 94 | 17.15 |
| Nx | 1 | 0.18 |
| Metastatic |
|
|
| M0 | 408 | 74.45 |
| M1 | 77 | 14.05 |
| Mx | 55 | 10.04 |
|
Unidentified | 8 | 1.46 |
| Cancer type |
|
|
| Colon
adenocarcinoma | 452 | 82.48 |
| Rectal
adenocarcinoma | 96 | 17.52 |
ABCD3 mRNA expression is decreased in
CRC tissues
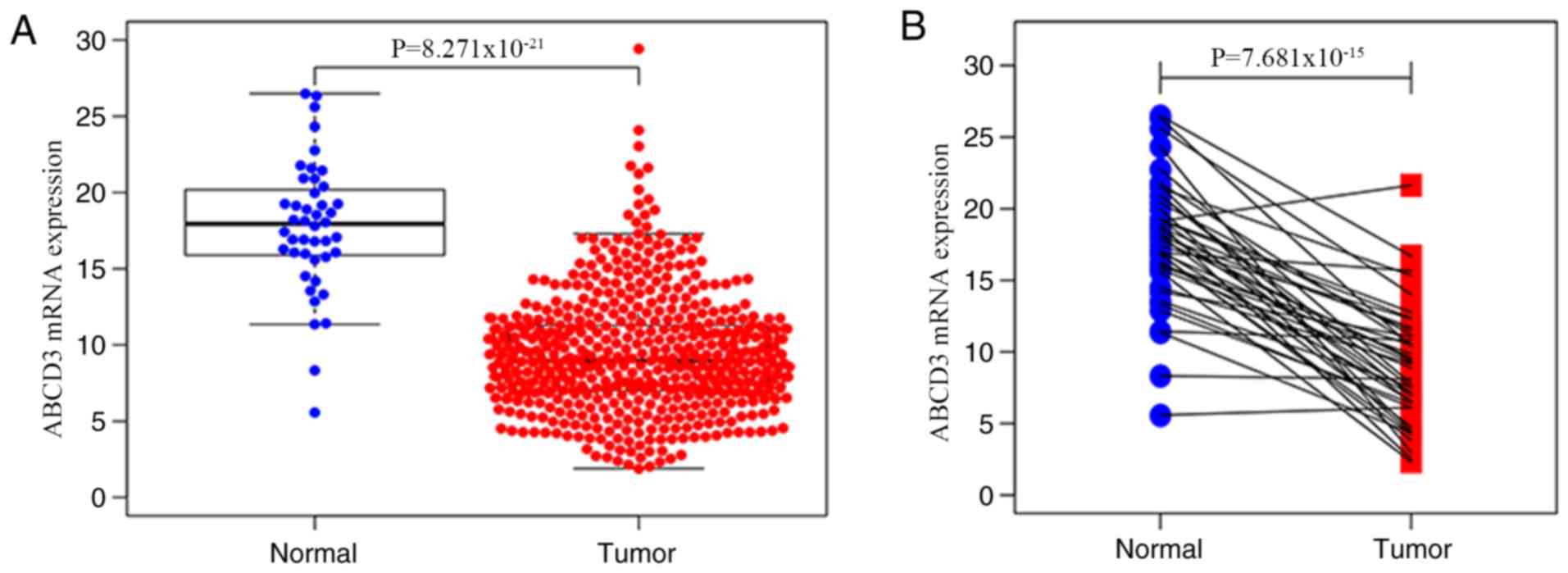
The results demonstrated that ABCD3 mRNA expression
was significantly decreased in CRC tissues compared with normal
tissues (P<0.001; Fig. 1A).
Furthermore, analysis of ABCD3 mRNA expression in 44 matched tumor
tissues and normal tissues demonstrated that ABCD3 mRNA expression
was significantly decreased in tumor tissues compared with normal
tissues (P<0.001; Fig. 1B). In
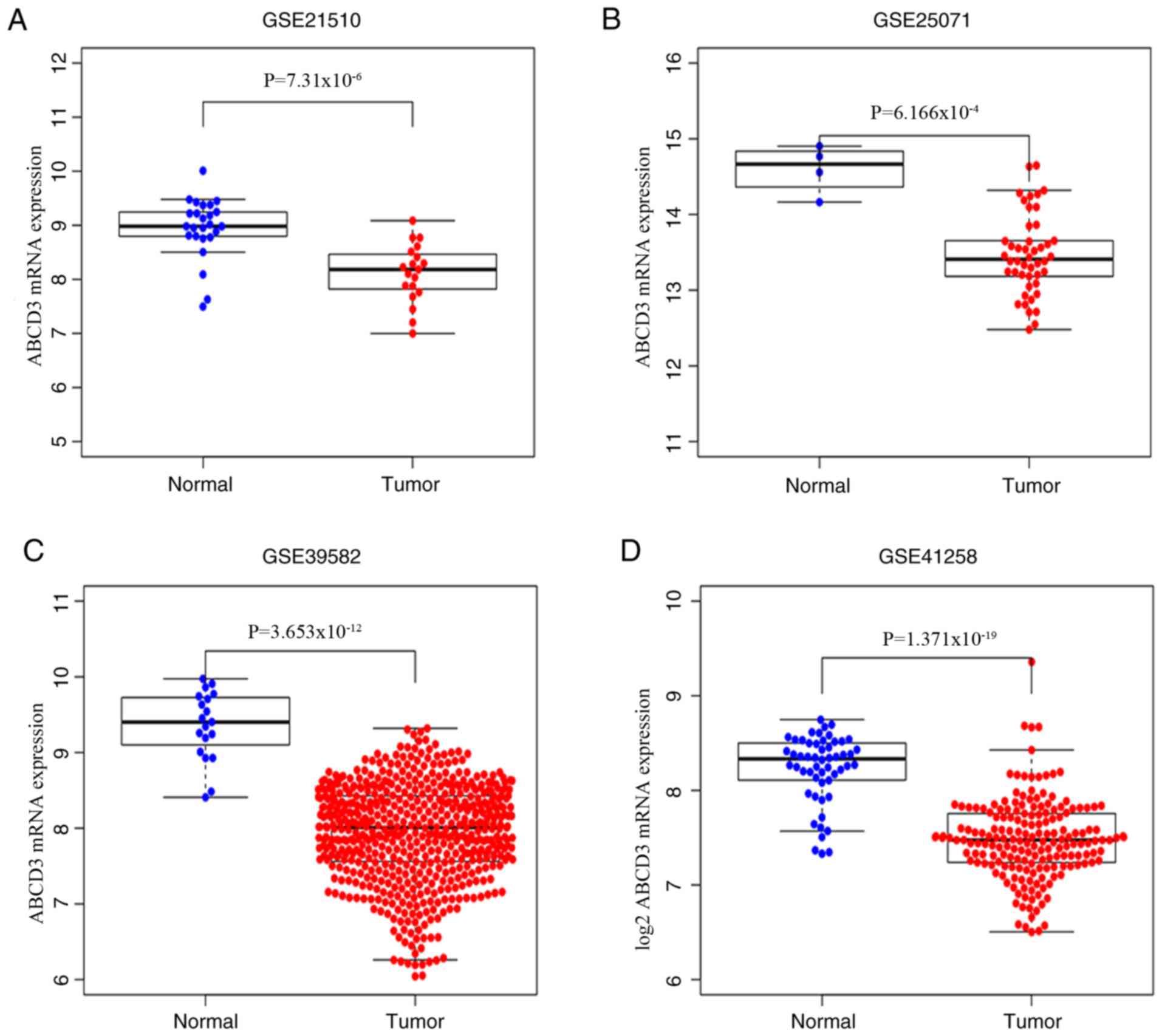
datasets GSE21510, GSE25071, GSE41258 and GSE39582 from the GEO
database; same analyses were performed, and the results
demonstrated that ABCD3 mRNA expression was decreased in the tumor
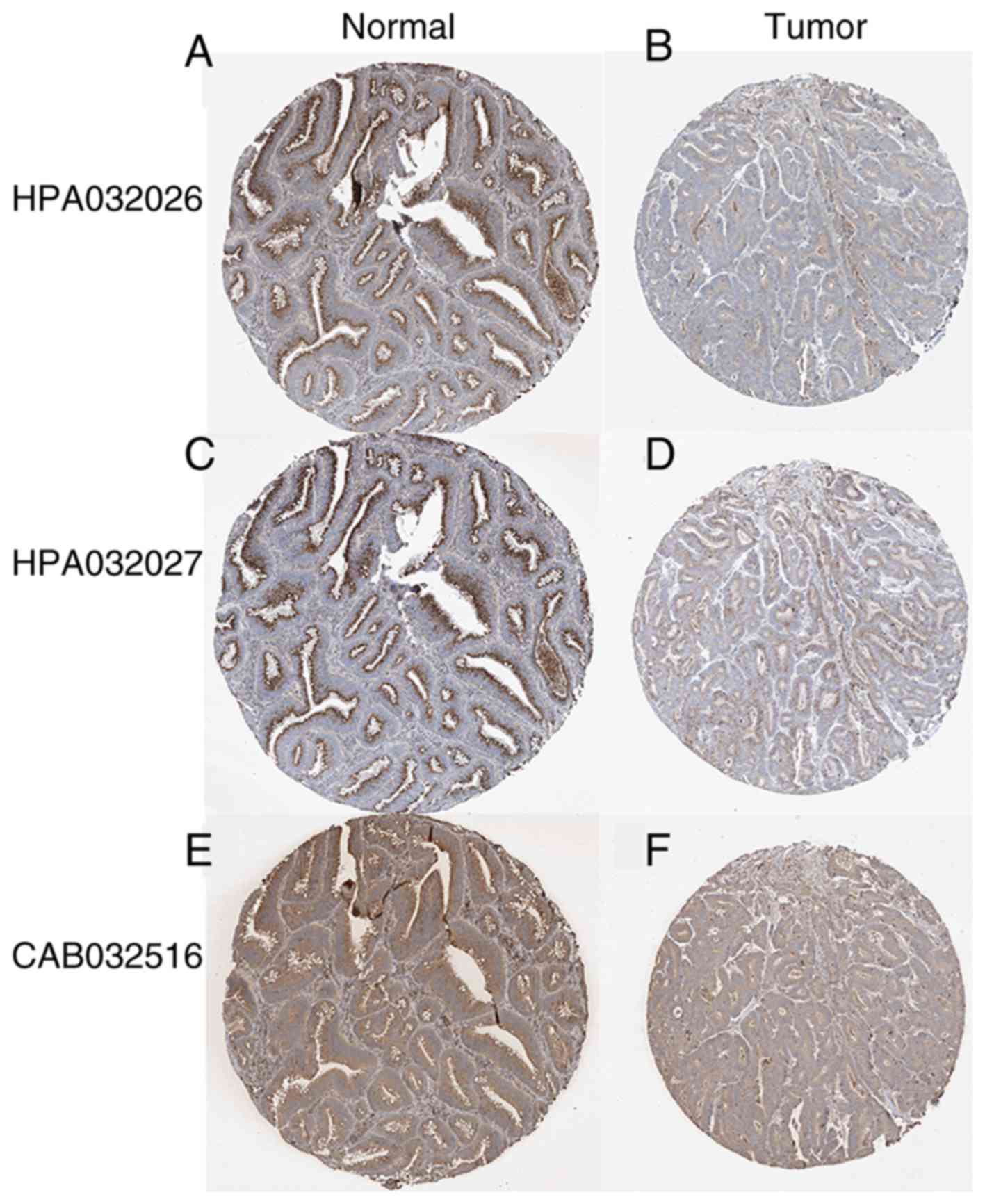
group compared with the normal group (P<0.001; Fig. 2A-D). In addition, representative
images from the Human Protein Atlas demonstrated that ABCD3 protein
expression determined by using three different antibodies was
higher in normal colon tissues compared with colon adenocarcinoma
(Fig. 3).
Diagnostic capability of ABCD3 mRNA
expression in CRC
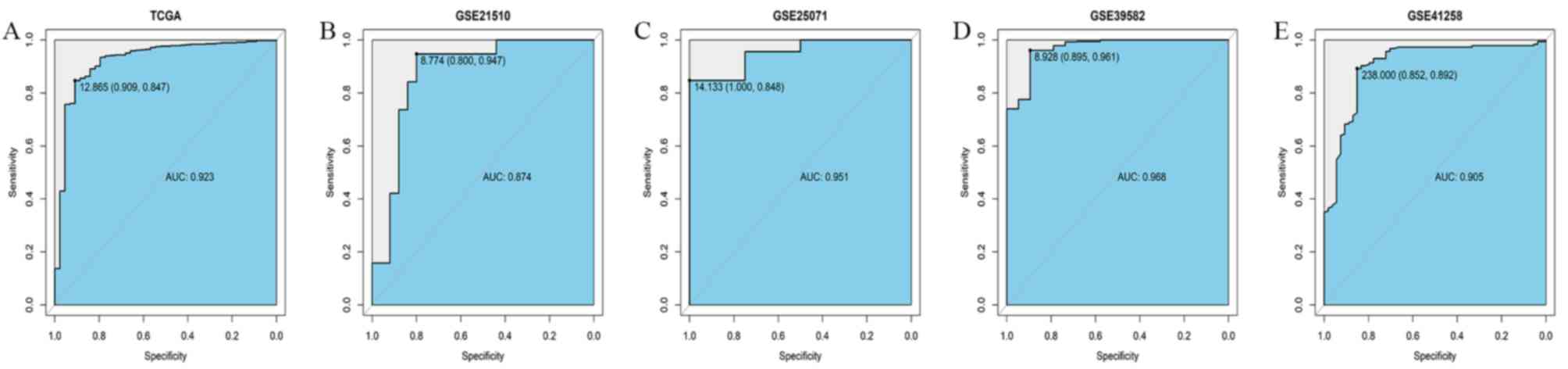
To evaluate the diagnostic value of ABCD3 mRNA
expression, a ROC curve was designed based on ABCD3 mRNA expression
data in CRC and normal tissues from TCGA database. The area under
the ROC curve (AUC) was 0.923 with the sensitivity and specificity
of 0.909 and 0.847, respectively, which indicated a modest
diagnostic value of ABCD3 mRNA expression (Fig. 4A). Similar analysis was performed in
the GSE21510, GSE25071, GSE39582 and GSE41258 datasets from GEO
database. The AUC in GSE21510 dataset was 0.874, with sensitivity
and specificity of 0.800 and 0.947, respectively (Fig. 4B). The AUC in GSE225071 dataset was
0.951, with sensitivity and specificity of 1.000 and 0.848,
respectively (Fig. 4C). The AUC in
GSE9582 dataset was 0.968, with sensitivity and specificity of
0.895 and 0.961 (Fig. 4D). The AUC
in GSE41258dataset was 0.905, with sensitivity and specificity of
0.852 and 0.892 (Fig. 4E). These
results suggested that ABCD3 mRNA expression may have a diagnostic
value in CRC.
Association between ABCD3 mRNA
expression and clinicopathological characteristics of patients with
CRC
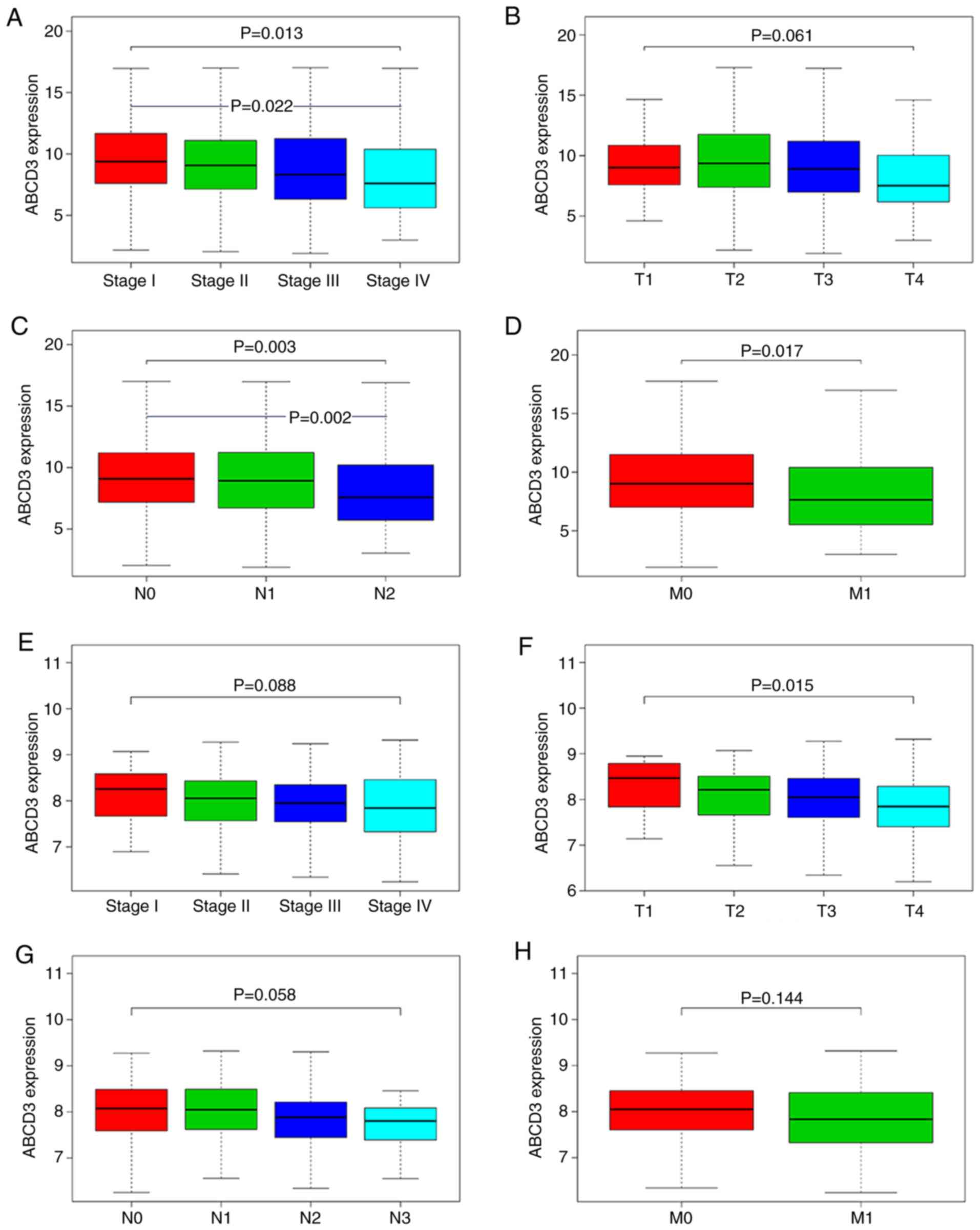
The results demonstrated that the ABCD3 mRNA levels
in tumor tissues were significantly different in different clinical
stages (P=0.013), T stages (P=0.061), lymph node metastasis
statuses (P=0.003) by using Kruskal-Wallis test, and in distant
metastasis statuses (P=0.017) by using Wilcoxon rank sum test.
Furthermore, the use of Bonferroni test demonstrated that ABCD3
mRNA expression in Stage IV vs. Stage I (P=0.022) and N2 vs. N0
(P=0.002) was statistically significant (Fig. 5A-D).
Furthermore, in the GSE39582 dataset, the ABCD3 mRNA
levels in tumor tissues were only significantly different between
T1 and T4 stages (P=0.015; Fig.
5F).
Logistic regression analysis demonstrated that ABCD3
mRNA expression in CRC tissues was significantly associated with
stage (OR=0.51 for stage IV vs. stage I; P=0.031); T stage (OR=0.50
for T4 vs. T2; P=0.035), lymph node status (OR=0.6 for N1+2 vs. N0;
P=0.036). However, it was not significantly associated with distant
metastasis status (OR=0.64 for M1 vs. M0; P=0.081; Table II). The results from χ2
test demonstrated that only stage (P=0.040) and lymph-node status
(P=0.049) were associated with ABCD3 mRNA expression (Table III). These findings suggested that
ABCD3 mRNA expression may serve a tumor suppressor role in CRC.
 | Table II.Association between ABCD3 mRNA
expression and the clinicopathological characteristics of patients
with colorectal cancer from The Cancer Genome Atlas (logistic
regression). |
Table II.
Association between ABCD3 mRNA
expression and the clinicopathological characteristics of patients
with colorectal cancer from The Cancer Genome Atlas (logistic
regression).
| Clinical
characteristics | Total, n | OR in ABCD3
expression (range) | P-value |
|---|
| Stage (IV vs.
I) | 170 | 0.51
(0.27–0.94) | 0.031 |
| T stage (T4 vs.
T2) | 156 | 0.50
(0.26–0.95) | 0.035 |
| Lymph node status
(N1+2 vs. N0) | 540 | 0.69
(0.49–0.98) | 0.036 |
| Distant metastasis
(M1 vs. M0) | 478 | 0.64
(0.39–1.05) | 0.081 |
 | Table III.Association between ABCD3 mRNA
expression and the clinicopathological characteristics of patients
with colorectal cancer from The Cancer Genome Atlas. |
Table III.
Association between ABCD3 mRNA
expression and the clinicopathological characteristics of patients
with colorectal cancer from The Cancer Genome Atlas.
|
|
| ABCD3 |
|
|
|---|
|
|
|
|
|
|
|---|
| Variables | Number | Low expression | High
expression | χ2
value | P-value |
|---|
| Sex |
|
|
|
|
|
|
Female | 243 | 120 | 123 | 0.054 | 0.816 |
|
Male | 204 | 103 | 101 |
|
|
| Age at diagnosis,
years |
|
|
|
|
|
|
>60 | 307 | 148 | 159 | 1.106 | 0.293 |
|
≤60 | 140 | 75 | 65 |
|
|
| T stage |
|
|
|
|
|
|
T1+2 | 93 | 42 | 51 | 1.05 | 0.306 |
|
T3+4 | 354 | 181 | 173 |
|
|
| Metastasis |
|
|
|
|
|
| M0 | 375 | 180 | 195 | 3.32 | 0.068 |
| M1 | 72 | 43 | 29 |
|
|
| Lymph node
status |
|
|
|
|
|
| N0 | 267 | 123 | 144 | 3.872 | 0.049 |
|
N1-2 | 180 | 100 | 80 |
|
|
| Clinical stage |
|
|
|
|
|
|
I+II | 118 | 140 | 258 | 4.207 | 0.040 |
|
III+IV | 105 | 84 | 189 |
|
|
Role of ABCD3 mRNA expression in
OS
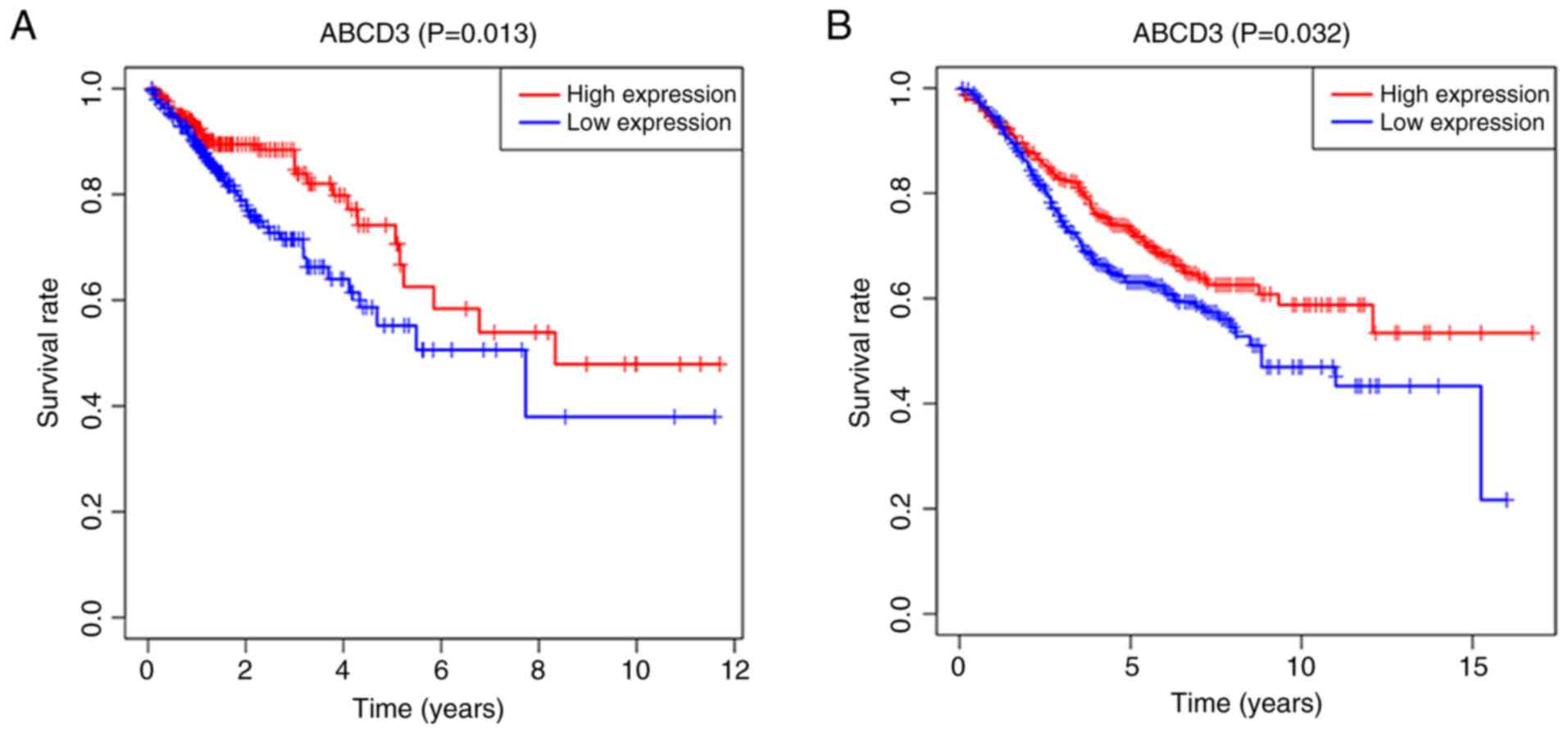
Kaplan-Meier survival analysis demonstrated that
patients from the TCGA database with low ABCD3 mRNA expression had
a poorer OS compared with patients with high ABCD3 mRNA expression
(P=0.013; Fig. 6A). Furthermore,
similar analysis in patients from the GSE39582 dataset demonstrated
that OS was significantly decreased in patients with low ABCD3 mRNA
expression compared with those with high ABCD3 mRNA expression
(P=0.032; Fig. 6B).
Univariate analysis of clinicopathological
characteristics demonstrated that ABCD3 mRNA level was a predictor
of OS [P=0.0028; HR=0.89; 95% CI (0.84–0.96)], which was also the
case for age(P=0.008; HR=1.03; 95% CI, 1.01–1.05), clinical stage
(P<0.001; HR=2.59; 95% CI, 1.99–3.36), T stage (P<0.001;
HR=3.27; 95% CI, (2.09–5.12), lymph nodes status (P<0.001;
HR=2.24; 95% CI, 1.72–2.93) and distant metastasis status
(P<0.001; HR=5.27; 95% CI, 3.33–8.34) (Table IV). Following multivariate analysis,
ABCD3 mRNA expression remained independently associated with OS
[P=0.016; HR=0.92; 95% CI (0.86–0.92)], as well as age (P<0.001;
HR=1.04; 95% CI, 1.02–1.07) and T stage (P=0.007; HR=2.01; 95% CI,
1.21–3.32) (Table IV). These
findings suggested that ABCD3 mRNA expression may be considered as
an independent prognostic factor for patients with CRC, and that
decreased ABCD3 mRNA expression may be associated with a poorer
OS.
 | Table IV.Univariate cox regression and
multivariate cox regression analyses in patients with colorectal
cancer from The Cancer Genome Atlas. |
Table IV.
Univariate cox regression and
multivariate cox regression analyses in patients with colorectal
cancer from The Cancer Genome Atlas.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinicopathologic
variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (continuous
variable) | 1.03 | 1.01–1.05 | 0.008 | 1.04 | 1.02–1.07 | <0.001 |
| Sex | 0.95 | 0.61–1.49 | 0.833 |
|
|
|
| Stage | 2.59 | 1.99–3.36 | <0.001 | 1.80 | 0.83–3.90 | 0.136 |
| T stage | 3.27 | 2.09–5.12 | <0.001 | 2.01 | 1.21–3.32 | 0.007 |
| Metastasis | 5.27 | 3.33–8.34 | <0.001 | 1.54 | 0.54–4.42 | 0.422 |
| Lymph node
status | 2.24 | 1.72–2.93 | <0.001 | 1.13 | 0.71–1.77 | 0.611 |
| ABCD3
expression | 0.90 | 0.84–0.96 | 0.003 | 0.92 | 0.86–0.98 | 0.016 |
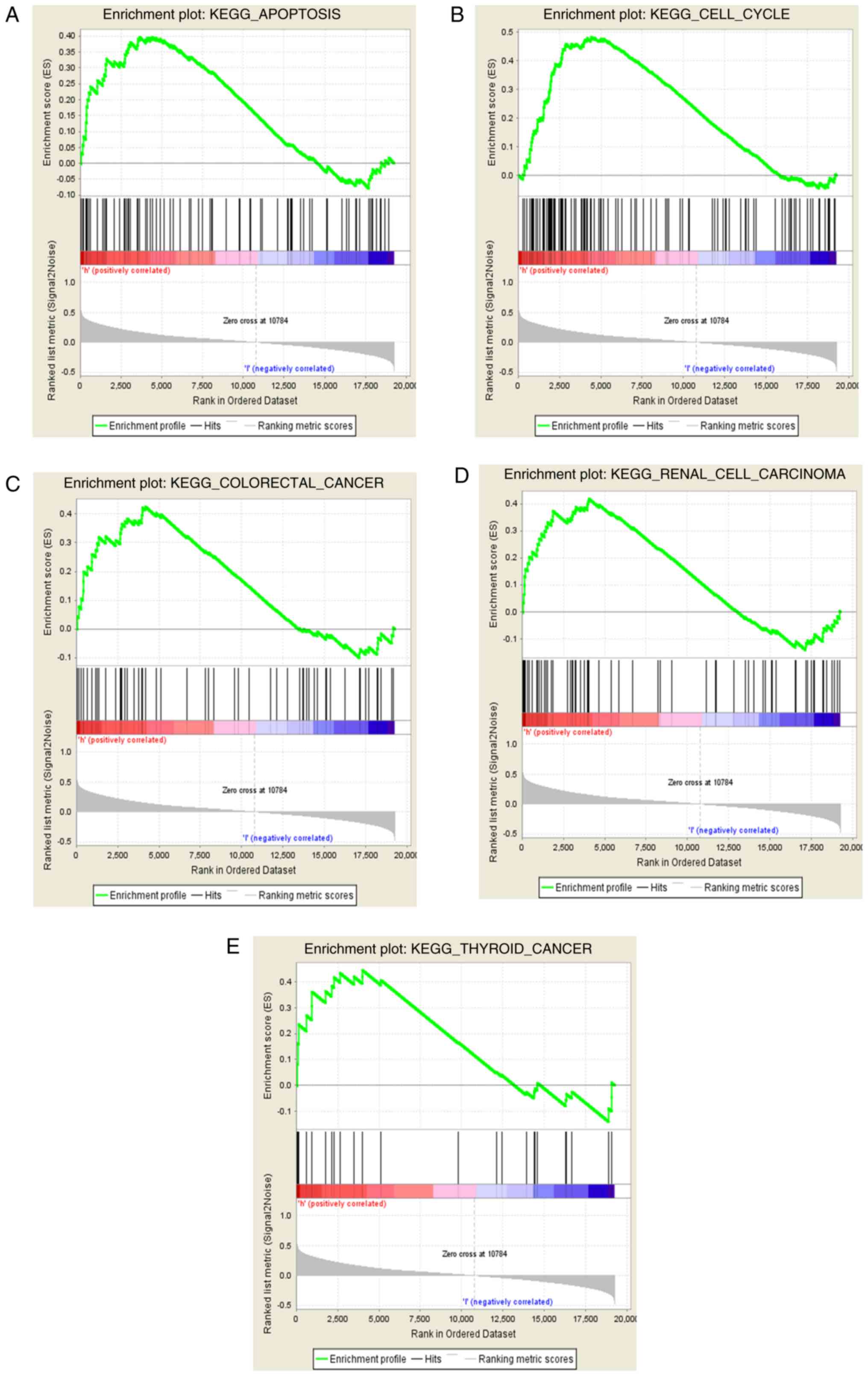
Screening of signaling pathways
associated with ABCD3
GSEA method was used to determine the signaling
pathways in which high and low ABCD3 expression levels are
identified in the enrichment of MSigDB Collection
(c2.cp.v6.2.symbols.gmt). The results demonstrated that five
signaling pathways, including cell apoptosis, cell cycle, renal
cell carcinoma, thyroid cancer, and CRC were significantly enriched
in ABCD3 high mRNA expression (FDR values, 0.216, 0.178, 0.214,
0.214 and 0.188, respectively; NOM P-values, 0.032, 0.037, 0.040,
0.025 and 0.006, respectively; and NES, 1.586, 1.696, 1.563, 1.595
and 1.699, respectively; Fig. 7A-E).
These findings suggested that ABCD3 may be involved in the
progression of CRC.
Discussion
At present, there are only a few studies on the role
and underlying mechanism of ABCD3 in tumors, such as prostate tumor
(16), ovarian cancer (15,17),
colon cancer (14), chronic myeloid
leukemia (25), melanoma (26) and retinoblastoma (27). However, the aforementioned studies
only investigated the difference of ABCD3 mRNA or protein
expression in tumors. For instance, ABCD3 protein expression is
significantly decreased in colon adenocarcinoma tissues compared
with adjacent normal colon tissues (28). In addition, Yasui et al
(29) demonstrated that ABCD3 is
amplified in 19 resistant cancer cell lines. To the best of our
knowledge, no parameters of CRC have been used to evaluate the
correlation between ABCD3 mRNA expression and CRC before. The
present study investigated the difference in ABCD3 mRNA expression
between CRC and normal tissues. In addition, to the best of our
knowledge, this study was the first to evaluate the diagnostic and
prognostic values of ABDC3 mRNA expression.
In the present study, ABCD3 mRNA data of patients
with CRC from TCGA and GEO databases were collected, and ABCD3
protein expression was obtained from the Human Protein Atlas. The
results demonstrated that the mRNA and protein expression of ABCD3
was downregulated in CRC tissues compared with normal tissues.
Furthermore, the high ABCD3 mRNA expression in CRC tissues was
negatively associated with clinical stage, T stage, lymph node
status and distant metastasis, but was associated with better
prognosis. In addition, results from GSEA demonstrate that high
ABCD3 mRNA expression was enriched in cell apoptosis, cell cycle,
renal cell carcinoma, thyroid cancer, and CRC, suggesting that
ABCD3 mRNA may be considered as a new therapeutic target and
diagnostic and prognostic biomarker in CRC.
ABCD3, which is a member of the superfamily of ABC
transporters, participates in the peroxisomal import of fatty acids
and/or fatty acyl-CoAs in the organelle (11). Increasing evidence demonstrated that
ABC transporters affect the progression of chemoresistance by
regulating the efflux of anticancer agents outside cancer cells
(26,30,31).
This common feature could be due to a subpopulation of slow-cycling
cancer stem cells, which show enhanced multidrug resistance and
tumorigenic potential (32).
Although the underlying mechanisms of ABCD3 remain unknown, other
molecules from the ABC transporters family have been studied. For
example, elevated expression of ABCF1 enhances drug efflux and
chemoresistance, and accelerates colony and spheroid formation,
cell migration and epithelial-mesenchymal transition in
hepatocellular carcinoma (33). In
addition, the molecule Guajadial can suppress drug resistance that
could be mediated by the repression of ABC transporter expression
and by the PI3K/Akt pathway in drug-resistant breast cancer cells
(34). A previous study similar to
the present one demonstrated that ABCB9 mRNA expression is
decreased in ovarian cancer tissues compared with normal ovarian
tissues, and that decreased ABCB9 mRNA expression is associated
with poor OS in patients. These results suggested that ABCB9 might
be considered as an independent prognostic indicator in ovarian
cancer (35). Similarly, the
findings form the present study suggested that ABCD3 may serve a
crucial role in drug resistance and may affect the OS of patients
with CRC.
The present study demonstrated that ABCD3 was
involved in cell apoptosis, cell cycle, renal cell carcinoma,
thyroid cancer and CRC according to results from GSEA. However,
these predictions must be further investigated and confirmed.
Although the present study suggested some
associations between ABCD3 mRNA expression and CRC, it presented
some limitations. Firstly, to fully elucidate the crucial role of
ABCD3 in the progression of CRC, numerous clinical factors,
including recurrence, histological grade and treatment situation,
should be considered. Secondly, the number of tumor samples
differed significantly from the number of normal samples, and
further investigation using a higher sample size is therefore
required. Thirdly, the GSE21510, GSE25071, GSE41258 and GSE39582
datasets were not analyzed by same group or individuals, therefore
the testing standards may have differed between these datasets and
so no systematic meta-analysis was performed. Fourthly, the results
from the present study were only based on the bioinformatics
analysis of multiple databases. Future study will therefore include
experimental in vitro results. However, despite the
limitations of the present study, the present bioinformatics
analysis did provide novel insight into the function of ABCD3 in
CRC, including target molecules screening, gene function analysis
and identification of molecular signaling pathways.
In conclusion, the present study demonstrated that
low ABCD3 mRNA expression in patients with CRC was associated with
poor OS. In addition, ABCD3 was enriched in signaling pathways such
as apoptosis, cell cycle, renal cell carcinoma, thyroid cancer, and
CRC, therefore ABCD3 may function in CRC progression. The present
study partially revealed the function of ABCD3 in CRC and
demonstrated that it may be considered as a diagnostic and
prognostic biomarker in patients with CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the TCGA database (https://mirrors.tuna.tsinghua.edu.cn/CRAN/) and GEO
database (https://www.ncbi.nlm.nih.gov/gds/).
Authors' contributions
GY and JY conceived and designed the study. GY
performed the bioinformatics analysis. YuZ analyzed the data. GY
and YaZ wrote the manuscript. JY reviewed the manuscript. JW
participated in the collection of data and the bioinformatics
analysis. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ABCD3
|
ATP binding cassette subfamily D
member 3
|
|
CI
|
confidence interval
|
|
CRC
|
colorectal cancer
|
|
GEO
|
Gene Expression Omnibus
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
HR
|
hazard ratio
|
|
OR
|
odds ratio
|
|
ROC
|
receiver operating characteristic
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Landreau P, Drouillard A, Launoy G,
Ortega-Deballon P, Jooste V, Lepage C, Faivre J, Facy O and Bouvier
AM: Incidence and survival in late liver metastases of colorectal
cancer. J Gastroenterol Hepatol. 30:82–85. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Asano H, Kojima K, Ogino N, Fukano H,
Ohara Y and Shinozuka N: Postoperative recurrence and risk factors
of colorectal cancer perforation. Int J Colorectal Dis. 32:419–424.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Al Bandar MH and Kim NK: Current status
and future perspectives on treatment of liver metastasis in
colorectal cancer (Review). Oncol Rep. 37:2553–2564. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gires O: Lessons from common markers of
tumor-initiating cells in solid cancers. Cell Mol Life Sci.
68:4009–4022. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Locker GY, Hamilton S, Harris J, Jessup
JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF and Bast RC Jr:
ASCO: ASCO 2006 update of recommendations for the use of tumor
markers in gastrointestinal cancer. J Clin Oncol. 24:5313–5327.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Engstrom PF, Arnoletti JP, Benson AB 3rd,
Chen YJ, Choti MA, Cooper HS, Covey A, Dilawari RA, Early DS,
Enzinger PC, et al: NCCN Clinical practice guidelines in oncology:
Colon cancer. J Natl Compr Canc Netw. 7:778–831. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rose J, Augestad KM and Cooper GS:
Colorectal cancer surveillance: What's new and what's next. World J
Gastroenterol. 20:1887–1897. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Labianca R, Nordlinger B, Beretta GD,
Brouquet A and Cervantes A; ESMO Guidelines Working Group, :
Primary colon cancer: ESMO Clinical practice guidelines for
diagnosis, adjuvant treatment and follow-up. Ann Oncol. 21 (Suppl
5):v70–v77. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schmoll HJ, Van Cutsem E, Stein A,
Valentini V, Glimelius B, Haustermans K, Nordlinger B, van de Velde
CJ, Balmana J, Regula J, et al: ESMO Consensus Guidelines for
management of patients with colon and rectal cancer. a personalized
approach to clinical decision making. Ann Oncol. 23:2479–2516.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kawaguchi K and Morita M: ABC transporter
subfamily D: Distinct differences in behavior between ABCD1-3 and
ABCD4 in subcellular localization, function, and human disease.
Biomed Res Int. 2016:67862452016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cole SP, Bhardwaj G, Gerlach JH, Mackie
JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM and
Deeley RG: Overexpression of a transporter gene in a
multidrug-resistant human lung cancer cell line. Science.
258:1650–1654. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Holla VR, Wu H, Shi Q, Menter DG and
DuBois RN: Nuclear orphan receptor NR4A2 modulates fatty acid
oxidation pathways in colorectal cancer. J Biol Chem.
286:30003–30009. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu T, Zhang H and Qi H: Transcriptome
profiling analysis reveals biomarkers in colon cancer samples of
various differentiation. Oncol Lett. 16:48–54. 2018.PubMed/NCBI
|
|
15
|
Seborova K, Vaclavikova R, Soucek P,
Elsnerova K, Bartakova A, Cernaj P, Bouda J, Rob L, Hruda M and
Dvorak P: Association of ABC gene profiles with time to progression
and resistance in ovarian cancer revealed by bioinformatics
analyses. Cancer Med. 8:606–616. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Reams RR, Jones-Triche J, Chan OT,
Hernandez BY, Soliman KF and Yates C: Immunohistological analysis
of ABCD3 expression in Caucasian and African American prostate
tumors. Biomed Res Int. 2015:1329812015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Elsnerova K, Mohelnikova-Duchonova B,
Cerovska E, Ehrlichova M, Gut I, Rob L, Skapa P, Hruda M, Bartakova
A, Bouda J, et al: Gene expression of membrane transporters:
Importance for prognosis and progression of ovarian carcinoma.
Oncol Rep. 35:2159–2170. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hlavata I, Mohelnikova-Duchonova B,
Vaclavikova R, Liska V, Pitule P, Novak P, Bruha J, Vycital O,
Holubec L, Treska V, et al: The role of ABC transporters in
progression and clinical outcome of colorectal cancer. Mutagenesis.
27:187–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsukamoto S, Ishikawa T, Iida S, Ishiguro
M, Mogushi K, Mizushima H, Uetake H, Tanaka H and Sugihara K:
Clinical significance of osteoprotegerin expression in human
colorectal cancer. Clin Cancer Res. 17:2444–2450. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Agesen TH, Berg M, Clancy T, Thiis-Evensen
E, Cekaite L, Lind GE, Nesland JM, Bakka A, Mala T, Hauss HJ, et
al: CLC and IFNAR1 are differentially expressed and a global
immunity score is distinct between early- and late-onset colorectal
cancer. Genes Immun. 12:653–662. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sheffer M, Bacolod MD, Zuk O, Giardina SF,
Pincas H, Barany F, Paty PB, Gerald WL, Notterman DA and Domany E:
Association of survival and disease progression with chromosomal
instability: A genomic exploration of colorectal cancer. Proc Natl
Acad Sci USA. 106:7131–7136. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marisa L, de Reyniès A, Duval A, Selves J,
Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D,
Ayadi M, et al: Gene expression classification of colon cancer into
molecular subtypes: Characterization, validation, and prognostic
value. PLoS Med. 10:e10014532013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trojani A, Pungolino E, Dal Molin A,
Lodola M, Rossi G, D'Adda M, Perego A, Elena C, Turrini M, Borin L,
et al: Nilotinib interferes with cell cycle, ABC transporters and
JAK-STAT signaling pathway in CD34+/lin- cells of patients with
chronic phase chronic myeloid leukemia after 12 months of
treatment. PLoS One. 14:e02184442019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Heimerl S, Bosserhoff AK, Langmann T,
Ecker J and Schmitz G: Mapping ATP-binding cassette transporter
gene expression profiles in melanocytes and melanoma cells.
Melanoma Res. 17:265–273. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hendig D, Langmann T, Zarbock R, Schmitz
G, Kleesiek K and Gotting C: Characterization of the ATP-binding
cassette transporter gene expression profile in Y79: A
retinoblastoma cell line. Mol Cell Biochem. 328:85–92. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lauer C, Völkl A, Riedl S, Fahimi HD and
Beier K: Impairment of peroxisomal biogenesis in human colon
carcinoma. Carcinogenesis. 20:985–989. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yasui K, Mihara S, Zhao C, Okamoto H,
Saito-Ohara F, Tomida A, Funato T, Yokomizo A, Naito S, Imoto I, et
al: Alteration in copy numbers of genes as a mechanism for acquired
drug resistance. Cancer Res. 64:1403–1410. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fletcher JI, Williams RT, Henderson MJ,
Norris MD and Haber M: ABC transporters as mediators of drug
resistance and contributors to cancer cell biology. Drug Resist
Updat. 26:1–9. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schuetz JD and Ishikawa T: ABC
transporters and cancer. Preface. Adv Cancer Res. 125:xv–xvii.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Begicevic RR and Falasca M: ABC
Transporters in cancer stem cells: Beyond chemoresistance. Int J
Mol Sci. 18:E23622017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fung SW, Cheung PF, Yip CW, Ng LW, Cheung
TT, Chong CC, Lee C, Lai PB, Chan AW, Tsao GS, et al: The
ATP-binding cassette transporter ABCF1 is a hepatic oncofetal
protein that promotes chemoresistance, EMT and cancer stemness in
hepatocellular carcinoma. Cancer Lett. 457:98–109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Y, Zhai Z, Li H, Wang X, Huang Y and Su
X: Guajadial reverses multidrug resistance by inhibiting ABC
transporter expression and suppressing the PI3K/Akt pathway in
drug-resistant breast cancer cells. Chem Biol Interact. 305:98–104.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hou L, Zhang X, Jiao Y, Li Y, Zhao Y, Guan
Y and Liu Z: ATP binding cassette subfamily B member 9 (ABCB9) is a
prognostic indicator of overall survival in ovarian cancer.
Medicine (Baltimore). 98:e156982019. View Article : Google Scholar : PubMed/NCBI
|