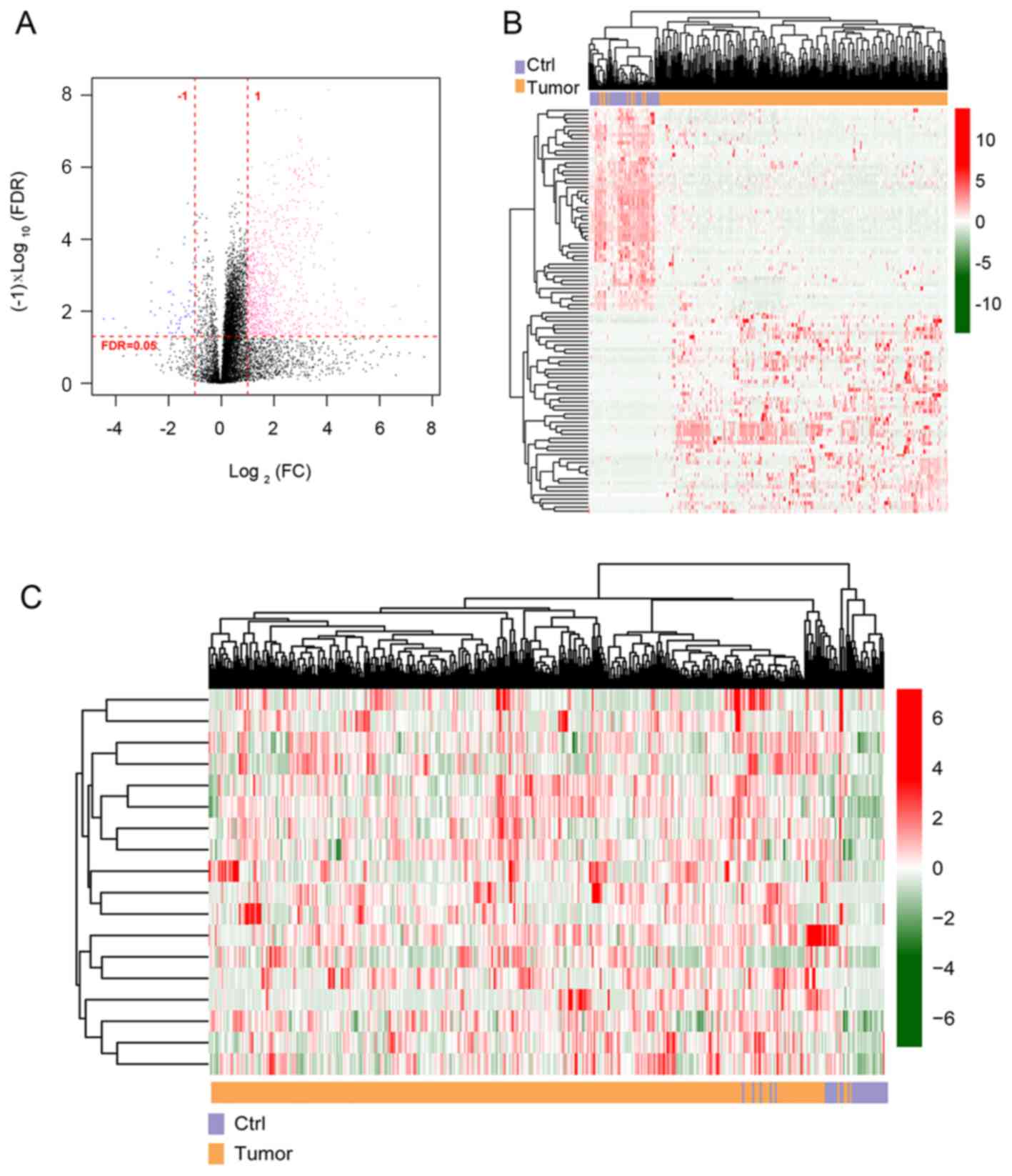
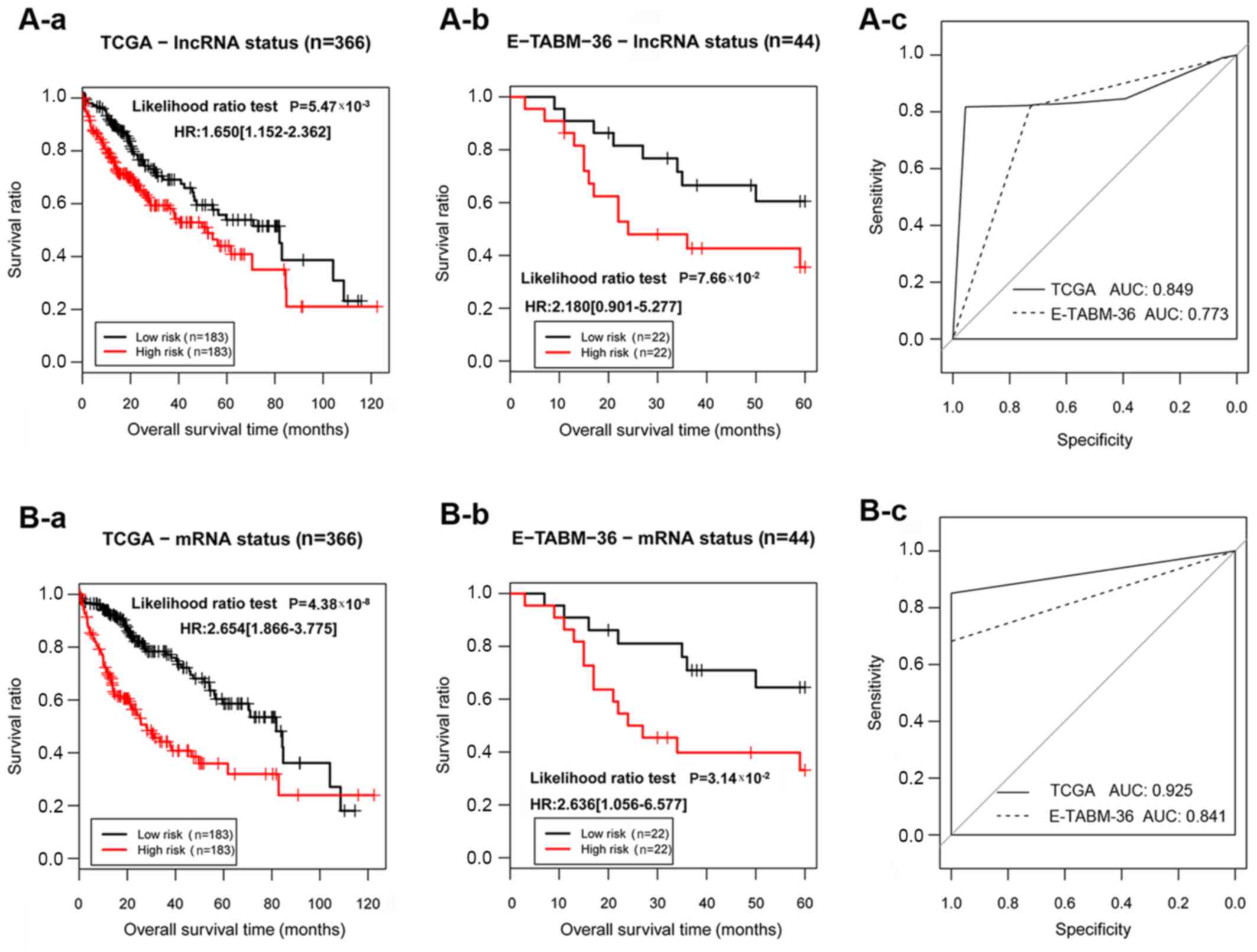
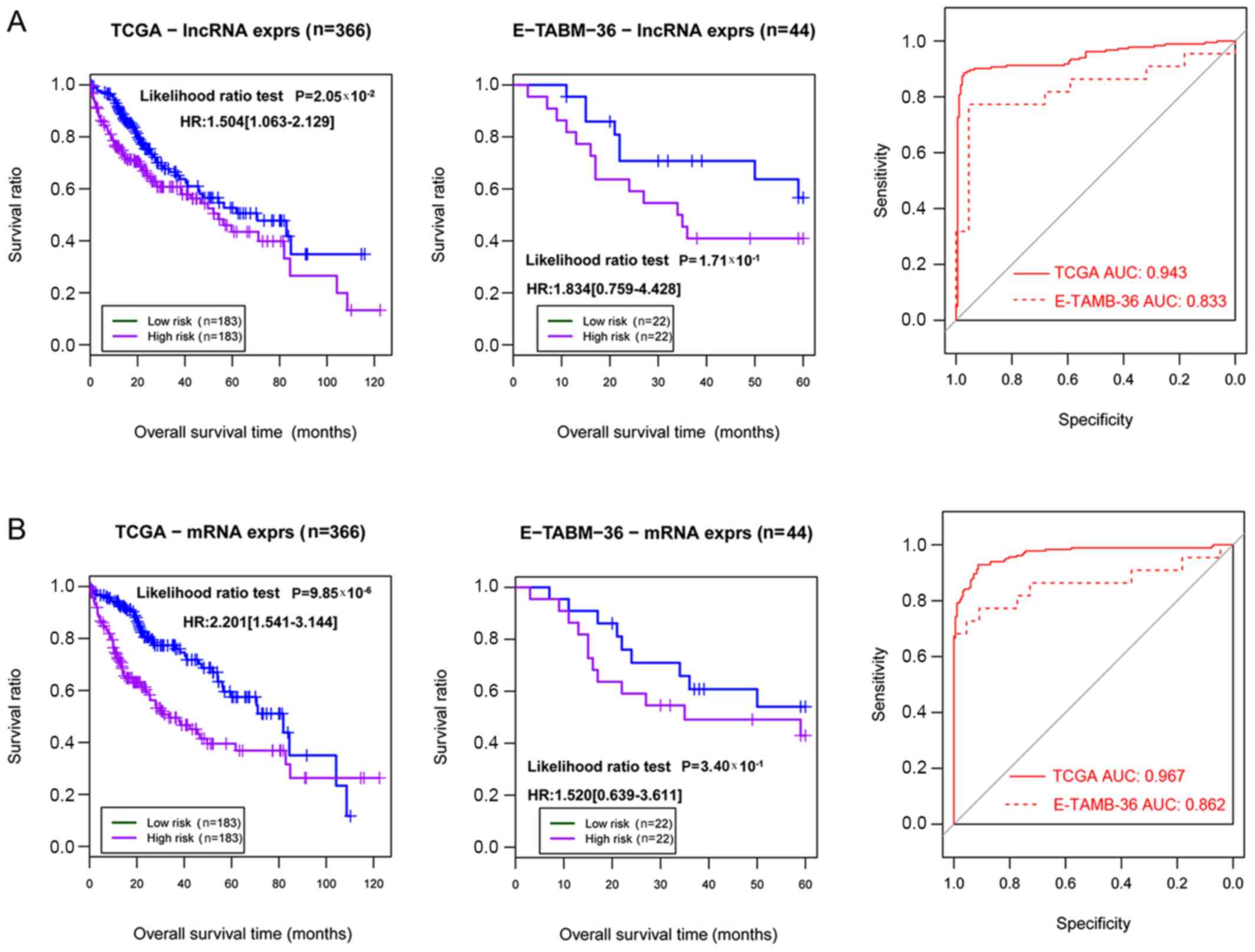
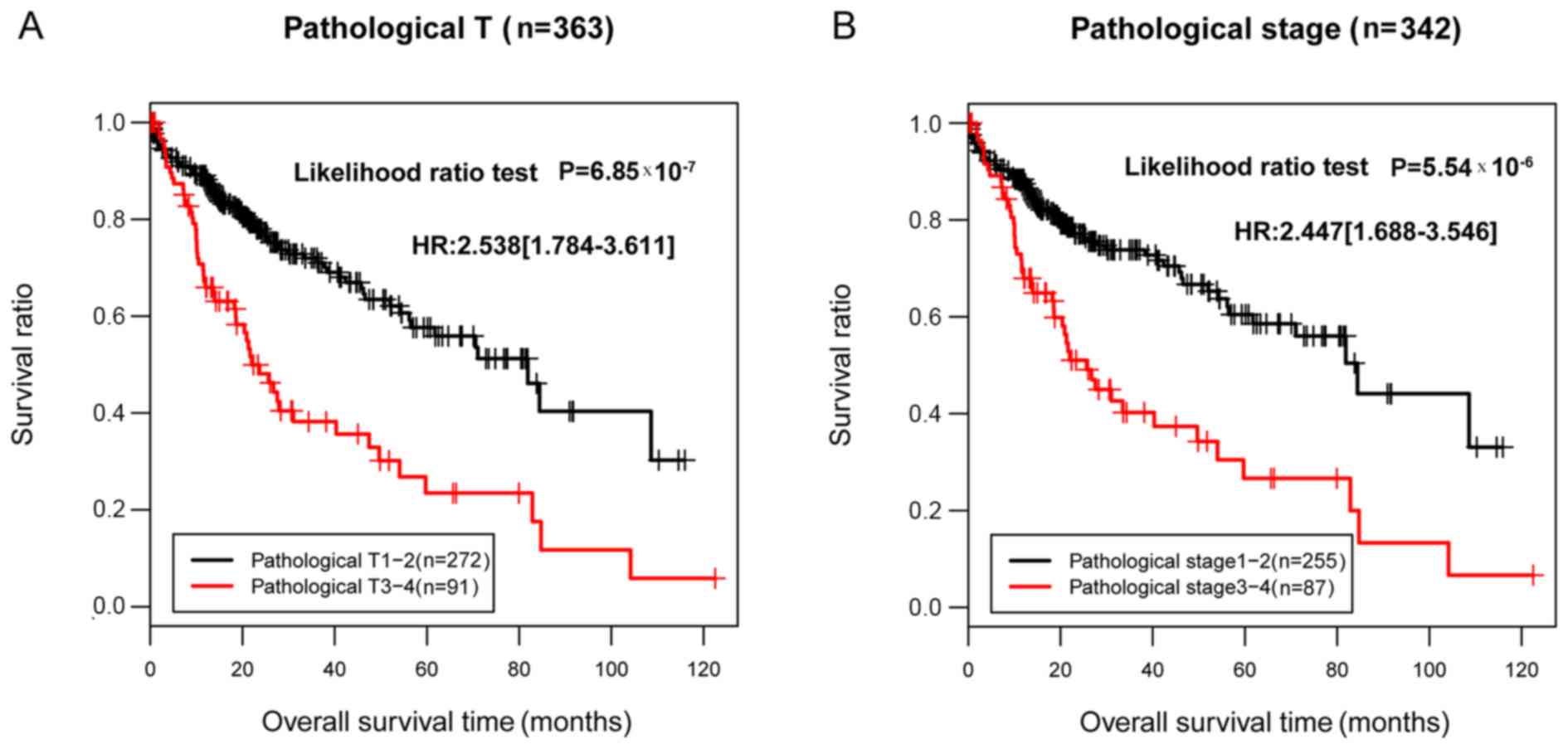
|
GOTERM_BP_DIRECT | GO:0006260~DNA
replication | 43 |
P=1.95×10−14 |
7.41×10−11 | CLSPN, BLM, DBF4,
TIPIN, POLA1, CHEK1, POLA2, MCM10, CDT1, CDC45, POLE2, ORC6, EGF,
ORC1, RTEL1, EXO1, RECQL4, CDC7, CDK1, CDC6, GINS2, CCDC88A, DTL,
GINS3, GINS4, BRIP1, TREX1, MCM2, WRN, CDC25C, MCM4, BRCA1, CDC25A,
RBBP8, MCM6, POLD3, DNA2, RFC4, POLD1, RRM2, CHAF1B, BARD1,
DSCC1 |
|
GOTERM_BP_DIRECT | GO:0051301~cell
division | 67 |
P=2.18×10−13 |
4.14×10−10 | CDK19, KIFC1,
CDC14A, KNTC1, PTTG1, SPICE1, CCNE2, KIF2C, CCNE1, NDE1, CDCA8,
OIP5, CCNA2, CDCA4, CDCA3, KIF2A, KIF14, ANAPC1, CDC7, CDK1, CDC6,
ARHGEF2, KIF11, CCNF, TPX2, UBE2C, TACC3, NCAPD3, RBBP8, NCAPD2,
CDK3, MAD2L1, SPAG5, ZWINT, BUB1B, FBXL7, UBE2S, HAUS6, NEK2,
TIPIN, SPC25, NCAPH, NCAPG2, NCAPG, BUB1, FBXO5, SKA1, ZWILCH,
HELLS, ERCC6L, PARD6B, PSRC1, CENPF, KIF18B, NDC80, CENPE, CDC20,
BIRC5, CDC25C, SMC2, CENPJ, CDC25A, SMC4, CCNB1, CCNB2, KIF20B,
CDK20 |
|
GOTERM_BP_DIRECT | GO:0006270~DNA
replication initiation | 18 |
P=5.76×10−12 |
7.28×10−9 | CDC7, CDC6, GINS4,
POLA1, POLA2, MCM2, MCM10, MCM4, RPA4, MCM6, PRIM1, CCNE2, CCNE1,
CDC45, POLE2, PRIM2, ORC6, ORC1 |
|
GOTERM_BP_DIRECT | GO:0000082~G1/S
transition of mitotic cell cycle | 31 |
P=1.08×10−11 |
1.03×10−8 | BCAT1, DBF4, POLA1,
PKMYT1, POLA2, MCM10, RPA4, CDT1, CCNE2, PRIM1, CCNE1, TYMS, CDC45,
CDKN2A, POLE2, PRIM2, FBXO5, ORC6, ORC1, CDC7, CDC6, CDK1, GPR132,
MCM2, CDKN3, MCM4, CDC25A, RBBP8, CDK3, MCM6, RRM2 |
|
GOTERM_BP_DIRECT | GO:0007067~mitotic
nuclear division | 47 |
P=1.71×10−9 |
1.30×10−6 | HAUS6, NEK2, TIPIN,
KNTC1, PKMYT1, CEP55, AURKB, PTTG1, SPC25, KIF2C, NCAPG2, OIP5,
INCENP, TTYH1, BUB1, FBXO5, SKA1, ZWILCH, CCNA2, ASPM, TUBB3,
HELLS, CDCA3, ERCC6L, ANAPC1, CDK1, CDC6, ARHGEF2, KIF11, CCNF,
KIF15, TPX2, CENPF, NDC80, CDC20, BIRC5, PBK, CDC25C, CDC25A,
RBBP8, CDK3, CCNB2, PLK1, KIF20B, BUB1B, FBXL7, CIT |
|
GOTERM_BP_DIRECT | GO:0007062~sister
chromatid cohesion | 28 |
P=1.99×10−9 |
1.26×10−6 | KNTC1, AURKB,
SPC25, KIF2C, CDCA8, NDE1, DDX11, CENPA, INCENP, BUB1, SKA1,
ZWILCH, KIF2A, ERCC6L, CENPO, CENPM, CENPQ, KIF18A, CENPF, NDC80,
BIRC5, CDC20, CENPE, CENPI, MAD2L1, PLK1, ZWINT, BUB1B |
|
GOTERM_BP_DIRECT | GO:0000731~DNA
synthesis involved in DNA repair | 16 |
P=4.77×10−9 |
2.59×10−6 | EXO1, XRCC3,
RAD51AP1, XRCC2, BLM, POLA1, BRCA2, BRIP1, WRN, BRCA1, RAD51,
RBBP8, POLD3, DNA2, POLD1, BARD1 |
|
GOTERM_BP_DIRECT | GO:0000732~strand
displacement | 14 |
P=4.99×10−9 |
2.36×10−6 | EXO1, XRCC3,
RAD51AP1, XRCC2, BLM, BRCA2, BRIP1, WRN, BRCA1, RAD51, RBBP8, DNA2,
RTEL1, BARD1 |
|
GOTERM_BP_DIRECT | GO:0006271~DNA
strand elongation involved in DNA replication | 11 |
P=7.05×10−9 |
2.97×10−6 | POLD3, GINS1,
PRIM1, GINS2, RFC4, POLD1, GINS3, GINS4, PRIM2, POLA1, POLA2 |
|
GOTERM_BP_DIRECT |
GO:0051726~regulation of cell cycle | 30 |
P=8.79×10−9 |
3.34×10−6 | CDK19, E2F2, E2F5,
FOXM1, PRR11, PKMYT1, MYBL2, SRC, CCNE2, CCNE1, OVOL2, PLCB1, MX2,
MAK, DTL, CCNF, L3MBTL1, RBL1, CENPF, CDC25C, MECOM, CDKL5, CDC25A,
CDK3, CCNB1, TRIM36, CCNB2, CDKL1, PLK1, CDK20 |
|
GOTERM_BP_DIRECT | GO:0000086~G2/M
transition of mitotic cell cycle | 29 |
P=3.32×10−7 |
1.14×10−6 | CEP72, HAUS6, NEK2,
FOXM1, PKMYT1, CHEK1, HMMR, NDE1, CDKN2B, CEP250, CEP290, PLCB1,
CDK1, CEP135, CEP192, TPX2, BIRC5, SFI1, CDC25C, CEP152, CENPJ,
CDC25A, CCNB1, PLK4, CCNB2, PLK1, FBXL7, CIT, MELK |
|
GOTERM_BP_DIRECT | GO:0000070~mitotic
sister chromatid segregation | 12 |
P=4.05×10−7 |
1.28×10−4 | KIFC1, MAD2L1,
CENPA, PLK1, NEK2, SPAG5, ZWINT, NUSAP1, KIF18B, NDC80, ESPL1,
SMC4 |
|
GOTERM_BP_DIRECT | GO:0000722~telomere
maintenance via recombination | 13 |
P=9.64×10−7 |
2.81×10−4 | POLD3, PRIM1, DNA2,
XRCC3, RFC4, POLE2, POLD1, PRIM2, POLA1, BRCA2, POLA2, WRN,
RAD51 |
|
GOTERM_BP_DIRECT |
GO:0007018~microtubule-based movement | 20 |
P=3.01×10−6 |
8.15×10−4 | KIF14, KIF23,
KIFC1, KIF4A, KIF24, KIF11, KIF5A, KIF15, KIF18A, KIF18B, CENPE,
RACGAP1, KIF3C, KIF2C, DYNC2H1, KIF20B, KIF21B, KIF26B, KIF20A,
KIF2A |
|
GOTERM_BP_DIRECT | GO:0043547~positive
regulation of GTPase activity | 69 |
P=1.39×10−5 |
3.52×10−3 | EZH2, BTC,
ARHGAP22, TIAM2, GIT2, CSF2RB, ARHGAP11A, AGAP1, DOCK10, PLCB1,
CSF2RA, ARHGEF2, ARHGEF18, GRIN2A, RAB3IL1, ARTN, SIPA1L3, DEPDC1,
ECT2, CDKL5, ANKRD27, NCK2, PLCE1, SH2D3A, ARHGAP33, OPHN1, RIN1,
SEMA4D, DENND3, RAPGEFL1, EPS8L1, SPTB, FGFR2, FGFR1, CSF2,
RAP1GAP, CYTH4, ASAP1, KITLG, LLGL1, ADAP1, RASAL1, DOCK2, CCL20,
GMIP, PLEKHG6, IL2RG, EGF, RASA3, RAP1GAP2, FGD6, IQSEC2, PIK3R2,
OBSCN, IL2RA, ABR, VAV3, ARHGEF38, RGS17, RACGAP1, VAV1, RGS20,
RGS1, CHML, RGS5, JAK3, XCL1, SH3BP1, ARAP2 |
|
GOTERM_BP_DIRECT |
GO:0000083~regulation of transcription
involved in G1/S transition of mitotic cell cycle | 10 |
P=1.51×10−5 |
3.57×10−3 | CCNE1, CDK1, CDC6,
TYMS, CDC45, RRM2, POLA1, FBXO5, ORC1, CDT1 |
|
GOTERM_BP_DIRECT | GO:0008283~cell
proliferation | 49 |
P=3.00×10−5 |
6.67×10−3 | BCAT1, STIL,
CDK5R1, IL9R, CDC14A, E2F8, NR6A1, UCHL1, POLA1, KITLG, EHF, AURKB,
FER, MCM10, TRAIP, SRC, TGFB2, GPC4, EDNRA, TYMS, KIF2C, BUB1,
TGFA, BHLHE41, IL1A, OCA2, SYK, CDK1, IL2RA, MKI67, DLGAP5, KIF15,
TPX2, SIX2, CENPF, ITGA2, TNFSF9, TACC3, CDC25C, CDC25A, CDK3,
SH2D2A, PLCE1, S100B, PLK1, SIX1, BUB1B, FBXL7, MELK |
|
GOTERM_BP_DIRECT | GO:0032508~DNA
duplex unwinding | 13 |
P=4.04×10−5 |
8.49×10−3 | RECQL4, GINS1,
GINS2, DNA2, CDC45, DDX11, BLM, GINS4, BRIP1, RAD54B, WRN, RTEL1,
CHD3 |
|
GOTERM_BP_DIRECT | GO:0007165~signal
transduction | 119 |
P=4.66×10−5 |
9.26×10−3 | IL9R, CRABP2, TLR1,
TNFSF15, IL15, TLR6, ITPKA, ADORA1, CASP8AP2, UNC5B, ANK2, PDE4A,
CHRNA5, CSF3R, IL1B, TYRO3, STK24, PIK3CD, LRRC1, DEPDC1, PDE4C,
PLAUR, GABRR2, LILRB1, TNFAIP6, CCR6, IGSF1, LRP12, LILRB4, PDE5A,
OPHN1, MST1R, CLEC5A, PTK7, OPN1SW, SOX9, SRC, RASAL1, VDR, NPHP4,
IL12RB1, EGF, TRAF5, NPHP1, GABRE, ABR, GNRH1, SPHK1, CD300C,
SALL2, RGS1, HIVEP3, XCL1, GRB7, CHRNG, GPR85, CLDN4, TAS2R4,
RASSF8, PDCD1, SIRPB1, EDNRA, ARHGAP22, TNFRSF11A, PTGES, CSF2RB,
MICAL1, CNTNAP1, ARHGAP11A, DLG5, PLCB1, LTA, MPP2, LIMK1, MPP3,
ARTN, CDS1, SH2D2A, HUNK, DOK1, NCK2, TNFRSF10C, ACVR2B, ARHGAP33,
CLIC3, ZDHHC13, RIN1, PDE9A, INPP4A, GNB3, RAP1GAP, CXCL5, GULP1,
CTNND2, TNFRSF8, KITLG, SFN, TRAIP, IGF1R, CCL20, FCGR1A, DGKG,
BCL11A, ARR3, IL2RG, SH2B2, DKKL1, RASA3, HPGDS, PIK3R2, EPO,
TNFSF4, LRRN2, TNFSF9, RACGAP1, LRP8, SH3BP1, ARAP2, CHRNA10 |
|
GOTERM_BP_DIRECT |
GO:0001932~regulation of protein
phosphorylation | 11 |
P=8.80×10−5 |
1.65×10−2 | PHIP, CCDC88A,
CCDC88C, ARR3, EZH2, CELSR3, FER, TLR7, KDM4D, TLR8, RAD51 |
|
GOTERM_BP_DIRECT |
GO:0007059~chromosome segregation | 15 |
P=2.62×10−4 |
4.63×10−2 | KIF11, NEK2, CENPF,
NDC80, CENPE, BRCA1, SPC25, NDE1, HJURP, SPAG5, OIP5, INCENP, SKA1,
TOP2A, TOP3B |
| KEGG_PATHWAY | hsa04110:Cell
cycle | 38 |
P=2.06×10−14 |
5.08×10−12 | E2F1, E2F2, E2F3,
CDC14A, E2F5, DBF4, PKMYT1, TTK, CHEK1, SFN, PTTG1, TGFB2, CCNE2,
CCNE1, CDC45, CDKN2A, CDKN2B, BUB1, ORC6, CCNA2, ORC1, CDC7,
ANAPC1, CDK1, CDC6, RBL1, ESPL1, CDC20, MCM2, CDC25C, MCM4, CDC25A,
MCM6, CCNB1, MAD2L1, CCNB2, PLK1, BUB1B |
| KEGG_PATHWAY | hsa03030:DNA
replication | 13 |
P=4.28×10−6 |
5.26×10−4 | POLD3, PRIM1, DNA2,
RFC4, POLE2, POLD1, PRIM2, POLA1, MCM2, POLA2, MCM4, RPA4,
MCM6 |
| KEGG_PATHWAY | hsa03440:Homologous
recombination | 11 |
P=1.95×10−5 |
1.60×10−3 | POLD3, XRCC3,
XRCC2, BLM, POLD1, BRCA2, RAD54B, RAD54L, RPA4, RAD51, TOP3B |
| KEGG_PATHWAY |
hsa04640:Hematopoietic cell lineage | 19 |
P=3.55×10−5 |
2.18×10−3 | CSF2, IL9R, IL2RA,
KITLG, ITGA2, CD1B, CD1A, ITGA3, ITGA4, ITGAM, FCGR1A, CD22, IL1B,
CSF3R, CD24, IL1A, CSF2RA, EPO, CD7 |
| KEGG_PATHWAY |
hsa04060:Cytokine-cytokine receptor
interaction | 35 |
P=1.20×10−4 |
5.87×10−3 | CSF2, IL9R, CXCL5,
IL21R, TNFSF15, TNFRSF8, IL15, TGFB2, LIF, TNFRSF11A, IL12RB1,
IL17B, CCL20, CLCF1, CCR10, CSF3R, CSF2RB, IL1B, IL2RG, LTA,
CSF2RA, IL1A, EPO, TNFSF4, IL2RA, TNFSF9, CCR8, AMH, TNFRSF10C,
ACVR2B, CCR6, IL20RA, IL12A, EDA, XCL1 |
| KEGG_PATHWAY | hsa05200:Pathways
in cancer | 48 |
P=3.17×10−4 |
1.29×10−2 | E2F1, FGFR2, E2F2,
FGFR1, E2F3, ADCY7, MMP9, ARNT2, MITF, EGLN3, KITLG, LPAR2, BDKRB1,
MMP1, TGFB2, GLI1, EDNRA, CCNE2, CCNE1, IGF1R, CDKN2A, CDKN2B,
PAX8, SLC2A1, TGFA, CSF3R, EGF, PLCB1, TRAF5, CSF2RA, PIK3R2,
CTBP2, PTGER4, MSH2, PIK3CD, CBL, FZD1, ITGA2, BRCA2, ITGA3, BIRC5,
FZD2, MECOM, FZD7, RAD51, COL4A5, LAMC2, GNB3 |
| KEGG_PATHWAY |
hsa04914:Progesterone-mediated oocyte
maturation | 16 |
P=1.21×10−3 |
4.19×10−2 | ANAPC1, CDK1,
ADCY7, PIK3CD, PKMYT1, CDC25C, CDC25A, CCNB1, IGF1R, CCNB2, MAD2L1,
PLK1, MAPK13, BUB1, CCNA2, PIK3R2 |