Introduction
Laryngeal carcinoma is a common head and neck
malignancy, and squamous cell carcinoma is the most frequent
histological type of laryngeal carcinoma, accounting for 98% of
cases (1,2). The main risk factors for laryngeal
carcinoma are tobacco smoking and alcohol consumption, and the
roles of these risk factors have been reported by a large number of
studies (3,4). While much is known regarding laryngeal
carcinoma, the molecular mechanism of the disease is yet to be
elucidated.
Insulin-like growth factor 1 (IGF-1) signaling is
mediated via the IGFR1 receptor (IGF1R), which is an important
growth regulatory pathway, and the enhanced activation of this
pathway is indicated to serve a crucial role in cancer cell
proliferation, migration and apoptosis (5–7).
Furthermore, IGF1R is aberrantly expressed in numerous types of
cancer cells, including laryngeal carcinoma (8).
IGF1R antisense imprinted non-protein coding RNA
(IRAIN) is a newly reported antisense imprinted non-coding RNA in
the IGF1R locus, which has an important role in the regulation of
IGF-1 signaling (9). IRAIN is
transcribed from an intronic promoter with antisense orientation
and with a parent-of-origin genetic manner (9,10). The
present study examined a single nucleotide polymorphism (SNP) of
IRAIN (rs8034564), and revealed the ‘A’ genotype was favorably
imprinted over the ‘G’ genotype, resulting in imbalanced expression
in the two parental alleles (10).
IRAIN has been shown to be downregulated in leukemia cell lines and
fresh blood cells of patients with high-risk acute myeloid leukemia
(AML) (9), and is downregulated in
breast cancer as an imprinted gene (10). Another previous study reported that
IRAIN was significantly increased in non-small cell lung carcinoma
(NSCLC) tissues, was related to tumor size and smoking status and
that downregulation of IRAIN suppressed the proliferation of NSCLC
cells via blocking cells in G1 phase (11). Pashaiefar et al demonstrated,
in 64 de novo non-M3 patients with AML compared with 51
healthy controls that poor prognosis with low IRAIN expression
tended to have a higher white blood cell count and blast count, and
had markedly shorter overall survival (OS) and relapse-free
survival (RFS; P=0.044 and 0.009, respectively); in addition, the
study found that patients with a refractory response to
chemotherapy and those with subsequent relapse, had lower initial
IRAIN expression and multivariate analysis identified IRAIN
transcript expression as an independent prognostic factor for OS
and RFS (12). Previous studies have
reported that the IGF1R pathway is frequently dysregulated in
laryngeal carcinoma (8,13); however, it has not been determined
whether IRAIN serves a role in laryngeal carcinoma. Therefore, the
aim of the present study was to investigate whether IRAIN was
differently expressed in laryngeal carcinoma tissue, paracancerous
tissue and healthy pharynx tissue.
Materials and methods
Patients
Patients with laryngeal carcinoma treated at The
Second Affiliated Hospital of Jilin University between August 2014
and August 2017 were recruited for this study. The study was
approved by the Research Ethics Committee of The Second Affiliated
Hospital of Jilin University, and written informed consent was
obtained from all patients.
Laryngeal carcinoma tissue and adjacent
non-malignant tissue (distance from the malignant tissue, 0.5–1.5
cm) was collected from 37 patients who received laryngectomy (some
tissues were too small to cover all the assays, thus the sample
number varied in each assay). The patient cohort consisted of 33
males and 4 females with an age range of 43–79 years old and a mean
age of 60.2 years. In addition, healthy pharynx tissue was
collected from 6 healthy individuals who received
uvulopalatopharynoplasty. All tissue specimens were collected
before patients had any radiotherapy and chemotherapy. After
excision, all tissue samples were immediately stored at −80°C until
use.
Histological type and TNM classifications were
determined according to the National Comprehensive Cancer Network
(NCCN) criteria (14). All malignant
tissue samples were examined by a pathologist, and determined to be
epithelial squamous cell carcinoma. The clinical and pathological
characteristics of the patients are presented in Table I.
 | Table I.Basic characteristics and
pathological features of patients (n=37) with laryngeal carcinoma
involved in the experiment. |
Table I.
Basic characteristics and
pathological features of patients (n=37) with laryngeal carcinoma
involved in the experiment.
| Characteristic | Patients |
|---|
| Age, years | 61 (43–79) |
| Sex |
|
|
Male | 33 (89.2%) |
|
Female | 4 (10.8%) |
| TNM stage |
|
| T1-T2
stage | 15 (40.5%) |
| T3-T4
stage | 22 (59.5%) |
| Histological
type |
|
| Highly
differentiation | 12 (32.4%) |
| Medium
and low differentiation | 25 (67.6%) |
| Lymph nodes
metastasis |
|
|
Yes | 11 (29.7%) |
| No | 26 (70.3%) |
| Clinical
classification |
|
| Glottic
carcinoma | 19 (51.4%) |
|
Supraglottic carcinoma | 18 (48.6%) |
Evaluation of genomic imprinting
Genomic DNA (gDNA) PCR product gene
sequencing
gDNA was extracted using the Animal Tissue Genomic
DNA Extraction kit (Beijing Zoman Biotechnology Co., Ltd.),
following the manufacturer's instructions. Primers were designed
according to the known SNP sites (10) (rs8034564; A/G) in the IRAIN
expression region reported in the National Center for Biotechnology
Information (9,10). The IRAIN forward (JH248) and reverse
(JH781) sequences (Table II) were
selected for target fragment amplification. Target fragments were
obtained using 2X Taq PCR MasterMix (Tiangen Biotech Co., Ltd.),
according to the manufacturer's instructions. PCR reaction
condition was as follows: Initial denaturation at 94°C for 3 min,
then IRAIN DNA was amplified for 30 cycles at 94°C for 30 sec, 55°C
for 30 sec of annealing and 72°C for 1 min of extension, with a
final extension at 72°C for 5 min. PCR products (5 µl/well) from
each sample in sequence underwent electrophoresis for 20 min at 120
V on 2% agarose gel in an electrophoresis tank containing 1X TAE
electrophoresis solution (Beijing Zoman Biotechnology Co., Ltd.);
ethidium bromide was used to image the agarose gels. After
electrophoresis, the gels were observed under a Tanon-4200 Gel
Imaging System (Tanon Science and Technology Co., Ltd.). A bright
electrophoresis band with clear edges was visible at ~450 bp, and
the target strip was excised from the gel and purified using a
SGMag Gel DNA Purification kit (Sangon Biotech Co., Ltd.), and sent
to the Shanghai Sangon Biotech Co., Ltd. for gene sequencing.
Complementary DNA (cDNA) PCR product gene sequencing was performed
if the SNP site in the sequencing results was A/G.
 | Table II.List of all primers. |
Table II.
List of all primers.
| A, Examination of
genomic imprinting |
|---|
|
|---|
| Forward primer no.,
sequence | Reverse primer no.,
sequence |
|---|
| IRAIN JH248,
5′-CGACACATGGTCCAATCACTGTT-3′ | IRAIN JH249,
5′-AGACTCCCCTAGGACTGCCATCT-3′ |
| IRAIN JH780,
5′-GTTTCCGCAGTAGCCGCTGAT-3′ | IRAIN JH781,
5′-CTGCGGGTCTCCGAAGCTC-3′ |
|
| B, Reverse
transcription-quantitative PCR |
|
| Forward primer
no., sequence | Reverse primer
no., sequence |
|
| IRAIN LJ21,
5′-CGATGGATACACGTTCTAATGC-3′ | IRAIN LJ22,
5′-CAAACAATCGGGTAGGATGG-3′ |
| β-actin J880,
5′-AGATCAAGATCATTGCTCCTCCTGA-3′ | β-actin J881,
5′-ATACTCCTGCTTGCTGATCCACATC-3′ |
| IGF1R JH217,
5′-GAAGTCTGGCTCCGGAGGAGGGTC-3′ | IGF1R JH218,
5′-ATGTGGAGGTAGCCCTCGATCAC-3′ |
|
| C, DNA
methylation analysis |
|
| Forward primer
no., sequence | Reverse primer
no., sequence |
|
| IRAIN JH852,
5′-TYGGGGGATGGAGGGGTATTAGGGT-3′ | IRAIN JH853,
5′-ACAAACRTCTAAATATCCCCRTAAAAC-3′ |
cDNA PCR product gene sequencing
Total RNA was extracted from frozen tissue samples
using an AxyPrep Total RNA Extraction kit (Corning, Inc.) following
the manufacturer's protocol. For reverse transcription (RT), 500 ng
total RNA was converted to cDNA using the Universal RT-PCR kit
(Beijing Dingguo Changsheng Biotechnology Co., Ltd.) according to
the manufacturer's protocol. Allelic expression of IRAIN was
examined via PCR (Universal RT-PCR kit from Beijing Dingguo
Changsheng Biotechnology Co., Ltd., the DNA polymerase was part of
this kit.) in cDNA samples using primers specific for polymorphic
restriction enzymes; the target fragments IRAIN JH780 and JH781
were selected for amplification (Table
II). PCR reaction conditions and the electrophoresis method
were the same as those used for gDNA PCR products. A bright
electrophoresis band with clear edges was observed between 100–200
bp. The target strip was excised and purified according to the
experimental method described previously article (10), and sent to the Sangon Biotech Co.,
Ltd. for gene sequencing.
Gene expression and reverse-transcription
quantitative (q)PCR
Total RNA was extracted from frozen tissue samples
(laryngeal carcinoma tissues, paired adjacent non-malignant tissues
and healthy pharynx tissues) using an AxyPrep Total RNA Extraction
kit (Corning, Inc.) according to the manufacturer's protocol. The
TransScript® All-in-One First-Strand cDNA Synthesis
Super Mix for qPCR kit (Beijing Transgen Biotech Co., Ltd.) was
used to synthesize cDNA; for first-strand synthesis, RNA was
combined with 5X TransScript® All-in-One Super Mix, gDNA
Remover and RNase-free water and incubated at 42°C for 15 min and
then at 85°C for 5 sec. For qPCR, cDNA samples were amplified using
a RocheLightCycler 480II RT system (Roche Diagnostics) and
TransStart® Top Green qPCR SuperMix kit (Beijing
Transgen Biotech Co., Ltd.). According to manufacturer's protocols,
qPCR was performed as follows: Initial denaturation at 94°C for 30
sec, followed by 40 cycles at 94°C for 5 sec and 60°C for 30 sec.
Each sample was analyzed in triplicate and data are presented as
mean ± SD. mRNA expression levels of IRAIN and IGF1R were
quantitated via normalizing to β-actin housekeeping gene (Table II; forward primer, J880 and reverse,
J881) according to the method previously described in the present
study. The PCR primers used for qPCR were: i) IRAIN, forward LJ21
and reverse LJ22; ii) IGF1R, forward JH217 and reverse JH218; and
iii) β-actin, forward J880 and reverse J881 (Table II). The data was analyzed by using
2−ΔΔCq method (15).
DNA methylation analysis
gDNA was extracted from tumors using the Animal
Tissue Genomic DNA Extraction kit (Beijing Zoman Biotechnology Co.,
Ltd.). gDNA (2 µg) was treated with sodium bisulfite [130 µl CT
Conversion reagent of EZ DNA Methylation-Gold kit (Zymo Research
Corp.)], and unmethylated cytosine was converted into uracil, while
methylated cytosine remained as cytosine. Genomic DNA was used for
bisulpite conversion with an EZ DNA Methylation-Gold kit (Zymo
Research Corp.). The following steps were performed: 98°C for 10
min, 64°C for 2.5 h and then 4°C storage for upto 20 h according to
the manufacturer's instructions. Then, bisulfite sequencing PCR
(BSP) primers were designed, and during PCR amplification all
uracil was converted to thymine. PCR products were sequenced to
determine if there was methylation of CpG sites using the
BSP-direct sequencing method (10).
PCR reactions were performed using Kantaq1 DNA polymerase (Ab
Peptides, Inc.). BSP was used to analyze DNA methylation status.
The PCR conditions were: Initial denaturation at 97°C for 10 min,
followed by 35 cycles at 96°C for 20 sec, annealing at 64°C for 30
sec, extension at 72°C for 30 sec and completion of the reaction at
72°C for 10 min. The primers used for assessing target fragment DNA
methylation were forward JH852 and reverse JH853 (Table II). PCR products were separated via
2% agarose gel electrophoresis and purified with an Axygen DNA Gel
Extraction kit (Axygen; Corning, Inc.). The success rate of
sequencing can be improved by cloning PCR products into a vector
for sequencing (10). The PCR
products were cloned into a pUCm-T vector (Sangon Biotech Co.,
Ltd.) using a PCR Cloning kit (cat. no. B522213; Sangon Biotech
Co., Ltd.) and white colonies grown on IPTG/X-gal indicator plates
were selected and the sequenced for analysis of CpG methylation as
previously described (9,10). The status of DNA methylation varies,
depending on cell types, microenvironment, gene activity and drug
treatment (10). Thus, no known
positive controls could be added in the assay for laryngeal cancer
tissues.
Statistical analysis
Categorical variables are presented as absolute (n)
and relative (%) frequencies, while continuous variables are
presented as medians (min, max). Each sample was analyzed in
triplicate and data are presented as mean ± SD. All data
calculations and statistical testing was performed using SPSS
version 22.0 statistical software (IBM Corp.). Differences between
groups were compared using the Student t-test or the Wilcoxon test
and corrected by Bonferroni test, P<0.05 was considered to
indicate a statistically significant difference.
Results
Non-imprinted expression of IRAIN in
healthy pharynx tissue, laryngeal carcinoma tissue and
paracancerous tissue
To determine whether IRAIN uses a similar epigenetic
mechanism to regulate genes locally in laryngeal carcinoma, it was
examined if IRAIN lncRNA was mono-allelically expressed in healthy
pharynx tissue, laryngeal carcinoma and paracancerous tissue, which
are heterozygous for the polymorphic Nde1 restriction site. In
total, two alleles, termed ‘A’ allele and ‘G’ allele, were detected
in both gDNA and corresponding cDNA samples (Tables III and IV). This indicated that IRAIN lncRNA
expression was non-imprinted in healthy pharynx tissue, laryngeal
carcinoma tissue and paracancerous tissue.
 | Table III.Expression of ‘A’ and ‘G’ in
laryngeal cancer tissue samples and paracanerous tissues. |
Table III.
Expression of ‘A’ and ‘G’ in
laryngeal cancer tissue samples and paracanerous tissues.
| Cancer tissue
no. | A:G | Ratio | Paracancerous
tissue no. | A:G | Ratio |
|---|
| CA 1 | 640:547 | 1.170 | PC1 | 724:509 | 1.422 |
| CA 2 | 313:405 | 0.773 | PC2 | 321:408 | 0.787 |
| CA 3 | 661:501 | 1.320 | PC3 | 648:536 | 1.209 |
| CA 4 | 337:384 | 0.878 | PC4 | 713:446 | 1.599 |
| CA 5 | 724:544 | 1.331 | PC5 | 634:536 | 1.183 |
| CA 6 | 705:454 | 1.553 | PC6 | 667:539 | 1.237 |
| CA 7 | 544:667 | 0.816 | PC7 | 686:520 | 1.320 |
| CA 8 | 689:522 | 1.320 | PC8 | 640:522 | 1.226 |
| CA 9 | 610:610 | 1.000 | PC9 | 683:544 | 1.256 |
| CA 10 | 653:495 | 1.319 | PC10 | 302:405 | 0.746 |
| CA 11 | 770:48 | 1.598 | PC11 | 599:476 | 1.258 |
| CA 12 | 648:522 | 1.241 | PC12 | 637:541 | 1.177 |
| CA 13 | 898:495 | 1.814 | PC13 | 683:525 | 1.301 |
| CA 14 | 479:705 | 0.679 | PC14 | 672:571 | 1.177 |
| CA 15 | 618:566 | 1.092 | PC15 | 582:618 | 0.942 |
| CA 16 | 552:501 | 1.102 | PC16 | 571:482 | 1.185 |
| CA 17 | 321:419 | 0.766 | PC17 | 324:384 | 0.844 |
 | Table IV.Expression of ‘A’ and ‘G’ in healthy
pharynx tissues. |
Table IV.
Expression of ‘A’ and ‘G’ in healthy
pharynx tissues.
| Healthy pharynx
tissue no. | A:G | Ratio |
|---|
| N1 | 354:438 | 0.808 |
| N2 | 364:394 | 0.924 |
| N3 | 373:413 | 0.903 |
| N5 | 356:408 | 0.873 |
| N6 | 373:416 | 0.897 |
Then, the allelic expression of IRAIN lncRNA in
laryngeal carcinoma tissue and paracancerous tissue was
investigated using SNP rs8034564 to distinguish the two parental
alleles. As this SNP does not contain a restriction enzyme site,
PCR sequencing was used to determine the allelic expression of
IRAIN. The gDNAs of laryngeal carcinoma tissues that were
heterogeneous for this SNP, including both the ‘A’ and ‘G’ alleles,
were selected for analysis. The gDNA PCR products of 32 laryngeal
carcinoma tissues and 6 healthy pharyngeal mucosa tissues were
sequenced (sent to the Shanghai Sangon Biotech Co., Ltd. for gene
sequencing). The SNP site (rs8034564) of 17 laryngeal carcinoma
tissues and five healthy pharyngeal mucosa tissues were A/G
heterozygous. SNP sequencing was then performed on the cDNA of
laryngeal carcinoma tissue and paracancerous tissue of these
patients who were heterozygous. However, it was found that the ‘A’
and ‘G’ alleles were detected in all corresponding cDNA samples
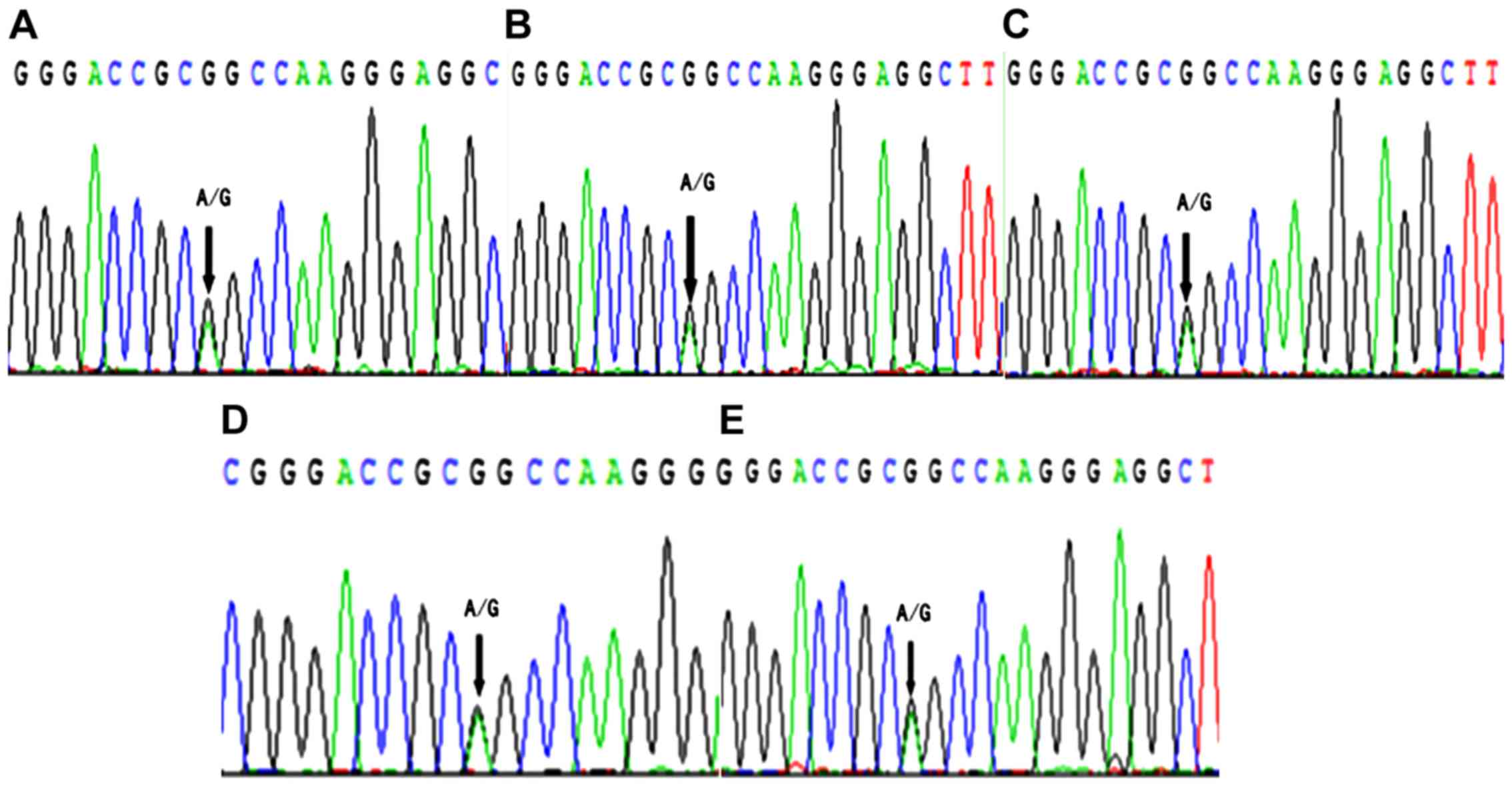
(Fig. 1). This finding suggested
that IRAIN was non-imprinted in laryngeal carcinoma tissue, which
was different compared with that in leukemia cells (9) and breast cancer tissue (10).
Allelic expression imbalance and
allelic-switch of IRAIN in laryngeal cancer
AEI was used to detect the expression difference of
two alleles in the same individual via a heterozygous SNP site. An
allelic expression ratio (AER) <0.8 or >1.2 are regarded as
evidence of AEI (16,17).
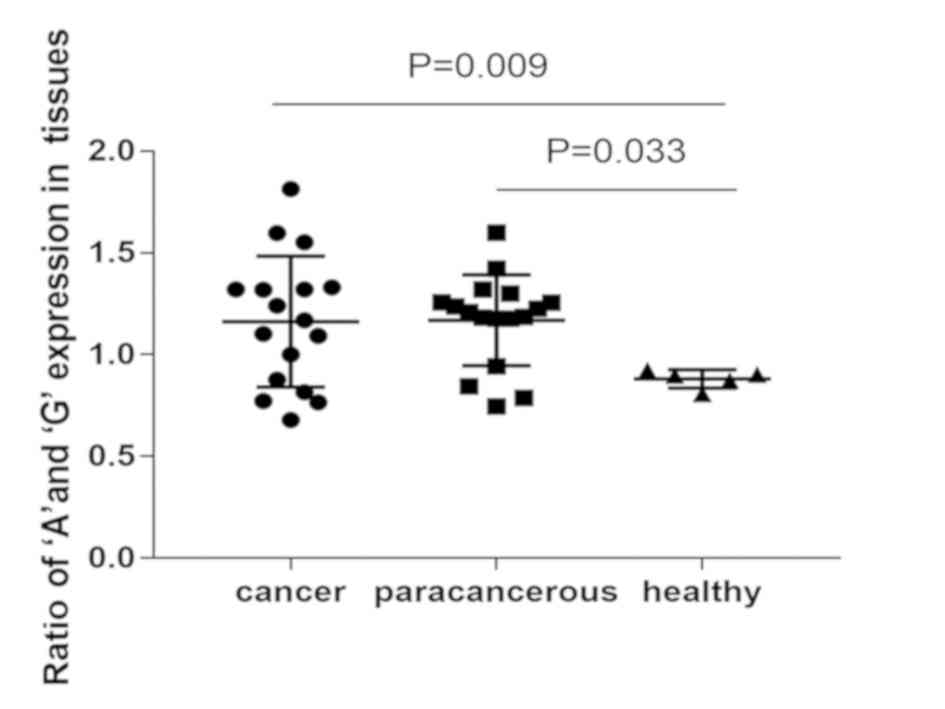
The results indicated that the expression of the ‘A’
allele was favored over the ‘G’ allele in laryngeal cancer tissue
samples, and their adjacent tissue counterparts (Table III). However, the expression of the
‘G’ allele was favored over the ‘A’ allele in healthy pharynx
tissue (Table IV). That suggested
IRAIN can cause A:G allelic switch in laryngeal cancer. The A/G
value between cancer tissue and healthy pharynx tissue,
paracancerous tissue and healthy pharynx tissue were significantly
different (P=0.009, P=0.033 respectively), whereas the value
between cancer tissue and paracancerous tissue not significantly
different (Fig. 2).
Decreased expression of IRAIN in
laryngeal cancer types
To evaluate the role of IRAIN in laryngeal cancer,
31 paired laryngeal carcinoma tissues and paracancerous tissue were
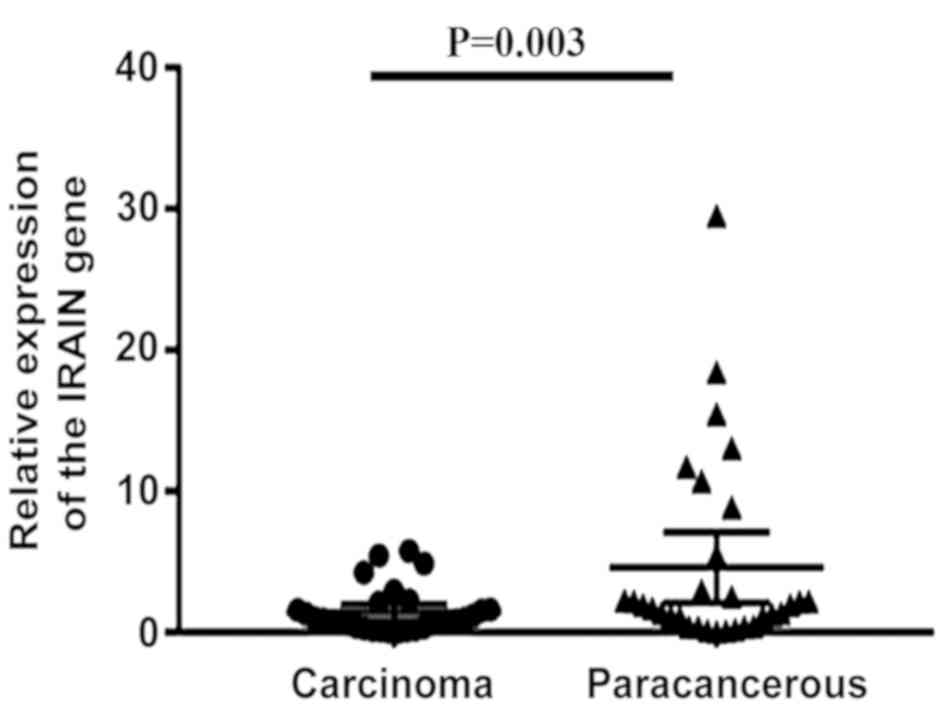
analyzed for IRAIN expression using RT-qPCR. It was identified that
IRAIN expression was significantly decreased in laryngeal cancer
tissue (P=0.003; Fig. 3). Next, the
association between IRAIN expression and clinicopathological
parameters, including age, histological type, tumor stage, tumor
grade and lymph node metastasis, was examined. The analysis
indicated that there was no association between IRAIN expression
and age, histological type, tumor stage, tumor grade and lymph node
metastasis (P>0.05; Table
III).
DNA methylation in the IRAIN
promoter
The IRAIN promoter is rich in CpG dinucleotides, and
in peripheral blood leucocytes, the promoter CpG islands are
semi-methylated (9). Compared with
peripheral blood leucocytes, Kang et al (10) reported there was aberrant DNA
methylation in the IRAIN promoter in breast cancer tissue
specimens. The present study analyzed DNA methylation in the IRAIN
promoter of laryngeal carcinoma tissue and paracancerous tissue. It
was found that the IRAIN promoter was almost completely
unmethylated in one paracancerous tissue sample (Fig. 4A). Moreover, the IRAIN promoter was
almost completely unmethylated in one laryngeal cancer sample
(Fig. 4D). In the other laryngeal
cancer samples (Fig. 4B and C), the
IRAIN promoter was totally unmethylated, as was seen in the
laryngeal carcinoma adjacent tissue. In the laryngeal cancer
tissues of three patients (Fig.
4B-D), which contained 13 genes CpG identified via bisulfite
sequencing method, it was found that the laryngeal cancer tissue
IRAIN promoter region was completely unmethylated or almost
completely unmethylated. Therefore, it was suggested that DNA
methylation was not involved in regulation of laryngeal cancer
tissues IRAIN genetic imprinting.
Discussion
lncRNAs are non-protein-coding RNA transcripts
>200 nucleotides, and are emerging as key regulators of diverse
cellular processes (18), such as
lncRNA-Xist, lncRNA-HOTAI, lncRNA-H19 and lncRNA-CRNDE. The
discovery of lncRNAs as biologically relevant molecules has led to
a rethinking of the central dogma of biology and revealed novel
cellular and molecular complexities; it is now known that numerous
genes can encode both mRNA and lncRNA (19).
Previous studies have reported that lncRNAs serve
important functions in the regulation of gene expression and other
biological processes, and are dysregulated in tumorigenesis
(20). For example, lncRNAs have
been identified to exert important roles in tumor recurrence,
including X inactive specific transcript, metastasis associated
lung adenocarcinoma transcript 1, HOX transcript antisense RNA,
heart and neural crest derivatives expressed 2 and LINC00675. In
addition, increased expression of lncRNA H19 imprinted maternally
expressed transcript (H19) has been observed in various primary and
metastatic tumors, including breast cancer (21), gastric cancer (22), liver cancer (23) and bladder cancer (24). Another previous study has reported
that lncRNA H19 promoted laryngeal squamous cell carcinoma
progression via microRNA (miR)-148a-3p and DNA methyltransferase 1
(25). It has also been shown that
colon cancer-associated transcript-1 (26), taurine upregulated 1 (27) and nuclear paraspeckle assembly
transcript 1 (28) are involved in
the progression of laryngeal squamous cell cancer, and are
associated with TNM stage, tumor size, overall survival and growth
regulation of laryngeal squamous cell cancer. Collectively,
previous studies have suggested that the identification and
investigation of cancer-associated lncRNAs may result in the
development of early diagnostic marker and new prognostic
biomarkers or treatments for various malignancies, including
laryngeal carcinoma.
Dysregulation of IGF1R has been implicated in
the progression of different malignancies, and resistance to
treatments, such as breast cancer (10), squamous-cell laryngeal cancer
(8) and hematopoietic malignancies
(9). A previous study developed and
validated a multigene predictor of recurrence in early laryngeal
cancer (29); the study identified a
panel of genes related to the IGF1R pathway that were able to
discriminate patients with a poor and a favorable prognosis
(P<0.0001 in the training set; P= 0.0001 in the validation set).
It has also been shown that IGF1R-α protein upregulation may serve
as an independent predictor of recurrence and survival in operable
laryngeal cancer (8). Another study
suggested that downregulation of miR-375 was one of the molecular
mechanisms responsible for the progression of laryngeal carcinoma
via targeting IGF1R directly, and affecting its downstream AKT
signaling pathways (13). IRAIN
is a novel lncRNA within the IGF1R locus that is
transcribed in an antisense direction from an intronic promoter,
with a full-length transcript of 5.4 kb (10). IRAIN has been revealed to serve a
role in AML (9–12), breast cancer (10), NSCLC (11), renal cell carcinoma (30) and pancreatic cancer (31).
In mice, the gene transcribing IGF2R is associated
with a long non-coding (lnc)RNA, antisense of IGF2R non-protein
coding RNA (Airn). These transcripts are reciprocally imprinted,
with Airn transcribed from the paternal allele only, and the
transcription of the antisense lncRNA Airn regulates in cis the
allelic expression of the IGF2R coding RNA (32–35). In
addition, Sun et al (9)
reported that in leukemia cells IRAIN was expressed solely from the
paternal allele.
In the present study, it was demonstrated that IRAIN
exhibited non-imprinted expression in laryngeal carcinoma tissue,
healthy pharynx tissue and paracancerous tissue. However, these
results are contrary to those of previous studies examining IRAIN
in AML and breast cancer (9,10). These different findings may be
because lncRNAs exhibits marked tissue-specific expression
patterns, as compared with protein-coding genes, and imprinted
genes develop acquired instability (36,37).
If a gene is expressed equally from both alleles
(bi-allelically), equivalent levels of each allele of the tracer
SNP will be detected in mRNA transcripts (16); this is referred to as ‘balanced
allelic expression’. When the expression of one allele is altered
due to a pathological defect, such as the presence of aberrant
promoter methylation or a frameshift mutation that results in
nonsense-mediated mRNA decay, the level of one allele will exceed
the other in the tracer SNP within the corresponding mRNA; this is
generally referred to as ‘allelic expression imbalance’ (AEI)
(16,17). AEI was used to detect the expression
difference of two alleles in the same individual using heterozygous
SNPs in the current study. SNPs within a gene of interest may be
used to distinguish between two genetic alleles, and investigate
their features in heterozygous individuals (16). With regards to cancer etiology and
development, identification of alleles and the detection of allelic
imbalances, such as transcriptional loss from one allele or
loss-of-heterozygosity due to deletion of one allele, within a
tumor are particularly useful (17).
Moreover, it has been reported that an AER <0.08 or >1.2 is
consistent with AEI (17,38). The present results indicated that the
lncRNA-IRAIN appeared to favor the ‘A’ genotype expression in
laryngeal cancer tissue and paracancerous tissue. The expression of
the ‘A’ allele was greater compared with the ‘G’ allele in 71% of
the 17 heterozygous laryngeal carcinoma tissue specimens examined.
The same result was found in the examination of paracancerous
tissue specimens (cA>G; rs8034564). However, the expression of
the ‘G’ allele was favored over the ‘A’ allele in healthy pharynx
tissue specimens (cA<G, rs8034564). These results demonstrated
that the expression of IRAIN lncRNA switches to the alternate
parental allele in laryngeal carcinoma tissue and paracancerous
tissue. However, the mechanisms underlying the AEI and
allele-switch in laryngeal carcinoma remains unknown. It was
hypothesized that the mechanism may be associated with the two
characteristics of lncRNA: lnc RNA expression possesses cell or
tissue type specificity and lncRNA is less conserved compared with
mRNA. Moreover, whether the AEI and aberrant allelic switch affects
the activity of the IGF1R signaling pathway in laryngeal carcinoma
is yet to be elucidated, and thus requires further research in the
future.
The current results indicated that IRAIN expression
was decreased in laryngeal carcinoma tissue, which is consistent
with the findings of previous studies of AML (9,12),
breast cancer (10) and renal cell
carcinoma (30). However, the
results are contrary to the findings of a study in NSCLC (11); this may be due to the fact that
lncRNAs have tissue-specific expression patterns, which is
different compared with the expression patterns of protein-coding
genes (36,37). The present study also identified no
associations between IRAIN expression and clinical characteristics,
such as age, histological type, tumor stage and grade and lymph
node metastasis. According these results, it was suggested that
IRAIN may exert an important role in laryngeal carcinoma
development and progression. Furthermore, IRAIN may serve as a
novel tumor suppressing lncRNA and as an early diagnostic marker in
laryngeal cancer.
Mammals are diploid organisms whose cells possess
two matched sets of chromosomes, one inherited from the mother and
one from the father (10,39). Thus, mammals have two copies of every
gene. Normally, both the maternal and paternal copy of each gene
have the same potential to be active in any cell (39). Genomic imprinting is an epigenetic
mechanism that changes this potential by restricting the expression
of a gene to one of the two parental chromosomes (9,10,39). DNA
methylation could potentially perform two different functions in
genomic imprinting (39); for
example, it could act as the imprinting mark via being acquired
de novo only by the chromosomes in one gamete (39). Moreover, DNA methylation could serve
to silence one of the parental alleles, as it is associated with
gene repression (40). In the
current study, it was demonstrated that the IRAIN promoter was
unmethylated in one paracancerous tissue specimen and in three
laryngeal carcinoma tissue specimens. Thus, the results indicated
that DNA methylation was not involved in gene imprinting expression
in the promoter region of IRAIN; however, these findings may
provide a reference for further studies regarding lncRNA and the
pathogenesis of laryngeal cancer.
In conclusion, the present results suggested that
IRAIN expression was significantly decreased in laryngeal carcinoma
tissue, and that IRAIN undergoes AEI and aberrant allelic switching
in laryngeal cancer. However, the present study had several
limitations. Firstly, the small sample size of laryngeal carcinoma
tissues and paracanerous tissues due to difficulty of collection.
Secondly, the number of individuals from whom healthy pharynx
tissue was obtained was lower compared to the patients from whom
laryngeal carcinoma and paracancerous tissues were obtained.
Thirdly, in the present study, the number of female patients were
lower compared to male patients leading to a sex bias. Additional
studies are required to further verify the current findings and
investigate the unanswered questions, including whether AEI and the
aberrant allele-switch of IRAIN lncRNA serve an important role in
the development of laryngeal cancer? If IRAIN lncRNA could be used
to predict the effectiveness of IGF1R targeted therapies? Or
whether IRAIN lncRNA can be used to as a potential therapeutic
target in laryngeal carcinoma treatment? Furthermore, it would be
also be of interest to investigate whether AEI and the aberrant
allelic switching of IRAIN lncRNA can be used to assess laryngeal
cancer risk.
Acknowledgements
Not applicable.
Funding
This study was funded by the Natural Science Funds
of Jilin Province, China (grant no. 20160101055JC).
Availability of data and materials
The datasets used in the present study are available
from the corresponding author upon reasonable request.
Authors' contributions
JW and MZ conceived and designed the research
methods, as well as drafted and revised the manuscript. JW and YM
performed the experiments. JW, DL and YG analyzed the data and
prepared the figures. JW assisted with the analysis of the data and
interpreted the results of the experiments. All authors approved
the final version of the manuscript, as well as agree to be
accountable for all aspects of the research and for ensuring the
accuracy and integrity of the study.
Ethics approval and consent to
participate
The experimental protocol was approved by the Ethics
Committee of the Second Hospital of Jilin University, and written
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Haapaniemi A, Koivunen P, Saarilahti K,
Kinnunen I, Laranne J, Aaltonen LM, Närkiö M, Lindholm P, Grénman
R, Mäkitie A, et al: Laryngeal cancer in Finland: A 5-year
follow-up study of 366 patients. Head Neck. 38:36–43. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Steuer CE, El-Deiry M, Parks JR, Higgins
KA and Saba NF: An update on larynx cancer. CA Cancer J Clin.
67:31–50. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gong H, Shi Y, Xiao X, Cao P, Wu C, Tao L,
Hou D, Wang Y and Zhou L: Alterations of microbiota structure in
the larynx relevant to laryngeal carcinoma. Sci Rep. 7:55072017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rudolph E, Dyckhoff G, Becher H, Dietz A
and Ramroth H: Effects of tumour stage, comorbidity and therapy on
survival of laryngeal cancer patients: A systematic review and a
meta-analysis. Eur Arch Otorhinolaryngol. 268:165–179. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bach LA: IGF-binding proteins. J Mol
Endocrinol. 61:T11–T28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jal L, Ram M, Akl R, van der Zee AGJ and
de Jong S: IGF system targeted therapy: Therapeutic opportunities
for ovarian cancer. Cancer Treat Rev. 60:90–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Winder T, Zhang W, Yang D, Ning Y, Bohanes
P, Gerger A, Wilson PM, Pohl A, Mauro DJ, Langer C, et al: Germline
polymorphisms in genes involved in the IGF1 pathway predict
efficacy of cetuximab in wild-type KRAS mCRC patients. Clin Cancer
Res. 16:5591–5602. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mountzios G, Kostopoulos I, Kotoula V,
Sfakianaki I, Fountzilas E, Markou K, Karasmanis I, Leva S,
Angouridakis N, Vlachtsis K, et al: Insulin-like growth factor 1
receptor (IGF1R) expression and survival in operable squamous-cell
laryngeal cancer. PLoS One. 8:e540482013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun J, Li W, Sun Y, Yu D, Wen X, Wang H,
Cui J, Wang G, Hoffman AR and Hu JF: A novel antisense long
noncoding RNA within the IGF1R gene locus is imprinted in
hematopoietic malignancies. Nucleic Acids Res. 42:9588–9601. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kang L, Sun J, Wen X, Cui J, Wang G,
Hoffman AR, Hu JF and Li W: Aberrant allele-switch imprinting of a
novel IGF1R intragenic antisense non-coding RNA in breast cancers.
Eur J Cancer. 51:260–270. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng J, Sun Y, Zhang EB, Lu XY, Jin SD and
Guo RH: A novel long noncoding RNA IRAIN regulates cell
proliferation in non small cell lung cancer. Int J Clin Exp Pathol.
8:12268–12275. 2015.PubMed/NCBI
|
|
12
|
Pashaiefar H, Izadifard M, Yaghmaie M,
Montazeri M, Gheisari E, Ahmadvand M, Momeny M, Ghaffari SH,
Kasaeian A, Alimoghaddam K and Ghavamzadeh A: Low expression of
long noncoding RNA IRAIN is associated with poor prognosis in
Non-M3 acute myeloid leukemia patients. Genet Test Mol Biomarkers.
22:288–294. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo J, Wu J, Li Z, Qin H, Wang B, Wong TS,
Yang W, Fu QL and Lei W: miR-375 suppresses IGF1R expression and
contributes to inhibition of cell progression in laryngeal squamous
cell carcinoma. Biomed Res Int. 2014:3745982014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
National Comprehensive Cancer Network
(NCCN), . NCCN Clinical Practice Guidelines in Oncology-Head and
Neck Cancers. Version 1.2018. NCCN, Plymouth Meeting. (PA).
2018.
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kwok CT and Hitchins MP: Allele
quantification Pyrosequencing® at designated SNP sites
to detect allelic expression imbalance and Loss-of-Heterozygosity.
Methods Mol Biol. 1315:153–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang ZM, Wang DZ and Shi JX: Detection and
application of allelic expression imbalance. Zhonghua Yu Fang Yi
Xue Za Zhi. 45:9–11. 2011.(In Chinese). PubMed/NCBI
|
|
18
|
Sun M and Kraus WL: From discovery to
function: The expanding roles of long noncoding RNAs in physiology
and disease. Endocr Rev. 36:25–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Grelet S, Link LA, Howley B, Obellianne C,
Palanisamy V, Gangaraju VK, Diehl JA and Howe PH: A regulated PNUTS
mRNA to lncRNA splice switch mediates EMT and tumour progression.
Nat Cell Biol. 19:1105–1115. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J,
Wei M, Xu C, Wu C, Zhang Z, et al: Long non-coding RNA metastasis
associated in lung adenocarcinoma transcript 1 derived miniRNA as a
novel plasma-based biomarker for diagnosing prostate cancer. Eur J
Cancer. 49:2949–2959. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Collette J, Le Bourhis X and Adriaenssens
E: Regulation of human breast cancer by the long Non-coding RNA
H19. Int J Mol Sci. 18:23192017. View Article : Google Scholar
|
|
22
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Conigliaro A, Costa V, Lo Dico A, Saieva
L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M,
et al: CD90+ liver cancer cells modulate endothelial cell phenotype
through the release of exosomes containing H19 lncRNA. Mol Cancer.
14:1552015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lv M, Zhong Z, Huang M, Tian Q, Jiang R
and Chen J: lncRNA H19 regulates epithelial-mesenchymal transition
and metastasis of bladder cancer by miR-29b-3p as competing
endogenous RNA. Biochim Biophys Acta Mol Cell Res. 1864:1887–1899.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu T, Qu L, He G, Tian L, Li L, Zhou H,
Jin Q, Ren J, Wang Y, Wang J, et al: Regulation of laryngeal
squamous cell cancer progression by the lncRNA
H19/miR-148a-3p/DNMT1 axis. Oncotarget. 7:11553–11566. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y and Hu H: Long non-coding RNA
CCAT1/miR-218/ZFX axis modulates the progression of laryngeal
squamous cell cancer. Tumour Biol.
39:10104283176994172017.PubMed/NCBI
|
|
27
|
Zhang Z, Wang X, Cao S, Han X, Wang Z,
Zhao X, Liu X, Li G, Pan X and Lei D: The long noncoding RNA TUG1
promotes laryngeal cancer proliferation and migration. Cell Physiol
Biochem. 49:2511–2520. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H,
Xuan L, Wang X, Tian L, Sun Y, et al: Long noncoding RNA NEAT1
promotes laryngeal squamous cell cancer through regulating
miR-107/CDK6 pathway. J Exp Clin Cancer Res. 35:222016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fountzilas E, Kotoula V, Angouridakis N,
Karasmanis I, Wirtz RM, Eleftheraki AG, Veltrup E, Markou K,
Nikolaou A, Pectasides D and Fountzilas G: Identification and
validation of a multigene predictor of recurrence in primary
laryngeal cancer. PLoS One. 8:e704292013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Z, Liu Q, Tieqiang L, Xiaodong L,
Guangwei Z, Yang L, Hao Z and Chaoyang Z: Abnormal expressed long
non-coding RNA IRAIN inhibits tumor progression in human renal cell
carcinoma cells. Open Life Sci. 11:200–205. 2016. View Article : Google Scholar
|
|
31
|
Lian Y, Wang J, Feng J, Ding J, Ma Z, Li
J, Peng P, De W and Wang K: Long non-coding RNA IRAIN suppresses
apoptosis and promotes proliferation by binding to LSD1 and EZH2 in
pancreatic cancer. Tumour Biol. 37:14929–14937. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu JF, Balaguru KA, Ivaturi RD, Oruganti
H, Li T, Nguyen BT, Vu TH and Hoffman AR: Lack of reciprocal
genomic imprinting of sense and antisense RNA of mouse insulin-like
growth factor II receptor in the central nervous system. Biochem
Biophys Res Commun. 257:604–608. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu JF, Pham J, Dey I, Li T, Vu TH and
Hoffman AR: Allele-specific histone acetylation accompanies genomic
imprinting of the insulin-like growth factor II receptor gene.
Endocrinology. 141:4428–4435. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Stöger R, Kubicka P, Liu CG, Kafri T,
Razin A, Cedar H and Barlow DP: Maternal-specific methylation of
the imprinted mouse Igf2r locus identifies the expressed locus as
carrying the imprinting signal. Cell. 73:61–71. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wutz A, Smrzka OW, Schweifer N,
Schellander K, Wagner EF and Barlow DP: Imprinted expression of the
Igf2r gene depends on an intronic CpG island. Nature. 389:745–749.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yan H, Yuan W, Velculescu VE, Vogelstein B
and Kinzler KW: Allelic variation in human gene expression.
Science. 297:11432002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Barlow DP and Bartolomei MS: Genomic
imprinting in mammals. Cold Spring Harb Perspect Biol.
6:a0183822014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li E and Zhang Y: DNA methylation in
mammals. Cold Spring Harb Perspect Biol. 6:a0191332014. View Article : Google Scholar : PubMed/NCBI
|