Introduction
Colon cancer is one of the most common malignant
tumors in the clinical practice. Its morbidity and mortality
worldwide are gradually increasing, posing a great threat to human
life (1). There is lack of specific
early stage symptoms and the digestive system signs appear in the
middle and late stages, which can cause serious adverse effects
(2,3). At present, the main treatment for
clinical colon cancer is still surgical resection, but due to the
lack of sensitive diagnostic indicators, most patients have already
developed lymph nodes or distant metastases at the time of
diagnosis, and the surgical resection rate and prognosis are not
ideal (4,5). Therefore, it is very important to find
a biomarker for early diagnosis of colon cancer.
It has been found that miRNAs are differentially
expressed in colon cancer cells (6).
They are closely related to the biological and clinical
characteristics of colon cancer, and play an important role in the
occurrence and progression of colon cancer (7). As studies have shown, miRNAs are
thought to have a regulator role in tumor suppression and
tumorigenesis. For example, miR-185 can inhibit the proliferation
and invasion of colon cancer cells by targeting Wnt1, and
regulating the level of miR-185 may have a therapeutic effect on
colon cancer patients (8).
miR-223-3p can promote the progression of colon cancer by
negatively regulating PRDM1 (9).
Based on these results, miRNAs are attracting attention as a
potential target for the treatment and diagnosis of colon cancer.
Recently, the role of miR-141 in tumor growth has been described.
Expression level of miR-141 in patients with colon cancer at stage
IV has increased, which can easily distinguish patients with
distant metastasis, other stages and healthy control, showing that
plasma miR-141 is a potential prognostic factor for predicting poor
survival of colon cancer patients (10). miR-34C is a tumor suppressor
regulator, which is down-regulated in most forms of cancer, and can
inhibit the growth of malignant tumors by inhibiting genes related
to proliferation, anti-apoptosis and migration (11). A study has shown that the expression
of miR-34C is down-regulated in colon cancer, and the loss of
expression is consistent with the data of colon cancer cell lines
(12).
There are few previous studies on the diagnosis of
colon cancer with serum miR-34c and miR-141 (13–15), and
the study on the role of miRNA expression in colon cancer provided
new diagnostic methods and thinking for the diagnosis and treatment
of colon cancer. In this study, by observing the expression of
miR-34c and miR-141 in the serum of colon cancer patients, the
diagnostic value of miR-34c and miR-141 in colon cancer and their
relationship with clinicopathological features were
investigated.
Patients and methods
General data
A total of 64 patients diagnosed with colon cancer
and treated in Hubei Cancer Hospital (Wuhan, China) were selected
as the experimental group, including 44 males and 20 females. The
patients were aged 25–65 years, with an average age of (46.5±8.4)
years. According to TNM staging system, there were 22 cases of
stage I+II, and 42 cases of stage III. There were 19 cases of lymph
node metastasis, and 15 cases of poor differentiation, 49 cases of
high and medium differentiation. Sixty-four healthy subjects were
included in the control group, including 38 males and 26 females.
The controls were aged 26–57 years, with an average age of
(44.5±7.9) years. The study was approved by the Ethics Committee of
the Hubei Cancer Hospital, and the subjects and/or their families
were informed and signed an informed consent.
Inclusion and exclusion criteria
Inclusion criteria were: Patients met NCCN colon
cancer tumor clinical practice guidelines (16). CT, color Doppler ultrasound and MRI
were performed to rule out distant metastasis. According to TNM
staging system there were Stage I, II and III. Patients did not
receive previous chemotherapy, or radiotherapy. Patients were
diagnosed for the first time, with detailed clinicopathological
data. Exclusion criteria were: Patients without other malignant
tumors, hematological diseases. Patients with severe complications
and immune system diseases. Patients with poor treatment compliance
caused by severe mental illness, and patients unwilling to
participate in the present study.
Main instruments and reagents
ABI PRISM 7500 quantitative PCR instrument (Beijing
Image Trading Co., Ltd.; cat. no. 100005). M-MLV reverse
transcription kit (Beijing Shengkeboyuan Biotechnology Co., Ltd.;
cat. no. RTP50). TRIzol extraction kit (Shanghai Xinfan Biological
Technology Co., Ltd.; cat. no. XFR1030). UV-visible
spectrophotometer (Bioteke corporation; cat. no. ND5000). microRNA
PCR premix kit (AcebioX; cat. no. PAMI000). The primers of miR-34c,
miR-141 and U6 were synthesized by Beijing Lvyuanbode Biotechnology
Co., Ltd. (Table I).
 | Table I.Primer sequences of miR-34c, miR-141
and U6. |
Table I.
Primer sequences of miR-34c, miR-141
and U6.
| Gene | Forward primers | Reverse primers |
|---|
| miR-34c |
5′-CGCGGATCCTCTATTTGCCATCGTCTA-3′ |
5′-CTGAAGCTTCAGGCAGCTCATTTGGAC-3′ |
| miR-141 |
5′-GCGAAGCATTTGCCAAGAA-3′ |
5′-CAATCACAGACCTGTTATTGC-3′ |
| U6 |
5′-CTCGCTTCGGCAGCACA-3′ |
5′-AACGCTTCACGAATTTGCGT-3′ |
RT-qPCR detection
Elbow venous blood (5 ml) of the subjects were
taken, after 10–15 min, the blood was centrifuged at 1,500 × g and
4°C for 10 min, and then stored at −70°C. The total RNA in the
serum was extracted using TRIzol kit (Takara), and the absorbance
values of RNA at 260 and 280 nm were measured by
ultraviolet-visible spectrophotometer. The RNA concentration and
purity were analyzed. Then, 2 µl of total RNA was taken to
reversely transcribe the first strand cDNA, according to the
reverse transcription kit. Reverse transcription reaction
conditions: 42°C for 30 min, 95°C for 5 min. The synthesized cDNA
sample was stored at −80°C. U6 was used as an internal reference.
The total volume of 20 µl includes 10 µl of PCR Premix, 2 µl of
upstream primer, 2 µl of downstream primer and dd water (Rnase and
Dnase free). PCR amplification cycle conditions were: 92°C for 5
min, 95°C for 5 sec, 65°C for 30 sec, 72°C for 5 sec, a total of 45
cycles. 2−∆ΔCq method was used to analyze the relative
expression of the target gene.
Statistical analysis
SPSS19.0 (IBM Corp.) statistical software was used
to analyze the data. Enumeration data in the group were represented
by numbers of case/percentage [n (%)] and were analyzed by
Chi-square test. Measurement data were expressed by mean ± standard
deviation. Independent sample t-test was used for comparison of
measurement data between two groups. ROC curve was used to assess
the diagnostic efficiency of miR-34c and miR-141 on colon cancer.
P<0.05, was considered statistically significant.
Results
General data
There was no difference between the experimental
group and the control group in gender, age, body mass index (BMI),
smoking history, drinking history, residence, educational level or
other general clinical data (P>0.05) (Table II).
 | Table II.General data in the experimental group
and control group [n (%), mean ± SD]. |
Table II.
General data in the experimental group
and control group [n (%), mean ± SD].
| Class | Experimental group
(n=64) | Control group
(n=64) | t/χ2 | P-value |
|---|
| Sex |
|
| 0.131 | 0.717 |
| Male | 44 (68.75) | 38 (59.38) |
|
|
|
Female | 20 (31.25) | 26 (40.62) |
|
|
| Age (years) | 46.5±8.4 | 44.5±7.9 | 1.388 | 0.168 |
| BMI
(kg/m2) | 22.94±4.06 | 22.49±3.72 | 0.654 | 0.514 |
| Smoking history |
|
| 0.283 | 0.595 |
| Yes | 36 (56.25) | 33 (51.56) |
|
|
| No | 28 (43.75) | 31 (48.44) |
|
|
| Drinking history |
|
| 1.647 | 0.199 |
| Yes | 44 (68.75) | 37 (57.81) |
|
|
| No | 20 (31.25) | 27 (42.19) |
|
|
| Place of
residence |
|
| 0.142 | 0.707 |
| City | 20 (31.25) | 22 (34.38) |
|
|
|
Country | 44 (68.75) | 42 (65.62) |
|
|
| Education level |
|
| 2.050 | 0.152 |
|
>Senior high school | 33 (51.56) | 41 (64.06) |
|
|
| ≤Senior
high school | 31 (48.44) | 23 (35.94) |
|
|
|
|
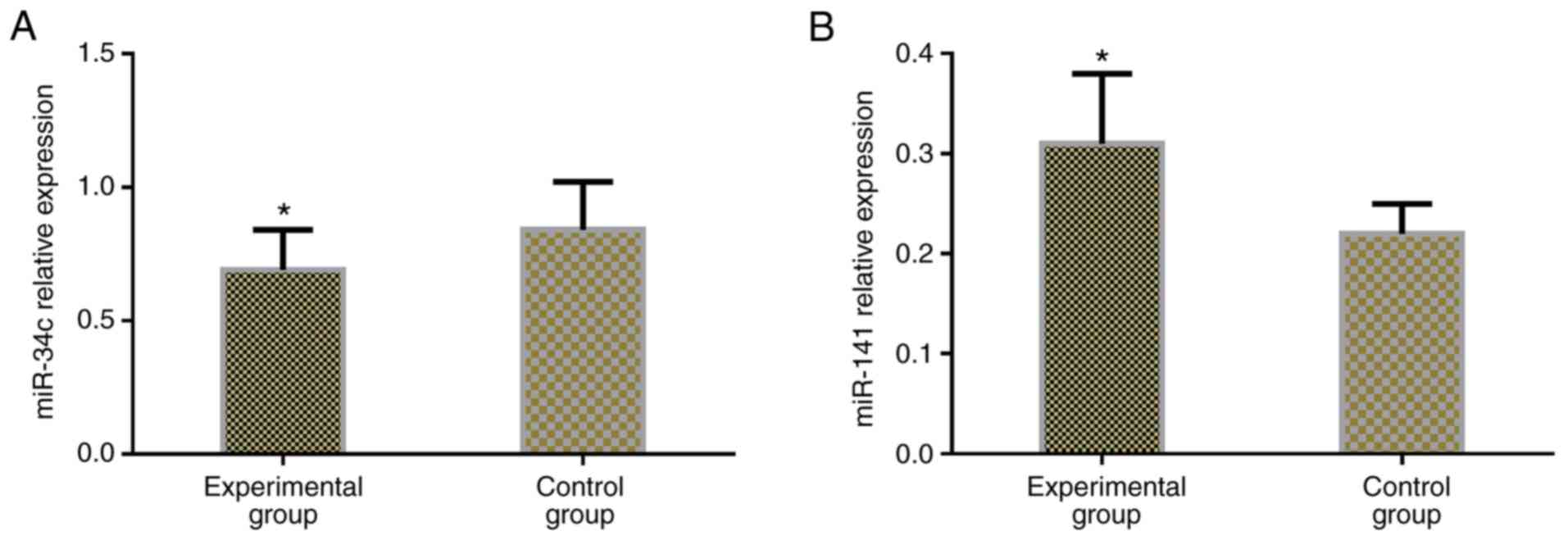
Expression levels of miR-34c and
miR-141 in colon cancer
The relative expression of miR-34c and miR-141 in
the serum of subjects in the two groups were detected, and it was
found that the serum level of miR-34c in the experimental group was
significantly lower than that in the control group (P<0.05), and
the relative expression of miR-141 in serum of patients in the
experimental group was significantly higher than that in the
control group (P<0.05) (Table
III and Fig. 1).
 | Table III.Expression levels of miR-34c and
miR-141 in serum of colon cancer (mean ± SD). |
Table III.
Expression levels of miR-34c and
miR-141 in serum of colon cancer (mean ± SD).
| Groups | n | miR-34c | miR-141 |
|---|
| Experimental
group | 64 | 0.69±0.15 | 0.31±0.07 |
| Control group | 64 | 0.84±0.18 | 0.22±0.03 |
| t | – |
5.121 |
9.454 |
| P-value | – | <0.001 | <0.001 |
Association of the expression of
miR-34c and miR-141 with clinicopathologic features of patients
with colon cancer
The expression of miR-34c and miR-141 in serum of
colon cancer was not associated with the clinicopathological
parameters such as age, gender, local tumor invasion, vascular
invasion, degree of differentiation, and neural invasion
(P>0.05), but was associated with tumor diameter, lymph node
metastasis, carcinoembryonic antigen, and TNM staging (P<0.05)
(Table IV).
 | Table IV.Correlation of miR-34c and miR-141
expression levels with the clinicopathologic features of patients
with colon cancer (mean ± SD). |
Table IV.
Correlation of miR-34c and miR-141
expression levels with the clinicopathologic features of patients
with colon cancer (mean ± SD).
| Clinicopathologic
features | n | miR-34c (n=64) | t | P-value | miR-141 (n=64) | t | P-value |
|---|
| Sex |
|
| 1.577 | 0.120 |
| 1.783 | 0.079 |
|
Male | 44 | 0.65±0.12 |
|
| 0.32±0.08 |
|
|
|
Female | 20 | 0.71±0.18 |
|
| 0.28±0.09 |
|
|
| Age, years |
|
| 0.712 | 0.479 |
| 1.445 | 0.154 |
|
<50 | 21 | 0.67±0.13 |
|
| 0.30±0.09 |
|
|
|
≥50 | 43 | 0.70±0.17 |
|
| 0.33±0.11 |
|
|
| Tumor diameter,
cm |
|
| 3.177 | 0.002 |
| 1.612 | 0.112 |
|
<5 | 47 | 0.74±0.13 |
|
| 0.33±0.12 |
|
|
| ≥5 | 17 | 0.61±0.18 |
|
| 0.33±0.16 |
|
|
| TNM stage |
|
| 4.512 | <0.001 |
| 3.438 | 0.018 |
| Grade
I+II | 22 | 0.81±0.18 |
|
| 0.29±0.05 |
|
|
| Grade
III | 42 | 0.64±0.12 |
|
| 0.34±0.09 |
|
|
| Grade of
differentiation |
|
| 1.862 | 0.673 |
| 0.904 | 0.369 |
| High
and moderate differentiation | 49 | 0.70±0.12 |
|
| 0.32±0.07 |
|
|
| Poor
differentiation | 15 | 0.63±0.15 |
|
| 0.30±0.09 |
|
|
| Lymph node
metastasis |
|
| 8.204 | <0.001 |
| 2.318 | 0.024 |
|
With | 19 | 0.43±0.12 |
|
| 0.34±0.07 |
|
|
|
Without | 45 | 0.80±0.18 |
|
| 0.30±0.06 |
|
|
| Local tumor
invasion |
|
| 1.842 | 0.703 |
| 1.592 | 0.116 |
|
With | 33 | 0.71±0.17 |
|
| 0.33±0.08 |
|
|
|
Without | 31 | 0.64±0.13 |
|
| 0.30±0.07 |
|
|
| Vascular
invasion |
|
| 0.533 | 0.596 |
| 1.500 | 0.139 |
|
With | 35 | 0.68±0.14 |
|
| 0.33±0.04 |
|
|
|
Without | 29 | 0.70±0.16 |
|
| 0.30±0.11 |
|
|
| Neural
invasion |
|
| 1.033 | 0.306 |
| 1.013 | 0.315 |
|
With | 40 | 0.67±0.15 |
|
| 0.34±0.08 |
|
|
|
Without | 24 | 0.71±0.15 |
|
| 0.32±0.07 |
|
|
| Carcinoembryonic
antigen, µg/l |
|
| 3.968 | 0.002 |
| 2.667 | 0.009 |
|
≥3.4 | 34 | 0.61±0.17 |
|
| 0.34±0.07 |
|
|
|
<3.4 | 30 | 0.77±0.15 |
|
| 0.29±0.08 |
|
|
The relative expression of serum
miR-34c and miR-141 in the diagnosis efficiency of colon
cancer
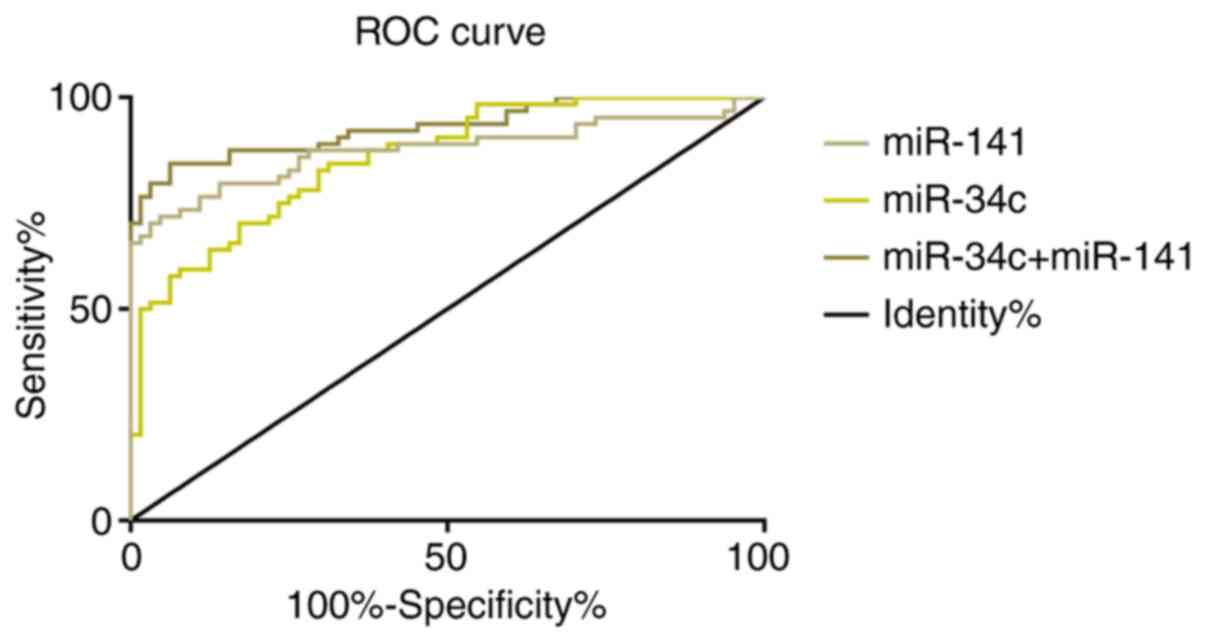
ROC curve of relative expression of serum miR-34c
and miR-141 for the diagnosis of colon cancer was drawn. AUC of
serum miR-34c for the diagnosis of colon cancer was 0.857 (95% CI:
0.795-0.919), its cut-off value was 0.800; diagnostic sensitivity
was 84.38%, and specificity was 68.75%. AUC of serum miR-141 for
diagnosis of colon cancer was 0.876 (95% CI: 0.810-0.941), its
cut-off value was 0.282; diagnostic sensitivity was 70.31%, and
specificity was 96.88%. ROC curve for the diagnosis of colon cancer
combined with serum miR-34c and miR-141 was further drawn. The
combined AUC of serum miR-34c and miR-141 for the diagnosis of
colon cancer was 0.929 (95% CI: 0.884~0.974), the cut-off value was
0.566; diagnosis sensitivity was 84.38% and the specificity was
93.75%. More details are shown in Table
V and Fig. 2.
 | Table V.Relative expression of serum miR-34c
and miR-141 in the diagnostic efficiency of colon cancer. |
Table V.
Relative expression of serum miR-34c
and miR-141 in the diagnostic efficiency of colon cancer.
| Diagnostic
indicator | AUC | 95% CI | Standard error | Cut-off value | Sensitivity
(%) | Specificity
(%) |
|---|
| miR-34c | 0.857 | 0.795-0.919 | 0.032 | 0.800 | 84.38 | 68.75 |
| miR-141 | 0.876 | 0.810-0.941 | 0.033 | 0.282 | 70.31 | 96.88 |
|
miR-34c+miR-141 | 0.929 | 0.884-0.974 | 0.023 | 0.566 | 84.38 | 93.75 |
Discussion
Colon cancer is a malignant lesion in mucosal
epithelium caused by a variety of carcinogenic factors. It is
mainly related to a high-fat, high-protein and low-fiber diet, and
is one of the common malignant tumors (17). The onset of colon cancer is insidious
and most of its onset is slow. In the early stage, there is no
obvious symptoms, patients are often diagnosed in the middle or
late stage, thus losing the best time for treatment (18). Therefore, how to improve the
diagnostic rate of colon cancer and overall survival is a major
problem for clinicians. Surgery is an early treatment method for
colon cancer. In recent years, the treatment of colon cancer has
improved to a certain extent, but the high postoperative
complication rate of colon cancer has not changed (19). Hence, actively looking for indicators
with high sensitivity is of great significance for the early
diagnosis and improvement of prognosis of colon cancer.
Colonoscopy is the current choice for clinical
screening of colon cancer (20,21). Due
to its high cost and invasive characteristics, colonoscopy is not
widely used in clinical practice. The other choice, fecal occult
blood test, has extremely high dietary requirements for patients
due to its low sensitivity (22), so
invasive biomarkers are urgently needed to detect colon cancer.
miRNAs can form specific inclusion bodies and exosomes, which are
suitable for detection in body fluids, serum and feces (23), making it possible to detect miRNAs in
serum as biomarkers for nasopharyngeal cancer.
Micro non-coding microRNAs (miRNAs) contribute to
the development and progression of cancer and are differentially
expressed in normal tissues and cancer (24). miRNAs have a ‘dual identity’ of
oncogene and cancer suppressor gene in the process of tumor
progression, which is closely related to the occurrence,
progression and metastasis of tumors (25). By regulating different signaling
pathways, miR-141 indirectly regulates physiological and
pathological conditions and plays an important role in the
progression of the disease. It has been shown to be low expressed
in a variety of tumors, and to be significantly decreased in tumor
tissues, lymph nodes, and sera of colon cancer patients. Studies
have revealed that the expression of miRNA-141 is down-regulated in
colon cancer, which improves the expression level of MAP4K4,
changes the anti-tumor response, and further increases tumor
proliferation, invasion and metastasis (26). miR-34c is down-regulated in many
different malignancies. Previous studies have revealed that the
down-regulation of miR-34c in endometrial cancer may be an
important factor for poor prognosis. Its overexpression can inhibit
cell proliferation, colony formation, invasion, metastasis and
apoptosis, and plays an important role in the formation, occurrence
and progression of a variety of tumors (27). For example, in the study of Yang
et al (28), the silencing of
tumor suppressors such as miR-34c was related to the development of
colorectal cancer, and showed that the overexpression of miR-34c
induced apoptosis by silencing its target cytokines and inhibited
the invasion and proliferation of colorectal cancer cells,
indicating that miR-34c was a promising target. In the study of Gao
et al (29), the
dysregulation of miR-141 depended on the type of cancer. It played
a dual role in tumorigenicity, and regulated cell movement and
controlling ‘dryness’. This phenomenon strongly suggested that
miR-141 was an oncogene or tumor suppressor gene and provided new
options for targeting cancer therapies. In this study, the relative
expression of miR-34c and miR-141 in serum of patients in the two
groups were detected. The serum level of miR-34c in the
experimental group was significantly lower than that in the control
group, while the expression of miR-141 was significantly higher
than that of the control group, and was associated with tumor
diameter, carcinoembryonic antigen, lymph node metastasis and TNM
staging (P<0.05), indicating that miR-34c and miR-141 might be
involved in the occurrence and progression of colon cancer, and
miR-34c might act as a tumor suppressor gene in colon cancer, and
miR-141 might act as an oncogene.
It has been reported that miR-34c and miR-141 can be
used as biomarkers in a variety of tumors. For example, in the
study of Tao et al (30),
miR-34c played a key role in ovarian cancer and could be used as a
potential diagnostic biomarker and a powerful therapeutic target
for ovarian cancer. Wang et al (31) reported that the overexpression of
miR-141 was not only closely related to the classification and size
of osteosarcoma, but also inhibited the growth and metastasis of
osteosarcoma cells by regulating epidermal growth factor and
affecting downstream pathway proteins. The results of this study
showed that the sensitivity and specificity of serum miR-34c for
the diagnosis of colon cancer were 84.38 and 68.75%, respectively,
and the sensitivity and specificity of serum miR-141 for the
diagnosis of colon cancer were 70.31 and 96.88%, respectively. The
sensitivity and specificity of the combined diagnosis of colon
cancer were 84.38 and 93.75%, respectively. It suggested that
miR-34c and miR-141 could be used as biological indicators with
good sensitivity and specificity in the diagnosis of colon cancer,
and their combination could improve the sensitivity in the
diagnosis of colon cancer. Therefore, miR-34c and miR-141 might
play important roles in the occurrence, progression and prognosis
of colon cancer.
The research subjects were screened strictly in
accordance with the inclusion and exclusion criteria, and there
were no significant differences between the experimental group and
the control group in the general clinical baseline data of gender
and age, which ensured the preciseness and reliability of the
study. Although this study confirmed the role of miR-34c and
miR-141 in the occurrence, progression and prognosis of colon
cancer, the functions of miR-34c and miR-141 in the prognosis of
patients were not observed. These deficiencies needed to be further
supplemented in future studies to support the results of this
study.
In conclusion, miR-34c and miR-141 might be involved
in the occurrence and progression of colon cancer. Serum miR-34c
and miR-141 were detected to have a good sensitivity and
specificity in the diagnosis of colon cancer, and combined
detection could improve the sensitivity in the diagnosis of colon
cancer. The combination of the two might be a new biomarker for
colon cancer diagnosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW and HY conceived and designed the study,
acquired, analyzed and interpreted the experiment data, drafted the
manuscript, and revised the manuscript critically for important
intellectual content. Both authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Hubei Cancer Hospital (Wuhan, China). Signed informed consents were
obtained from the patients and/or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nakagawa-Senda H, Hori M, Matsuda T and
Ito H: Prognostic impact of tumor location in colon cancer: The
Monitoring of Cancer Incidence in Japan (MCIJ) project. BMC Cancer.
19:4312019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu J, Dong H, Zhang Q and Zhang S:
Combined assays for serum carcinoembryonic antigen and
microRNA-17-3p offer improved diagnostic potential for stage I/II
colon cancer. Mol Clin Oncol. 3:1315–1318. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paudel MS, Mandal AK, Shrestha B, Poudyal
NS, Kc S, Chaudhary S, Shrestha R and Goel K: Prevalence of organic
colonic lesions by colonoscopy in patients fulfilling ROME IV
criteria of irritable bowel syndrome. JNMA J Nepal Med Assoc.
56:487–492. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mayanagi S, Kashiwabara K, Honda M, Oba K,
Aoyama T, Kanda M, Maeda H, Hamada C, Sadahiro S, Sakamoto J, et
al: Risk factors for peritoneal recurrence in stage II to III colon
cancer. Dis Colon Rectum. 61:803–808. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Q, Zhang C, Ma JX, Ren H, Sun Y and
Xu JZ: Circular RNA PIP5K1A promotes colon cancer development
through inhibiting miR-1273a. World J Gastroenterol. 25:5300–5309.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brase JC, Wuttig D, Kuner R and Sültmann
H: Serum microRNAs as non-invasive biomarkers for cancer. Mol
Cancer. 9:3062010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeng M, Zhu L, Li L and Kang C: miR-378
suppresses the proliferation, migration and invasion of colon
cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 22:122017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sheng S, Xie L, Wu Y, Ding M, Zhang T and
Wang X: MiR-144 inhibits growth and metastasis in colon cancer by
down-regulating SMAD4. Biosci Rep. 39:BSR201818952019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang W, Sun Z, Su L, Wang F, Jiang Y, Yu
D, Zhang F, Sun Z and Liang W: miRNA-185 serves as a prognostic
factor and suppresses migration and invasion through Wnt1 in colon
cancer. Eur J Pharmacol. 825:75–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chai B, Guo Y, Cui X, Liu J, Suo Y, Dou Z
and Li N: MiR-223-3p promotes the proliferation, invasion and
migration of colon cancer cells by negative regulating PRDM1. Am J
Transl Res. 11:4516–4523. 2019.PubMed/NCBI
|
|
11
|
Cheng H, Zhang L, Cogdell DE, Zheng H,
Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR and Zhang W:
Circulating plasma MiR-141 is a novel biomarker for metastatic
colon cancer and predicts poor prognosis. PLoS One. 6:e177452011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hagman Z, Haflidadottir BS, Ansari M,
Persson M, Bjartell A, Edsjö A and Ceder Y: The tumour suppressor
miR-34c targets MET in prostate cancer cells. Br J Cancer.
109:1271–1278. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Roy S, Levi E, Majumdar AP and Sarkar FH:
Expression of miR-34 is lost in colon cancer which can be
re-expressed by a novel agent CDF. J Hematol Oncol. 5:582012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang Z, Li X, Liu S, Li C, Wang X and
Xing J: MiR-141-3p inhibits cell proliferation, migration and
invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys
Res Commun. 514:699–705. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wan Y, Shen A, Qi F, Chu J, Cai Q, Sferra
TJ, Peng J and Chen Y: Pien Tze Huang inhibits the proliferation of
colorectal cancer cells by increasing the expression of miR-34c-5p.
Exp Ther Med. 14:3901–3907. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Benson AB III, Venook AP, Cederquist L,
Chan E, Chen YJ, Cooper HS, Deming D, Engstrom PF, Enzinger PC,
Fichera A, et al: Colon cancer, version 1.2017, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
15:370–398. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schweiger MR, Hussong M, Röhr C and
Lehrach H: Genomics and epigenomics of colorectal cancer. Wiley
Interdiscip Rev Syst Biol Med. 5:205–219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Soler M, Estevez MC, Villar-Vazquez R,
Casal JI and Lechuga LM: Label-free nanoplasmonic sensing of
tumor-associate autoantibodies for early diagnosis of colorectal
cancer. Anal Chim Act. 930:31–38. 2016. View Article : Google Scholar
|
|
19
|
Arnarson Ö, Butt-Tuna S and Syk I:
Postoperative complication following colonic resection for cancer
is associated with impaired long-term survival. Colorectal Dis.
21:805–815. 2019.PubMed/NCBI
|
|
20
|
Baxter NN, Goldwasser MA, Paszat LF,
Saskin R, Urbach DR and Rabeneck L: Association of colonoscopy and
death from colorectal cancer. Ann Intern Med. 150:1–8. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lieberman DA, Weiss DG, Bond JH, Ahnen DJ,
Garewal H and Chejfec G: Use of colonoscopy to screen asymptomatic
adults for colorectal cancer. N Engl J Med. 343:162–168. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Burt RW: Colon cancer screening.
Gastroenterology. 119:837–853. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ahmed FE, Ahmed NC, Gouda M and Vos P:
MiRNAs for the diagnostic screening of early stages of colon cancer
in stool or blood. Surg Case Rep Rev. 1:1–19. 2017. View Article : Google Scholar
|
|
24
|
Baffa R, Fassan M, Volinia S, O'Hara B,
Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM and
Rosenberg A: MicroRNA expression profiling of human metastatic
cancers identifies cancer gene targets. J Pathol. 219:214–221.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Osaki M, Okada F and Ochiya T: miRNA
therapy targeting cancer stem cells: A new paradigm for cancer
treatment and prevention of tumor recurrence. Ther Deliv.
6:323–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feng L, Ma H, Chang L, Zhou X, Wang N,
Zhao L, Zuo J, Wang Y, Han J and Wang G: Role of microRNA-141 in
colorectal cancer with lymph node metastasis. Exp Ther Med.
12:3405–3410. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li F, Chen H, Huang Y, Zhang Q, Xue J, Liu
Z and Zheng F: miR-34c plays a role of tumor suppressor in HEC-1-B
cells by targeting E2F3 protein. Oncol Rep. 33:3069–3074. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang S, Li W, Sun H, Wu B, Ji F, Sun T,
Chang H, Shen P, Wang Y and Zhou D: Resveratrol elicits
anti-colorectal cancer effect by activating miR-34c-KITLG in vitro
and in vivo. BMC Cancer. 15:9692015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao Y, Feng B, Han S, Zhang K, Chen J, Li
C, Wang R and Chen L: The roles of MicroRNA-141 in human cancers:
From diagnosis to treatment. Cell Physiol Biochem. 38:427–448.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tao F, Tian X, Lu M and Zhang Z: A novel
lncRNA, Lnc-OC1, promotes ovarian cancer cell proliferation and
migration by sponging miR-34a and miR-34c. J Genet Genomics.
45:137–145. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang J, Wang G, Li B, Qiu C and He M:
miR-141-3p is a key negative regulator of the EGFR pathway in
osteosarcoma. Onco Targets Ther. 11:44612018. View Article : Google Scholar : PubMed/NCBI
|