Introduction
Triple-negative breast cancer (TNBC) accounts for
10–20% of all types of breast cancer, and by definition, the
expression of estrogen receptor (ER), progesterone receptor (PR)
and human epidermal growth factor receptor 2 (HER2/neu) has to be
absent. TNBC, histologically a high-grade neoplasia, exhibits an
aggressive biological behavior and usually relapses during the
first 3 years of disease (1–3). Previous studies have evaluated the
effectiveness of chemotherapy in TNBC, such as neoadjuvant therapy,
and have demonstrated that these tumor types can be very
chemosensitive, with 35–50% of the tumor types able to achieve high
rates of pathological complete responses (pCR) (4–6).
Tumor-infiltrating lymphocytes (TILs) reflect the
local immune response against tumor growth and metastasis. The
interaction between the different T lymphocyte subtypes serves an
important role in the immune response of breast cancer (7–9). The
majority of TILs in solid tumors are CD3+, which
includes CD4+ helper cells (Th1 and Th2 subtypes),
CD4+ regulatory T-cells and CD8+ cytotoxic T
lymphocytes. High CD3+ cell density has been reported to
be associated with a favorable outcome in oropharyngeal cancer, and
a low CD3+ count has been shown to predict a shorter
disease-free survival (DFS) in colon and cervical cancer (10,11). In
general, the high number of CD8+ TILs has been
associated with an increased DFS and overall survival (12,13).
Previous studies have suggested that an increased
percentage of TILs in breast cancer is correlated with an improved
prognosis. Loi et al (14)
analyzed TILs in a murine model of residual molecularly
characterized TNBCs following neoadjuvant chemotherapy (NAC), and
it was concluded that genetic or transcriptomic alterations in
Ras/MAPK signaling were significantly associated with a low TIL
percentage.
In addition, in patients with TNBC, a high T
lymphocyte percentage has been associated with pathological
complete responses (pCR), increased DFS and improved overall
survival (15–17). However, molecular analyses, such as
the expression profiles of genes regulating TILs, have not been
thoroughly conducted in the clinical context.
The aim of the present study was to analyze the
percentage, immunophenotype, and molecular gene expression of TILs
in patients with TNBC tumors.
Materials and methods
Sample collection
A total of 96 tumor samples from the Department of
Pathology of Hospital San José TecSalud (Monterrey, Mexico) were
collected for the present study. IRB approval was obtained from the
Ethics Committee for Research at Tecnológico de Monterrey, and the
National Bioethics Commission (code id:
CONBIOETICA19CE100820130520), and also was granted in accordance
with the Declaration of Helsinki. Informed consent was obtained
from each patient prior to tumor sample collection.
Tumor tissue from patients with breast cancer was
prospectively collected at the Breast Cancer Unit between August
2011 and August 2015. Demographic, familiar, clinical, tumor grade,
lymph node status, ER, PR, and HER-2/neu data were recorded.
Tumor samples were collected using ultrasound-guided
core needle biopsies. Tumor cylinders (4–6) were
obtained from each patient for hematoxylin-eosin (H&E) staining
and immunohistochemistry prior to NAC. Tumor sampling for
microarray studies was performed as previously described (18).
NAC
Prior to standard NAC, the center of the breast
tumor was marked by a charcoal suspension injection. If the tumor
was palpable, the injection was performed by a breast surgeon. The
injection in non-palpable tumors was performed by a radiologist
under ultrasound guidance. The charcoal suspension was used to
localize the tumor following NAC for pathological analysis.
The NAC regimen consisted of doxorubicin i.v. (≥60
mg/m2) + cyclophosphamide IV (600 mg/m2)
every 3 weeks for 4 doses, followed by paclitaxel 80
mg/m2 by i.v. infusion for 1 h weekly for 12 doses.
Pathological analysis
The histological grade of the core needle biopsies
was obtained prior to neoadjuvant therapy, using the
Bloom-Richardson score (19). The
stage of breast cancer was determined according to the American
Joint Committee on Cancer (AJCC) (20). The assessment of the percentage of
TILs was performed as per the recommendations of the International
TIL Working Group 2014 in breast cancer (21). The patients were divided into three
categories according to TIL percentage: 1–19%, low (20); <49%, intermediate; and ≥50%, high
(21).
All biopsy specimens were fixed in 10% neutral
buffered formalin between 6–24 h and embedded in paraffin at room
temperature. Tissue sections (4-µm thick) were obtained on coated
glass slides prior to NAC treatment and stained with H&E. The
H&E stain was performed by deparaffinizing sections, washing
twice with xylene for 10 min each. The sections were subsequently
rehydrated twice using a descending alcohol gradient (100, 95 and
70%). Next, the sections were briefly washed in distilled water,
stained in Harris hematoxylin solution for 8 min, washed in running
tap water for 5 min, treated with 1% acid alcohol for 30 sec and
washed with running tap water for 1 min at room temperature.
Lastly, the sections were treated with 0.2% ammonia water for 30
sec, rinsed in 95% alcohol (10 dips) and counterstained in
eosin-phloxine solution for 30–60 sec at room temperature. The
stromal component of TILs was evaluated within the borders of the
invasive carcinoma using a magnification of ×200 (ocular, ×10;
objective, X20). The following areas were excluded from the study:
Tumoral borders; areas around the in situ carcinoma; normal
breast lobules; and tumor areas exhibiting artifacts; necrosis and
hyalinization. The cancer immunophenotype experiments were
performed on the core needle biopsies, prior to neoadjuvant
therapy. The evaluation of the pathological response following NAC
was determined using the de Miller-Payne and MD Anderson Cancer
Center (Residual Cancer Burden) system (22).
Immunohistochemistry
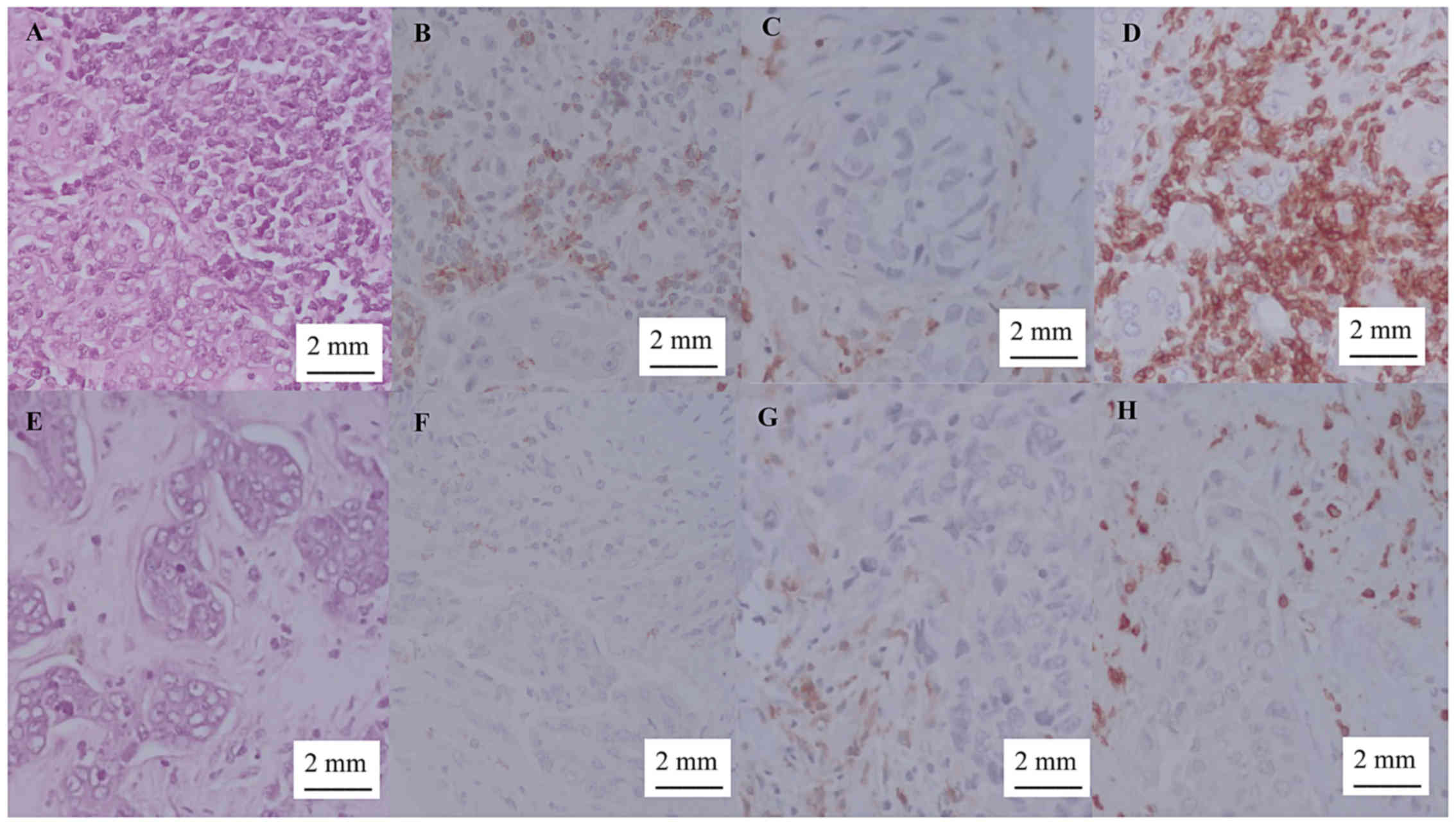
ER, PR, HER-2, Ki67, CD3+,
CD4+ and CD8+ IHC analysis were performed
with a Ventana BenchMarck GX® autostainer (Roche
Diagnostics). Sections of 5-mm were obtained, paraffin slides were
deparaffinized using 2 changes of xylene for 10 min each and
hydrated using graded alcohol and distilled water (2 changes of
100% ethanol, 2 changes of 95% ethanol and 2 changes of distilled
water) for 10 min each at room temperature (15–25°C). Heat-induced
epitope retrieval with citrate buffer was performed. Slides were
then cooled and rinsed with distilled water, rinsed in tris
buffered saline with 20 ml of Tween-20 for 5 min. Slides were then
rinsed with 3% hydrogen peroxide, followed by a rinse with a wash
buffer and covered with 300 µl of protein block for 5 min at room
temperature. Slides were treated with Dako; Agilent Technologies
Inc. monoclonal primary antibodies to HER2/Neu (1:500; Clone 4B5
for 32 min at 37°C), ER (1:500; Clone SP1), PR (1:500; Clone 1E2),
Ki67 (1:100; Clone 30–9), CD3+ (1:500; clone 2GV6),
CD4+ (1:500; clone SP35) and CD8+ (1:500;
clone SP57). Except for HER2/Neu all the other primary antibodies
were incubated for 16 min at 37°C. Slides were then rinsed with
wash buffer and secondary antibody Dako Agilent Technologies Inc.
Envision labeled polymer HRP anti-rabbit (1:100; cat. no. K4002)
was applied for 5 min at 15–25°C. After the secondary reagent, DAB
was applied for 10 min and the slides were rinsed with distilled
water. Counterstaining was done with hematoxylin for 3 min at room
temperature and slides were washed in tap water at room
temperature. Slides were then blued in ammonia water, rinsed in tap
water, dehydrated in graded alcohol (95 and 100% ethanol), cleared
in xylene (2 changes) for 10 min each at room temperature and
coverslipped for microscopic examination. The immunohistochemistry
were performed following the American Society of Clinical
Oncology/College of American Pathologists guidelines (Tables I and II) (23).
 | Table I.ER, PR, and Her-2
immunohistochemistry antibodies. |
Table I.
ER, PR, and Her-2
immunohistochemistry antibodies.
|
| Receptor type |
|---|
|
|
|
|---|
| Antibodies | ER | PR | Her-2 |
|---|
| Clones | SPI (Rabbit
monoclonal primary antibody) | 1E2 Rabbit
monoclonal primary antibody) | 4B5 Rabbit
monoclonal primary antibody) |
| Supplier | Roche | Roche | Roche |
| Dilutions | The antibody is
diluted in 1:500 M Tris-HCl with 2% carrier protein, and 0.10%
ProClin 300, a preservative. | The antibody is
diluted in 1:500 M Tris-HCl with 2% carrier protein, and 0.1%
ProClin 300, a preservative. | The antibody is
diluted in 1:500 M Tris buffered saline, 0.01 M EDTA, 0.05% Brij-35
with 0.3% carrier protein and 0.05% sodium azide, a
preservative. |
| Thresholds | Nuclear positivity
>1% | Nuclear positivity
>1% | Overexpression must
meet threshold criteria for intensity of staining (≥2 on a scale of
0–3+) and percent positive tumor cells (>10%). Staining must
also localize to the cellular membrane. |
| Guidelines | ASCO/CAP | ASCO/CAP | ASCO/CAP |
 | Table II.CD3+, CD4+, and
CD8+ immunohistochemistry antibodies. |
Table II.
CD3+, CD4+, and
CD8+ immunohistochemistry antibodies.
|
| Antigen type |
|---|
|
|
|
|---|
| Antibodies |
CD3+ |
CD4+ |
CD8+ |
|---|
| Clones | 2GV6 Rabbit
monoclonal primary antibody | SP35 Rabbit
monoclonal primary antibody) | SP57 Rabbit
monoclonal primary antibody) |
| Supplier | Roche | Roche | Roche |
| Dilutions | The antibody is
diluted in 1:500 M Tris-HCI with 1% carrier protein and ProClin
300, a preservative. | The antibody is
diluted in 1:500 M Tris-HCL with 1% carrier protein and 0.10%
ProClin 300, a preservative. | The antibody is
diluted in 1:500 M Tris-HCL with 1% carrier protein and a
preservative |
| Thresholds | The cellular
staining pattern anti-CD3 (2GV6) antibody is membranous and/ or
cytoplasmic. | The cellular
staining pattern anti- CD4 (SP35) is membranous. | The staining
pattern anti-CD8 (SP57) is membranous. |
Microscopic evaluation of
CD3+, CD4+ and CD8+ TILs
TIL-positive immunohistochemical scoring was
performed by 2 breast pathologists from Hospital San José TecSalud,
Tecnológico de Monterrey (Monterrey, Mexico). CD3+,
CD4+ and CD8+ TILs were counted in 5 randomly
selected high-power fields using a Carl Zeiss® Axio Lab
A1 light microscope (magnification, ×40), from which the average
was calculated. TIL count in the stroma was rated as follows: +
(1–25 cells); ++ (26–50 cells); and +++ (≥51 cells); as previously
described (24). TIL cases with
<25 cells were considered as ‘low TIL count’, and those with
>25 cells (++, +++) as ‘high TIL count’ (Fig. 1) (24).
RNA extraction
RNA extraction of tissue biopsies was performed
according to the manufacturer's instructions, using RNeasy Fibrous
Tissue Mini kit (Qiagen, Inc.), which is used to obtain
high-quality RNA in small tissue biopsies. The RNA quality was
assessed by capillary electrophoresis using the Experion™ Automated
Electrophoresis Station (Bio-Rad Laboratories, Inc.). The mean RNA
Quality index was 7.84 (range, 6.1–9.8). The RNA concentration was
determined using a NanoDrop 8000 spectrophotometer (Thermo Fisher
Scientific, Inc.). The mean RNA concentration of the tested samples
was 269.02 ng/µl, (range, 38.30–999.13 ng/µl). If several core
tumor samples were collected during the procedure, 2–3 core samples
were used to isolated total RNA and another 2–3 for the
immunohistochemistry (IHC) assay.
Microarray analysis
Gene expression analysis was performed using frozen
fresh tissue tumor samples. The tumor tissue samples from the
biopsies were immediately treated with an RNase inhibitor preserver
solution (RNAlater; Thermo Fisher Scientific, Inc.) (25). Selected RNAs were used for microarray
hybridization and gene expression analysis, which were conducted
using the GeneChip 3′IVT Express kit (Thermo Fisher Scientific,
Inc.), following the manufacturer's protocol. A recommended
quantity of 100 ng total RNA input was used for each sample. Poly-A
controls (Thermo Fisher Scientific, Inc.), which are exogenous
positive controls that monitor the entire target preparation, were
used for all samples. The hybridization mixture was prepared and
applied to the GeneChip Human Genome U133 Plus 2.0 array (Thermo
Fisher Scientific, Inc.), measuring >43,000 transcripts that
represented >20,000 human genes. Washing and scanning processes
were performed in the Fluidics Station 400 and GeneChip Scanner
3000 7G, respectively. The preliminary data analysis was completed
using the Affymetrix Micorarray Suite software version 5.0.0.032
(Thermo Fisher Scientific, Inc.).
Microarray data processing
Normalization was performed using Robust Microarray
Analysis (RMA) and quantile normalization. One sample exhibited
poor quality and was removed from the analysis. The probes whose
mean expression (log scale) was <4 (in a logarithmic scale
resulting from RMA) were removed. A gene expression signature was
performed by t- and Kolmogorov tests with a multiple comparison
correction using the false discovery rate (FDR method (26). Probes with significant P-values in
both tests (FDRt and Kolmogorov test <0.05%) were considered
positive. Analyses were performed in R software [R Core Team.
(2014)] (27).
Gene Ontology (GO), Kyoto Encyclopedia
of Genes and Genomes (KEGG) and Gene network
Interaction analysis of the selected genes was
performed using GO (28,29) and KEGG (30–32)
databases through the online tool STRING: Functional protein
association networks version 11.0 (33). STRING analysis demonstrated
functional enrichments of selected genes: GO analysis of Biological
Process, Molecular Function, SCellular Component and KEEG Pathways.
The combined score was computed by combining the probabilities from
the different evidence channels and corrected for the probability
of randomly observing an interaction (34).
Statistical analysis
Categorical variables are presented as whole numbers
and percentages, while continuous variables are described using the
median and interquartile range. The χ2 test was used to
evaluate the association between pathological response and TILs.
Descriptive and association analysis was performed using GraphPad
Prism 6 for Windows version 6.01 software (GraphPad Software Inc.).
Due to the small sample size and lack of power, specific
association methods (exlogistic and firthlogit) were used.
Results
Demographics and clinical
characteristics
A total of 96 tumor samples were prospectively
collected at the Breast Cancer Unit of Hospital San José TecSalud
between August 2011 and August 2015. All patients were female. A
total of 26/96 samples were diagnosed as TNBC. These 26 patients
with TNBC comprised the study population.
The median age was 49-years old (range, 43–56
years); 80% of the tumors were poorly differentiated (grade 3) and
20% moderately differentiated (grade 2). Lymphovascular invasion
was present in 18 (73%) patients and ipsilateral lymph node
metastasis in 8 (30.7%) patients. Only 1 (3.4%) patient had
metastatic disease upon presentation.
Tumor staging according to the AJCC revealed that 5
patients had (19.2%) stage I, 15 (57.6%) stage II, 4 (15.3%) stage
III and 2 (7.6%) stage IV disease. All patients received standard
NAC. The clinicopathological characteristics of the cohort are
summarized in Table III.
 | Table III.Clinicopathological characteristics
of the patient cohort (n=26). |
Table III.
Clinicopathological characteristics
of the patient cohort (n=26).
|
Characteristics | n | % |
|---|
| Age at diagnosis,
years |
|
18-39 | 1 | 5 |
|
40-59 | 15 | 57 |
|
>60 | 10 | 38 |
| Sex |
|
Female | 26 | 100 |
|
Male | 0 | 0 |
| Sample collection
method |
| Core
needle biopsy | 26 | 100 |
| Treatment |
|
NAC | 26 | 100 |
|
Radiotherapy | 22 | 85 |
| Tumoral grade |
| 1 | 0 | 0 |
| 2 | 5 | 20 |
| 3 | 21 | 80 |
| Lymphovascular
invasion |
|
Present | 19 | 73 |
|
Absent | 7 | 27 |
| Lymph node
status |
|
Positive | 8 | 30.7 |
|
Negative | 18 | 69 |
| Metastasis |
|
Yes | 1 | 3.4 |
| No | 25 | 96.6 |
| Pathological
Stage |
| I | 5 | 19.2 |
| II | 15 | 57.6 |
|
III | 4 | 15.3 |
| IV | 2 | 7.6 |
Pathological response
The pathological response to NAC was assessed, and
it was identified that 4 (15.3%) patients had pNR, 16 (61.5%) pPR
and 6 (23%) pCR. TIL assessment demonstrated low, intermediate, and
high TIL counts in 8 (30.7%), 10 (38.4%), and 8 (30.7%) patients,
respectively. The CD3 count was high in 22 (85.6%) and low in 4
(15.3%) of the specimens. Notably, the CD4 count was low in 22
(85.6%) and high in 4 (15.3%) specimens. The CD8 count was low in
21 (80.7%) and high in 5 (19.2%) samples. The CD counts determined
by microscopy were associated with the number of lymphoid cells;
CD3 showed a higher TIL count of lymphoid cells (>25 lymphoid
cells), meanwhile CD4 and CD8 were associated with a low TIL count
(<25 lymphoid cells). With regard to pCR cases, 50% had a high
number of TILs, 16% an intermediate number, and 33% a low number.
With regard to the patients with pPR, 25% had a high number of
TILs, 50% an intermediate number and 25% a low number. Finally,
none of the patients with pNR exhibited a high number of TILs,
while 25% of patients with pNR had an intermediate number and 75%
pNR a low number (Table IV).
 | Table IV.Association between TIL count
(including CD3, CD4 and CD8 count) and pathological response. |
Table IV.
Association between TIL count
(including CD3, CD4 and CD8 count) and pathological response.
|
|
|
| CD3 | CD4 | CD8 |
|---|
|
|
|
|
|
|
|
|---|
| Disease
presentation | n | % | Low (n=4) | High (n=22) | Low (n=22) | High (n=4) | Low (n=21) | High (n=5) |
|---|
| Pathological
response |
|
Complete response | 6 | 23 | 16.7 | 83.3 | 100 | 0 | 100 | 0 |
| Partial
response | 16 | 61.5 | 6.2 | 93.8 | 87.5 | 12.5 | 75 | 25 |
| No
response | 4 | 15.3 | 50 | 50 | 50 | 50 | 75 | 25 |
| TILs |
|
High | 8 | 30.7 | 100 | 0 | 25 | 75 | 50 | 50 |
|
Intermediate | 10 | 38.4 | 20 | 80 | 100 | 0 | 90 | 10 |
|
Low | 8 | 30.7 | 25 | 75 | 75 | 25 | 100 | 0 |
Immunohistochemistry, gene expression
and analysis results
Using immunohistochemistry, CD3 and CD4 counts were
significantly associated with pPR and pCR (P=0.04), but CD8 was not
associated with pathological response (P=0.75) (Table V). On the contrary, microarray
analysis presented in a heat map demonstrated that CD3, CD4 and CD8
were significantly associated to pathological response (P<0.05;
Fig. 2). This difference may be due
to the heat map being an average of a global analysis that includes
all pathological responses (pCR, pPR, and pNR) (35). In addition, microarray analysis
determines the RNA messenger protein expression whereas IHC only
analyzes the protein expression (36). In addition, most pCR cases had a high
CD3 count (83.3%). No additional associations between TILs and
other clinicopathological parameters were identified in the study
cohort.
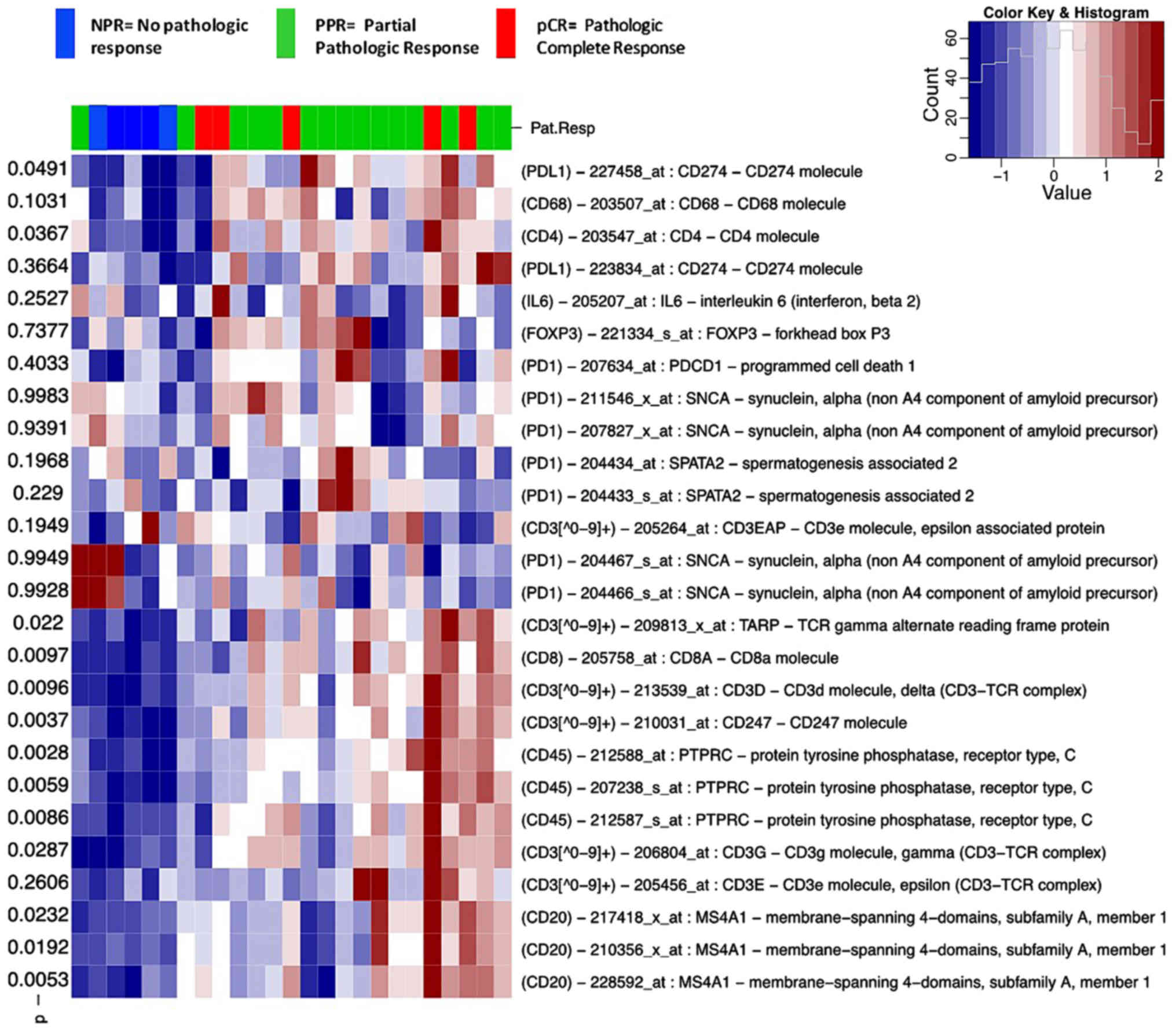 | Figure 2.Heat map of immunity-associated
genes with, P-values and color key. Heatmap of differential
gene expression of selected genes (PDL1, CD68, CD4, IL6, FOXP3,
PD1, CD3, CD8, CD3, CD45 and CD20) associated with
immunity. P-values ranging from 0.002–0.998 are presented on the
left side of the heat map and demonstrate the association of
pathological response and selected genes. The histogram in the
color key in columns presents the low gene expression from 0 to −1
in blue and high gene expression from 1–2 in red. nPR, no
pathologic response; pPR, partial pathologic response; pCR,
pathologic complete response. |
 | Table V.Association between TILs and
pathological response. |
Table V.
Association between TILs and
pathological response.
| Variable | No pathological
response | Pathological
response | χ2 |
|---|
| CD3 TILs |
|
Low | 2 | 2 | 0.04 |
|
High | 2 | 20 |
|
| CD4 TILs |
|
Low | 2 | 20 | 0.04 |
|
High | 2 | 2 |
|
| CD8 TILs |
|
Low | 3 | 18 | 0.75 |
|
High | 1 | 4 |
|
Gene expression analysis of selected genes [CD3,
CD4. CD8, CD20, CD45, forkhead box P3 (FOXP3),
interleukin 6, programmed cell death 1 and CD274 molecule] showed
that CD3, CD4, CD8, CD45 and CD20 exhibited a high
expression in the patients with pPR and pCR, whereas a low
expression of the aforementioned genes was observed in patients
with pNR (Fig. 2).
Gene interaction
GO analysis showed that the predicted target genes
exhibited a stronger interaction among CD68, IL6, FOXP3, PTPRC,
CD274, CD4, CD8A, PDCD1, CD3G, CD3D, CD247 and CD3E, a
weaker interaction with KRT20, MS4A1 and CD3EAP, and
no interaction with SPATA2 and SNCA. A total of 14
genes were involved in immune response, 7 in T-cell receptor
signaling pathway, 9 in lymphocyte activation, 8 in T-cell
activation, and 7 in T-cell differentiation (background gene,
131–1560; FDR, 2.90E-9-3.07E-10; Fig.
3A). Simultaneously, the KEGG database demonstrated 8 genes
that were involved in the T-cell receptor signaling pathway, 7
genes involved in hematopoietic cell lineage and TH17 cell
differentiation, and 5 genes involved in primary immunodeficiency
and Th1/Th2 cell differentiation (background gene 88–99; FDR, 1.33
E-7-7.39E-13; Fig. 3B).
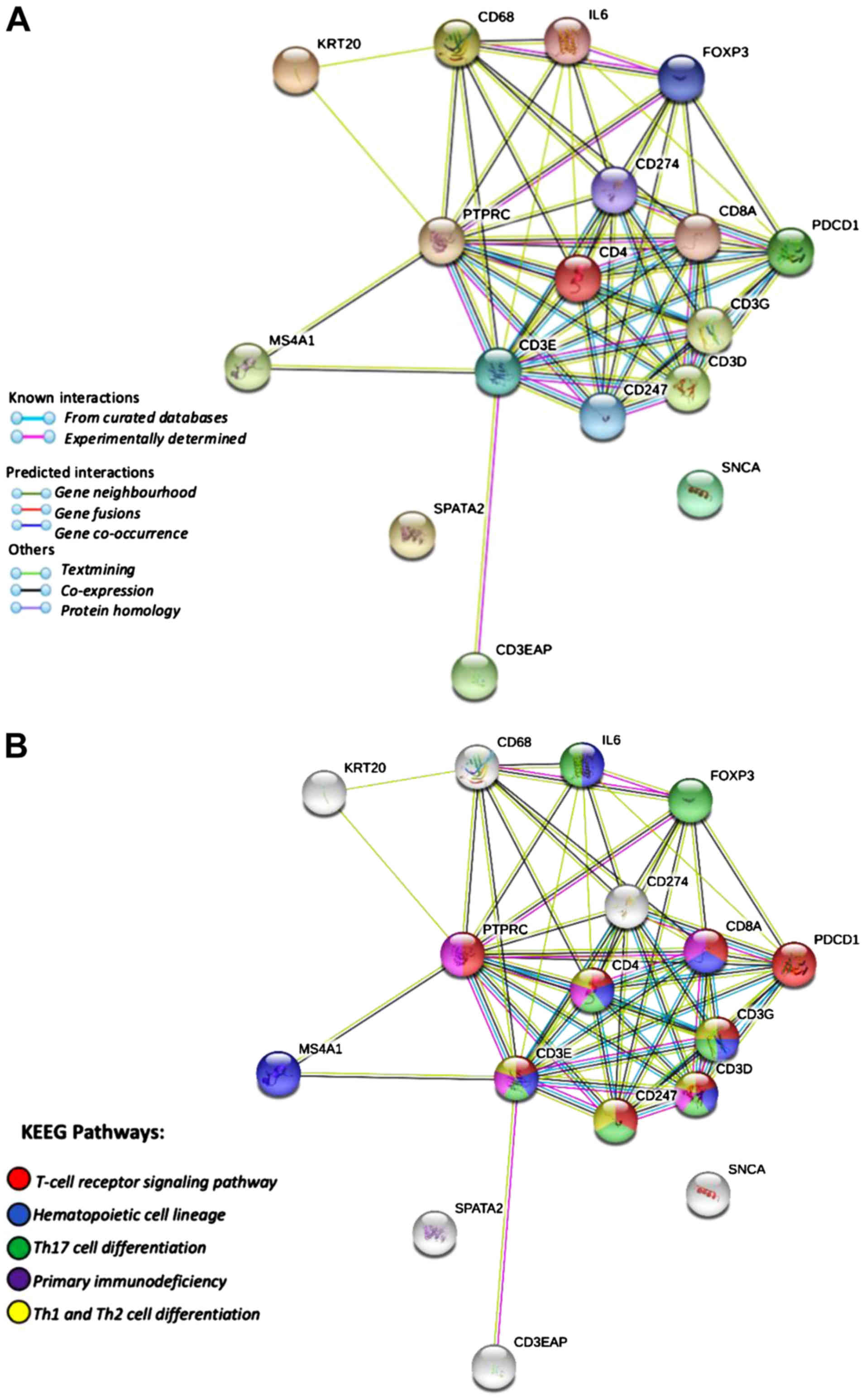 | Figure 3.Gene ontology terms and KEGG
pathways. Predicted target genes (CD68, IL6, FOXP3, PTPRC,
CD274, CD4, CD8A, PDCD1, CD3G, CD3D, CD247, CD3E, KRT20, MS4A1,
CD3EAP, SPATA2 and SNCA) and PPI networks of target
genes. (A) STRING PPI network of co-expressed and interacting
genes. The clusters of 17 genes represent proteins. The colored
nodes represent proteins, white nodes represent second-shell
interactions, filled nodes for 3D known or predicted structures and
edges represent protein-protein associations that did not
necessarily need to bind physically to each other. (B) KEGG
pathways associated with the proteins identified and reported in
the STRING analysis. Red represents the T-cell receptor pathway,
blue represents the hematopoietic cell lineage, green represents
Th17 cell differentiation, purple represents primary
immunodeficiency, yellow represents Th1 and Th2 cell
differentiation. KEGG, Kyoto Encyclopedia of Genes and Genomes;
PPI, protein-protein interactions network. |
Discussion
In the present study, the percentage of stromal TILS
on core needle biopsies of TNBC tumors, as well as their
immunophenotype prior to NAC, were evaluated. Moreover, the
association between TILs and pathological responses was examined.
Even though the pathological response of tumors to NAC has been
previously associated with a high percentage of TILs (37,38),
these have usually been classified into only two categories, low
and high (24). The present study
differs from previous studies in that it divided TILs into three
groups, to obtain a clearer understanding of their association with
the pathological response to NAC. Furthermore, prior studies
performed TIL counts using H&E staining, whereas, the count in
the present study was performed prior to staining. Notably, the
results demonstrated that patients with TNBC with pCR had an
increased CD3+ TIL population without increased
CD4+ and CD8+ TILs.
It is noteworthy that the vast majority of studies
performed in this field have included HER2/neu amplified cases,
which allowed for increased statistical power, but lacked
homogeneity within their sample cohort (15–18). On
the contrary, the present study used a purely TNBC cohort, thus
achieving a homogenous group, in which the majority of patients
with pCR exhibited a high percentage of TILs, consistent with prior
studies (4,5). Similarly, a high CD3+
expression was observed in the majority of pCR (83.3%) and pPR
(93.7%) cases. These data were consistent with the study by Rathore
et al (39), who reported an
association between high CD3+ expression and improved
survival, which has also been observed in cervical and epithelial
ovarian cancer (11,40). Clinically, the density of
CD3+ expression could be routinely measured to predict
pathological response in patients with TNBC.
Even though high CD4+ and CD8+
density have been consistently associated with improved overall
survival (12,13,18), in
the present cohort, CD4+ and CD8+ were
identified to be associated with a low TIL count in a patient with
pCR. This discrepancy may be due to the demographic data of the
patient and the size of the cohort; larger multicentric cohorts are
required to explore this association further.
Kim et al (41) reported a decreased CD8+
count in breast cancer with lymph node metastasis, high
proliferation rate and advanced tumor stage. However, in the
present cohort, no association was observed between TIL count and
vascular invasion or lymph node metastasis. Furthermore, the
majority of the patients included in the present study were
diagnosed with early-stage breast cancer (76.8% in stage I and II),
which could explain the absence of a correlation between TILs and
advance tumor stages herein. Nonetheless, a high percentage of
patients exhibited a high TIL count and CD3+
lymphocytes, which has been described previously (24,37).
Notably, patients with pCR and pPR exhibited high
gene expression levels of CD3, CD4, CD8, CD45 and
CD20. Conversely, patients with pNR exhibited a limited
expression of these genes, suggesting that there is limited
participation of both B (CD20) and T lymphocytes (CD3, CD4 and CD8)
in the antitumor cell response. This immune activity inevitably
affects the pathological response to NAC.
Levy et al (42) reported an immune signature of T-cell
infiltration in breast tumors through an exome study. They
described that exome reads mapping to the
complementarity-determining-region 3 (CDR3) of mature T-cell
receptor β can be used as an immune DNA signature. Exomes from the
CDR3 fraction of breast cancers in The Cancer Genome Atlas were
used to study Her2+ patients. Improved survival was
associated with increased TIL fraction, tumor purity, as well as
adaptive immunity gene expression signatures in the
Her2+ population. However, these differences were not
observed in patients with TNBC (42). In addition, gene profiling was not
performed.
The immune signature of metastatic breast cancer has
been studied (41), in which the
Her2+ expression and previous taxane treatment were
positively correlated with a high expression of 9 genes associated
with immune checkpoints: PDCD1 (PD-1); CD274 (PD-L1); CD276
(B7-H3); CTLA-4; IDO1; LAG3; VTCN1; HAVCR2; and TNFRSF4
(OX40) (41). Importantly,
these genes have interactions with each other.
Finally, quantitative immunofluorescence assay has
been performed to measure the stromal expression of CD3, CD8 and
CD20. It was identified that CD20 score predicted pCR independently
of age, size, nuclear grade, nodal status, ER, PR, HER2 and Ki-67.
CD3, CD8, and pathologist estimation did not demonstrate an
association with pathological response (18).
Even though the aforementioned studies have
performed important immune analyses, there is an absence of
literature examining immunophenotypic signatures in the context of
gene expression profiling of TILs in breast cancer. To the best of
our knowledge, the present study was the first to report an
association between immunophenotype and gene expression profiling
in patients with TNBC. This provides a new promising method for
assessing pathological response in TNBC.
The present study was not without limitations.
RT-qPCR experiments could not be performed to verify the results of
the GO and KEGG analyses, due to lack of tissue. However, our
previous study reported a gene expression signature for TNBC in a
previous study (25). Even though
the sample size was relatively small, the present study included a
cohort comprised solely of patients with TNBC. However, larger
multicentric studies are required to expand and confirm the present
results.
In conclusion, the results of the present study
demonstrated that TILs may predict the pathological response to NAC
in patients with TNBC. In addition, the results identified a more
accurate association between the high expression levels of CD3,
CD4, CD8, CD45 and CD20 genes and pPR and pCR, compared
with the association between the high expression of those genes and
immunohistochemistry alone.
Acknowledgements
The authors would like to thank Dr Paloma del C.
Monroig-Bosque (Pathology Department, Houston Methodist Hospital,
Houston, TX, USA) and Dr Tania Platero-Portillo (Northwell Health,
Department of Pathology, New York, NY, USA) for their contribution
in the interpretation of data and revision of the manuscript.
Funding
This work was partially supported by CONACYT-Mexico
through an approved grant (grant no.
SALUD-CONACYT-2011-C01-162301).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GSGM, revise the manuscript for intellectual
content, and gave final approval for the version to published.
GSGM, GMF, CALG, AD and SSF made contributions to the acquisition,
collection, interpretation of data and writing of the manuscript.
HDA, JVE, VTA, ROL and SCH made contributions to the data analysis
and funding. EEZ, MAAJ, CVG, and OPC made contributions for the
analysis and collection of data. All authors have read and approved
the manuscript.
Ethics approval and consent to
participate
IRB approval was obtained from the Ethics Committee
of Research at Tecnológico de Monterrey and the National Bioethics
Commission (code id: CONBIOETICA19CE100820130520), and was also
granted in accordance with the Declaration of Helsinki. Informed
consent was obtained from each patient prior to tumor sample
collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TILs
|
tumor-infiltrating lymphocytes
|
|
TNBC
|
triple-negative breast cancer
|
|
HER2/neu
|
human epidermal growth factor receptor
2
|
|
pCR
|
complete pathological responses
|
|
pPR
|
partial pathological response
|
|
pNR
|
no pathological response
|
|
DFS
|
disease-free survival
|
|
NAC
|
neoadjuvant chemotherapy
|
|
RMA
|
robust microarray analysis
|
|
FDR
|
false discovery rate method
|
|
CDR3
|
complementarity-determining-region
3
|
|
AJCC
|
American Joint Committee on Cancer
|
|
H&E
|
hematoxylin-eosin
|
References
|
1
|
Wang C, Kar S, Lai X, Cai W, Arfuso F,
Sethi G, Lobie PE, Goh BC, Lim LHK, Hartman M, et al: Triple
negative breast cancer in Asia: An insider's view. Cancer Treat
Rev. 62:29–38. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kumar P and Aggarwal R: An overview of
triple-negative breast cancer. Arch Gynecol Obstet. 293:247–269.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Ruijter TC, Veeck J, de Hoon JP, van
Engeland M and Tjan-Heijnen VC: Characteristics of triple-negative
breast cancer. J Cancer Res Clin Oncol. 137:183–192. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sharma P, López-Tarruella S, García-Saenz
JA, Khan QJ, Gómez HL, Prat A, Moreno F, Jerez-Gilarranz Y,
Barnadas A, Picornell AC, et al: Pathological response and survival
in triple-negative breast cancer following neoadjuvant carboplatin
plus docetaxel. Clin Cancer Res. 24:5820–5829. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nakashoji A, Matsui A, Nagayama A, Iwata
Y, Sasahara M and Murata Y: Clinical predictors of pathological
complete response to neoadjuvant chemotherapy in triple-negative
breast cancer. Oncol Lett. 14:4135–4141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nagao T, Kinoshita T, Hojo T, Tsuda H,
Tamura K and Fujiwara Y: The differences in the histological types
of breast cancer and the response to neoadjuvant chemotherapy: The
relationship between the outcome and the clinicopathological
characteristics. Breast. 21:289–295. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Adams S, Gatti-Mays ME, Kalinsky K, Korde
LA, Sharon E, Amiri-Kordestani L, Bear H, McArthur HL, Frank E,
Perlmutter J, et al: Current landscape of immunotherapy in breast
cancer: A review. JAMA Oncol. Apr 11–2019.(Epub ahead of print).
doi: 10.1001/jamaoncol.2018.7147. View Article : Google Scholar
|
|
8
|
Bayraktar S, Batoo S, Okuno S and Glück S:
Immunotherapy in breast cancer. J Carcinog. 18:22019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Basile D, Pelizzari G, Vitale MG, Lisanti
C, Cinausero M, Iacono D and Puglisi F: Atezolizumab for the
treatment of breast cancer. Expert Opin Biol Ther. 18:595–603.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sinicrope FA, Rego RL, Ansell SM, Knutson
KL, Foster NR and Sargent DJ: Intraepithelial effector
(CD3+)/regulatory (FoxP3+) T-cell ratio
predicts a clinical outcome of human colon carcinoma.
Gastroenterology. 137:1270–1279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ancuta E, Ancuţa C, Zugun-Eloae F,
Iordache C, Chirieac R and Carasevici E: Predictive value of
cellular immune response in cervical cancer. Rom J Morphol Embryol.
50:651–655. 2009.PubMed/NCBI
|
|
12
|
Seo AN, Lee HJ, Kim EJ, Kim HJ, Jang MH,
Lee HE, Kim YJ, Kim JH and Park SY: Tumour-infiltrating
CD8+ lymphocytes as an independent predictive factor for
pathological complete response to primary systemic therapy in
breast cancer. Br J Cancer. 109:2705–2713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu S, Lachapelle J, Leung S, Gao D,
Foulkes WD and Nielsen TO: CD8+ lymphocyte infiltration
is an independent favorable prognostic indicator in basal-like
breast cancer. Breast Cancer Res. 14:R482012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Loi S, Dushyanthen S, Beavis PA, Salgado
R, Denkert C, Savas P, Combs S, Rimm DL, Giltnane JM, Estrada MV,
et al: RAS/MAPK activation is associated with reduced
tumor-infiltrating lymphocytes in triple-negative breast cancer:
Therapeutic cooperation between MEK and PD-1/PD-L1 immune
checkpoint inhibitors. Clin Cancer Res. 22:1499–1509. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Denkert C, Loibl S, Noske A, Roller M,
Müller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R,
Hanusch C, et al: Tumor-associated lymphocytes as an independent
predictor of response to neoadjuvant chemotherapy in breast cancer.
J Clin Oncol. 28:105–113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamaguchi R, Tanaka M, Yano A, Tse GM,
Yamaguchi M, Koura K, Kanomata N, Kawaguchi A, Akiba J, Kanomata N,
et al: Tumor-infiltrating lymphocytes are important pathologic
predictors for neoadjuvant chemotherapy in patients with breast
cancer. Hum Pathol. 43:1688–1694. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ono M, Tsuda H, Shimizu C, Yamamoto S,
Shibata T, Yamamoto H, Hirata T, Yenemori K, Ando M, Tamura K, et
al: Tumor-infiltrating lymphocytes are correlated with response to
neoadjuvant chemotherapy in triple-negative breast cancer. Breast
Cancer Res Treat. 132:793–805. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brown JR, Wimberly H, Lannin DR, Nixon C,
Rimm DL and Bossuyt V: Multiplexed quantitative analysis of CD3,
CD8, and CD20 predicts response to neoadjuvant chemotherapy in
breast cancer. Clin Cancer Res. 20:5995–6005. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bloom HJ and Richardson WW: Histological
grading and prognosis in breast cancer. A study of 1409 cases of
which 359 have been followed for 15 years. Br J Cancer. 11:359–377.
1957. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
American Joint Committee on Cancer, . The
New Edition (7th) AJCC Staging System for Breast Cancer.
June;2010.http://cancerstaging.org/references-tools/deskreferences/Documents/AJCC%207th%20Ed%20Cancer%20Staging%20Manual.pdfOctober.
2019
|
|
21
|
Salgado R, Denkert C, Demaria S, Sirtaine
N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL,
Penault-Llorca F, et al: The evaluation of tumor-infiltrating
lymphocytes (TILs) in breast cancer: Recommendations by an
International TILs Working Group 2014. Ann Oncol. 26:259–271. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sahoo S and Lester SC: Pathology of breast
carcinomas after neoadjuvant chemotherapy: Overview with
recommendations on specimen processing and reporting. Arch Pathol
Lab Med. 133:633–642. 2009.PubMed/NCBI
|
|
23
|
Hammond ME, Hayes DF and Wolff AC:
Clinical Notice for American Society of Clinical Oncology-College
of American Pathologists guideline recommendations on ER/PgR and
HER2 testing in breast cancer. J Clin Oncol. 29:e4582011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rathore AS, Kurmar S, Konwar R, Makker A,
Negi MP and Goel MM: CD3+, CD4+,
CD8+ tumor infiltrating lymphocytes (TILs) are
predictors of favourable survival outcome in infiltrating ductal
carcinoma of breast. Indian J Med Res. 140:361–369. 2014.PubMed/NCBI
|
|
25
|
Santuario-Facio SK, Cardona-Huerta S,
Perez-Paramo YX, Trevino V, Hernandez-Cabrera F, Rojas-Martinez A,
Uscanga-Perales G, Martinez-Rodriguez JL, Martinez-Jacobo L,
Padilla-Rivas G, et al: A new gene expression signature for
triple-negative breast cancer using frozen fresh tissue before
neoadjuvant chemotherapy. Mol Med. 23:101–111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miyan M, Schmidt-Mende J, Kiessling R,
Poschke I and de Boniface J: Differential tumor infiltration by
T-cells characterizes intrinsic molecular subtypes in breast
cancer. J Transl Med. 14:2272016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
R Core Team. R, . A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: 2014, http://ww=w.R-project.org/October. 2017
|
|
28
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. Nat
Genet. 25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
The Gene Ontology Consortium, . The gene
ontology resource: 20 years and still going strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kanehisa M: Toward understanding the
origin and evolution of cellular organisms. Protein Sci.
28:1947–1951. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
von Mering C, Jensen LJ, Snel B, Hooper
SD, Krupp M, Foglierini M, Jouffre N, Huynen MA and Bork P: STRING:
Known and predicted protein-protein associations, integrated and
transferred across organisms. Nucleic Acids Res. 33:433–437. 2005.
View Article : Google Scholar
|
|
35
|
Cui X and Churchill GA: Statistical tests
for differential expression in cDNA microarray experiments. Genome
Biol. 4:2102003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee M, Tayyari F, Pinnaduwage D, Bayani J,
Bartlett JMS, Mulligan AM, Bull SB and Andrulis IL: Tumoral BRD4
expression in lymph node-negative breast cancer: Association with
T-bet+ tumor-infiltrating lymphocytes and disease-free
survival. BMC Cancer. 18:7502018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim ST, Jeong H, Woo OH, Seo JH, Kim A,
Lee ES, Shin SW, Kim YH, Kim JS and Park KH: Tumour-infiltrating
lymphocytes, tumour characteristics, and recurrence in patients
with early breast cancer. Am J Clin Oncol. 36:224–231. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mao Y, Qu Q, Zhang Y, Liu J, Chen X and
Shen K: The value of tumor infiltrating lymphocytes (TILs) for
predicting response to neoadjuvant chemotherapy in breast cancer: A
systematic review and meta-analysis. PLoS One. 9:e1151032014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rathore AS, Kumar S, Konwar R, Srivastava
AN, Makker A and Goel MM: Presence of CD3+ tumor
infiltrating lymphocytes is significantly associated with good
prognosis in infiltrating ductal carcinoma of breast. Indian J
Cancer. 50:239–244. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang L, Conejo-Garcia JR, Katsaros D,
Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H,
Schlienger K, Liebman MN, et al: Intratumoral T cells, recurrence,
and survival in epithelial ovarian cancer. N Engl J Med.
348:203–213. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim JY, Lee E, Park K, Park WY, Jung HH,
Ahn JS, Im YH and Park YH: Immune signature of metastatic breast
cancer: Identifying predictive markers of immunotherapy response.
Oncotarget. 8:47400–47411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Levy E, Marty R, Gárate Calderon V, Woo B,
Dow M, Armisen R, Carter H and Harismendy O: Immune DNA signature
of T cell infiltration in breast tumor exomes. Sci Rep.
6:300642016. View Article : Google Scholar : PubMed/NCBI
|