Introduction
Lung cancer is a highly metastatic type of cancer
with an increasing incidence rate worldwide every year with an
estimated 1.6 million mortalities in 2012 (1). The main subtypes of lung cancer are
small cell lung cancer (SCLC) (10–15%) and non-small cell lung
cancer (NSCLC) (80–85%), with the latter being the most common type
(2). Further investigation on the
treatment of NSCLC is therefore urgently needed. Platinum-based
drugs, including cis-dichlorodiamino platinum [cisplatin (DDP)],
carboplatin and lobaplatin, are currently the most potent
chemotherapeutic agents used to treat NSCLC (3). However, prolonged use of DDP may lead
to drug resistance, thus resulting in reduced clinical curative
effects, which in turn may lead to lung cancer recurrence and
distant metastasis (4). Previous
studies have demonstrated that the main mechanisms underlying DDP
resistance in cancer cells include increased cellular DDP
detoxification activity, inhibition of apoptosis and an enhanced
DNA damage repair capacity (5,6).
Investigating the enhancement of DDP sensitivity in DDP-resistant
cancer cells may therefore be of great importance.
Curcuminoids, which are yellow pigments extracted
from turmeric rhizomes, are used to treat a wide variety of
diseases, including neurodegenerative diseases, cardiovascular
diseases, metabolic disorders and lung fibrosis (7–10).
Furthermore, these compounds have been demonstrated to exhibit
potent anticancer activities at initial, promotion and progression
stages of tumor development in numerous cancers, including colon,
cervical, ovarian and gastric cancers (11). Curcuminoids consist of three main
bioactive components, curcumin (CMN), demethoxycurcumin (DMC) and
bisdemethoxycurcumin (BDMC) (12).
CMN is the most widely studied compound of the curcuminoids and is
used in the treatment of several solid tumors and hematological
malignancies, such as cancers of the digestive, lymphatic and
immune, pulmonary and the skin, due to its great antitumor
potential (13–15). However, several studies have
demonstrated that CMN exhibits poor oral bioavailability (16–18).
Quitschke (17) reported that DMC is
more stable in the blood compared with CMN. Chemically, DMC is very
similar to CMN. DMC only lacks the methoxy group that is linked to
the benzene ring. This minor difference in chemical structure
provides DMC with increased chemical stability and activity
(19,20). It has also been reported that DMC
modulates inhibition of cell survival, tumor suppression, and
activates mitochondrial and death receptor pathways (21–24). The
therapeutic potential of DMC in the adjuvant treatment of various
diseases should therefore be further investigated.
The present study aimed to investigate whether DMC
in combination with DDP could increase DDP sensitivity in
DDP-resistant A549 cells (A549/DDP), similar to taxanes, pemetrexed
and gemcitabine. The present study may provide a novel candidate
drug for combined chemotherapy following clinical lung cancer
surgery.
Materials and methods
Cell culture
Human alveolar basal epithelial adenocarcinoma cells
(A549 cells) and human lung fibroblasts (MRC-5 cells) were
purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences (cat. nos. SCSP-503 and SCSP-5040,
respectively). A549 cells were cultured in Ham's F-12K (Thermo
Fisher Scientific Inc.) medium (cat. no. 21127-022; Invitrogen;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.). MRC-5 cells were cultured in MEM
(cat. no. 11090081; Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
1X GlutaMAX (cat. no. 35050061; Invitrogen; Thermo Fisher
Scientific, Inc.), 1X non-essential amino acids (Invitrogen; Thermo
Fisher Scientific, Inc.; cat. no. 11140050) and 1 mM sodium
pyruvate solution (cat. no. 11360070; Invitrogen; Thermo Fisher
Scientific, Inc.). A549/DDP cells were obtained from Procell Life
Science & Technology Co., Ltd. (cat. no. CM-0519) and were
cultured in Ham's F-12K supplemented with 10% FBS, 2 µg/ml DDP and
1% penicillin/streptomycin. The medium was changed every two days
and cells were passaged when the cell density reached 80–90%.
MTT assay
All cells were seeded into 96-well plates at the
density of 5×103 cells/well (A549 and A549/DDP cells) or
1×104 cells/well (MRC-5 cells). A549 cells were treated
with DDP at various concentrations (1.25, 2.5, 5, 10, 20 and 40
µM). A549/DDP cells were treated with higher concentrations of DDP
(5, 10, 20, 40, 60, 80 and 100 µM). A549, A549/DDP and MRC-5 cells
were treated with DMC at doses of 5, 10, 15, 20, 25, 30 and 35 µM.
After culturing the cells for a specified period of time (24, 48,
72 or 96 h), 20 µl MTT (5 µg/ml; Sigma-Aldrich; Merck KGaA) was
added and the cells were cultured in the incubator for 1 h. Next,
the culture medium was discarded carefully, 100 µl DMSO was added
and the plate was agitated for 10 min on a shaker. The absorbance
was read at 550 nm on a microplate reader (Bio-Rad Laboratories
Inc.).
TUNEL assay
Coverslips were inserted into a 24-well plate and
then A549/DDP cells were seeded on the coverslips at the density of
5×104 cells per well. After 24 h, the medium was
replaced with medium containing drugs (10 µM DDP, 5 µM DMC or
both), and after 48 h incubation, TUNEL staining was performed.
Briefly, cells were washed gently with PBS to remove non-adherent
cells and were fixed with 4% paraformaldehyde for 30 min at room
temperature. Cells were then incubated with PBS containing 0.1%
Triton at 4°C for 10 min for permeabilization. The solution was
discarded and 50 µl of TUNEL detection solution (cat. no. C1090;
Beyotime Institute of Biotechnology) (2 µl of TdT enzyme with 48 µl
of fluorescent labeling solution) was added at 37°C for 60 min in
the dark. Finally, slides were mounted with an anti-fluorescence
quencher (cat. no. G1401; Wuhan Servicebio Technology Co., Ltd.)
and were kept at 4°C. Slides were observed under a fluorescence
microscope (magnification, ×200) within one week.
Western blotting
A549 cells were lysed in RIPA lysis buffer (cat. no.
P0013B; Beyotime Institute of Biotechnology) after 48 h of drug
treatment. Total proteins were extracted and were denatured at 95°C
for 10 min. The protein concentration was determined by the BCA
method, and protein concentration was subsequently normalized using
loading buffer. Proteins (30 µg) were separated by 10% SDS-PAGE and
were then transferred onto PVDF membranes. Membranes were blocked
with 5% skimmed milk in PBST for 2 h at room temperature and were
incubated with primary antibodies overnight at 4°C. Next, membranes
were washed three times with PBS for 15 min and were incubated with
secondary antibodies at room temperature for 2 h. Membranes were
washed three times with PBS for 15 min and were exposed to 200 µl
enhanced chemiluminescence substrate (Thermo Fisher Scientific,
Inc.). Bands were detected on a Bio-Rad instrument and relative
expression levels were normalized to endogenous control using
Image-Pro Plus software v.6.0 (National Institutes of Health). The
primary antibody against ERCC1 was obtained from Abcam (1:2,000;
cat. no. ab129267). The antibodies against Bcl-2 (cat. no. 4223),
Bax (cat. no. 5023), caspase 3 (cat. no. 9662), PARP (cat. no.
9532), cleaved caspase 3 (cat. no. 9664), cleaved PARP (cat. no.
5625) and β-actin (cat. no. 4970) were purchased from Cell
Signaling Technology, Inc. and were used at a dilution of 1:1,000.
The secondary goat anti-rabbit antibodies (cat. nos. 7074 and 7076;
Cell Signaling Technology, Inc.) were used at a dilution of
1:5,000.
Subcutaneous xenograft assay
A total of 16 male BALB/c nude mice (4–5 weeks,
20–24 g) were purchased from Beijing Vital River Laboratory Animal
Technology Co. The mice were housed under specific pathogen free
(SPF) conditions and fed with SPF standards of care, The mice were
housed with a 12 h light/dark cycle at 25±2°C and 50±10% humidity,
and were provided with sterile food and water. A549/DDP
(5×105) cells were subcutaneously injected into the left
axilla of 16 nude mice. When the maximum tumor volume reached 100
mm3, 16 mice were randomly divided into four groups of
four mice as follows: Control group, monotherapy groups (DDP or
DMC) and a combination treatment group. DDP was injected
intraperitoneally at a dose of 6.0 mg/kg every three days and DMC
was injected intraperitoneally at a dose of 30 mg/kg every day for
20 days. DDP and DMC were dissolved in DMSO (50 µl in DMSO per
mouse for injection). The control group was treated with DMSO (50
µl per mouse). The tumor volume was measured every 4 days until day
20 using the following formula: Tumor volume (units,
mm3)=width2 × length/2. The mice were
sacrificed on day 20. Mice were injected intraperitoneally with 300
mg/kg ketamine and 20 mg/kg xylazine for euthanasia, and the death
of mice was confirmed when the mice stopped breathing.
Immunohistochemistry (IHC)
Mouse subcutaneous tumors were collected following
euthanasia. Subsequently, tissue sections (4-µm) were dewaxed with
xylene (twice) for 10 min and hydrated using a descending alcohol
series (100% twice, 90% twice, 75% twice) for 3 min at room
temperature. Antigens were retrieved in a 95°C water bath for 20
min. Endogenous peroxidase activity was blocked using 3% hydrogen
peroxide for 20 min at room temperature. After blocking with 5% BSA
(Sigma-Aldrich; Merck KGaA), the sections were incubated with
primary antibody (Ki-67, 1:200; cat. no. 9027S; Cell Signaling
Technology, Inc.) at 4°C overnight. The next day, sections were
incubated with secondary antibody (1:200; cat. no. ZDR-5306;
OriGene Technologies Inc.) for 2 h at 37°C, followed treating with
DAB chromogen (1 drop of the DAB Chromogen per ml of substrate
buffer; Dako; Agilent Technologies, Inc.) and hematoxylin (cat. no.
C0107; Beyotime Institute of Biotechnology) for 2 min at room
temperature. Finally, the slides were dehydrated and fixed by resin
and observed with an Olympus inverted microscope (Olympus
Corporation; magnification, ×200).
Statistics analysis
Tumor size data were presented as mean ± SEM, all
other data were represented mean ± SD. Student's unpaired t-test
was used to analyze the statistical significance among two groups.
One-way ANOVA followed by the post hoc Bonferroni's test was used
to compare the differences among ≥3 groups. All statistical
analyses were performed with SPSS v.20.0 software (IBM Corp.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
A549/DDP cells are resistant to DDP
but not to DMC
A549 and A549/DDP cells were treated with different
concentrations of DDP for 48 h. The results from the MTT assay
demonstrated that the IC50 value of DDP in normal A549
cells was 3.8 µM, whereas it reached 47.67 µM in A549/DDP cells,
which is more than 10 times higher compared with that in normal
A549 cells (Fig. 1A). Furthermore,
DDP had almost no effect on A549/DDP cells at concentrations
ranging from 5–30 µM (Fig. 1B). This
finding indicated that A549/DDP cells were successfully induced. In
addition, A549 and A549/DDP cells were treated with different
concentrations of DMC. The IC50 values of DMC in A549
and A549/DDP cells were 19.88 and 20.83 µM, respectively (Fig. 1C and D), indicating that induced
A549/DDP cells were not resistant to DMC. It has been reported that
targeting ERCC1 may mediate therapeutic sensitivity to
platinum-based drugs (25).
Subsequently, to further explore the resistance of A549/DDP cells
to DDP, western blotting was performed. The results demonstrated
that the expression of ERCC1 was significantly increased in
A549/DDP cells compared with that in A549 cells (Fig. 1E).
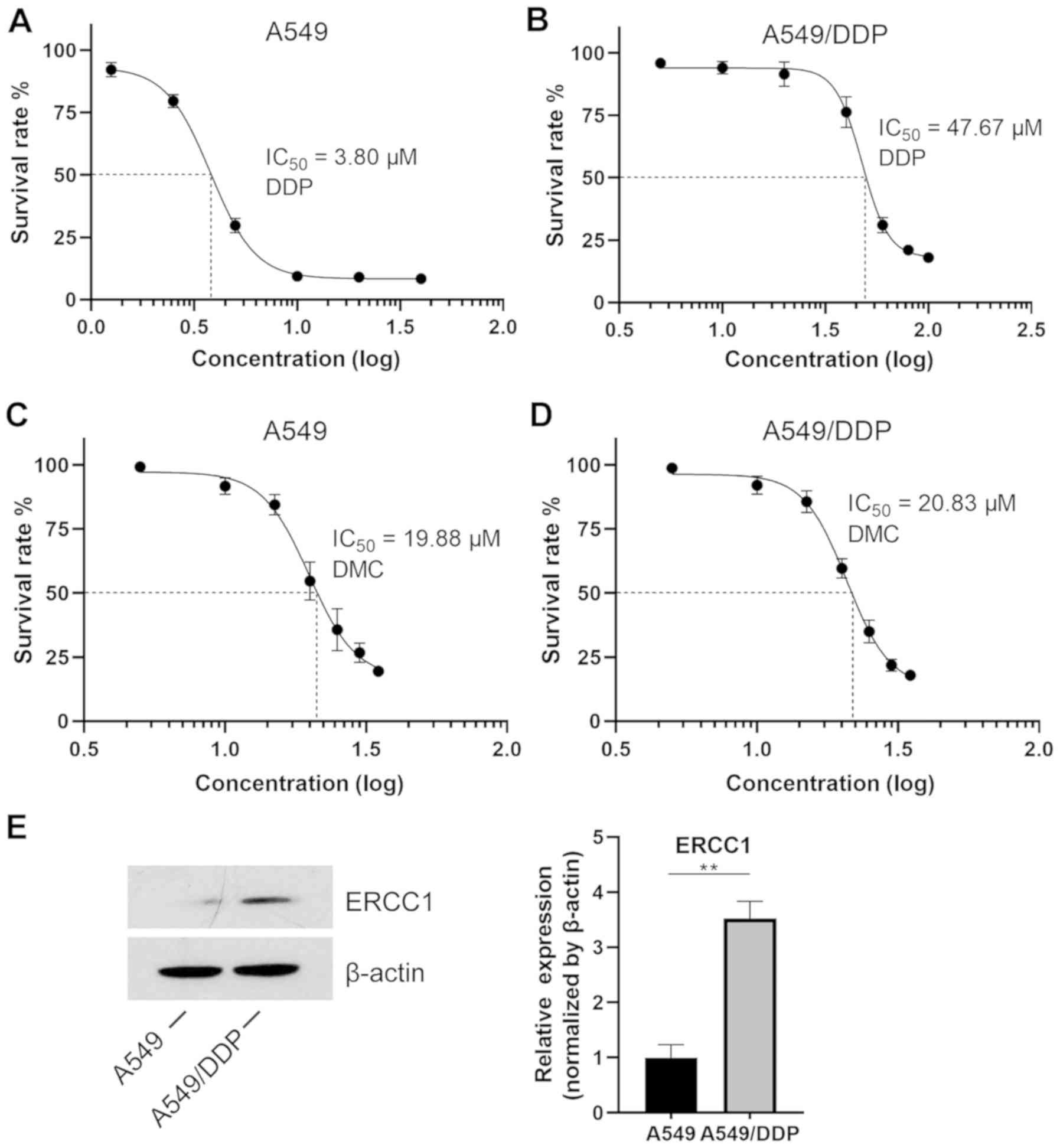 | Figure 1.IC50 values in A549 and
A549/DDP cells. (A) After 48 h, A549 cells were treated with DDP
and an MTT assay was performed. The concentrations of DDP were
1.25, 2.5, 5, 10, 20 and 40 µM. (B) A549/DDP cells were treated
with increasing concentrations of DDP (5, 10, 20, 40, 60, 80 and
100 µM). (C) A549 and (D) A549/DDP cells were treated with DMC at
the same doses. The drug doses were 5, 10, 15, 20, 25, 30 and 35
µM. (E) Expression of ERCC1 in both A549 and A549/DDP cell lines.
**P<0.01. DDP, cisplatin; DMC, demethoxycurcumin; ERCC1,
excision repair cross-complementary 1. |
Combination treatment with DMC
increases the sensitivity of A549/DDP cells to DDP
The efficiency of DMC and DDP combined treatment in
A549/DDP cells was determined and showed that all DMC
concentrations (5, 10 and 20 µM) could restore the cell sensitivity
to DDP in drug-resistant A549/DDP cells (Table I). Based on the concentration
screening results, the lowest DMC concentration (5 µM) was chosen
for further experiments. A549/DDP cells were treated with DDP (3.8
µM) in combination with DMC. The DDP concentration of 3.8 µM
corresponded to the IC50 value obtained in normal A549
cells. However, the combined drug group did not significantly
inhibit cell proliferation compared with the single drug groups
(Fig. 2A; combined drug group vs.
DDP, P=0.112; combined drug group vs. DMC, P=0.053). Therefore,
cells were subsequently treated with elevated concentrations of
DDP. The results demonstrated that combination treatment with 10
and 20 µM DDP significantly inhibited the proliferation of A549/DDP
cells (P<0.05; Fig. 2B and C).
Subsequently, normal human lung fibroblasts (MRC-5 cells) were
treated with high concentrations of DMC. The results from the MTT
assay revealed that DMC exhibited low toxicity in MRC-5 cells, even
at concentrations up to 80 µM (Fig.
2D). Furthermore, combination treatment with DMC and DDP did
not induce significantly greater toxicity in MRC-5 cells compared
with DDP monotherapy (Fig. 2D).
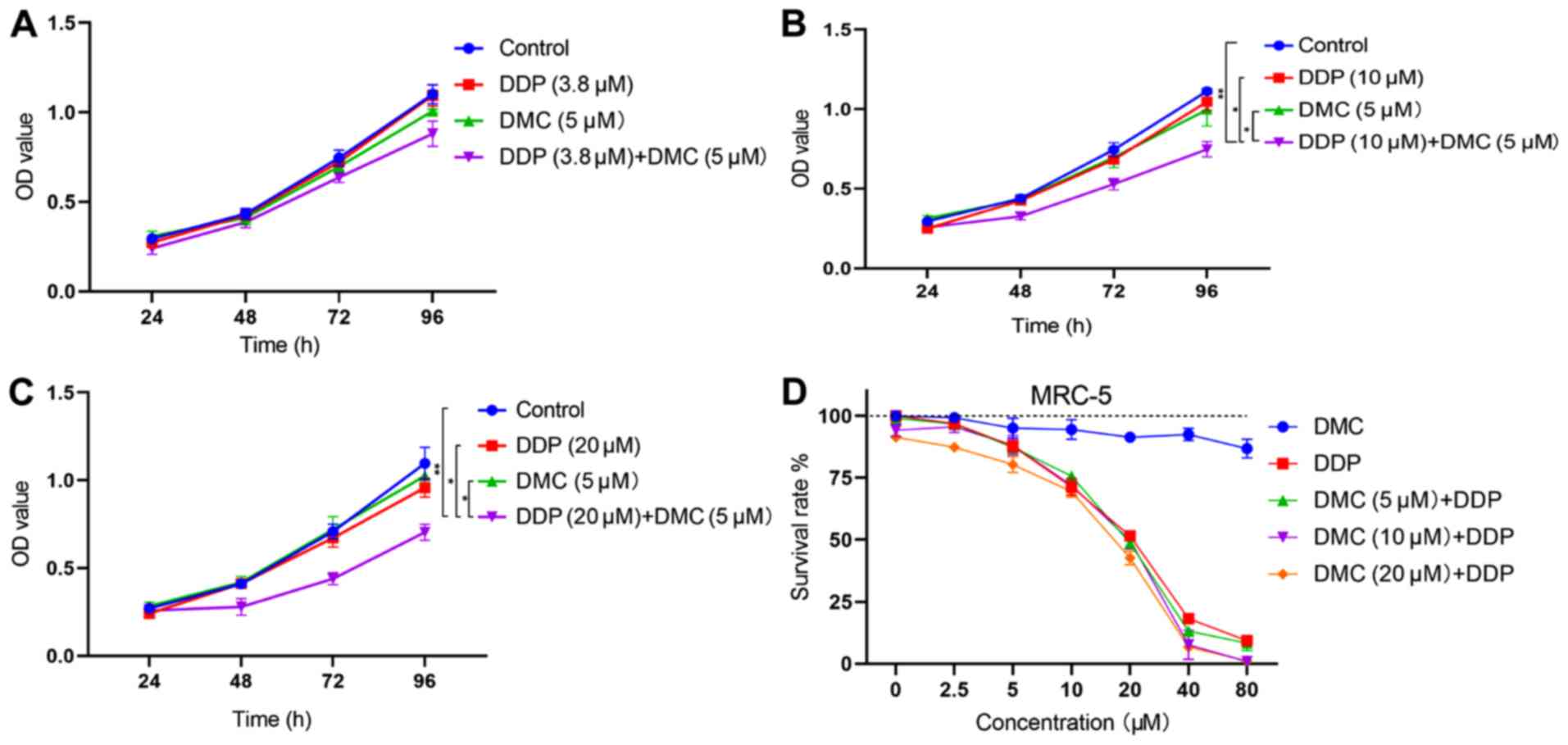 | Figure 2.Combined treatment with DMC increases
DDP sensitivity in A549/DDP cells. (A) Treatment of A549/DDP cells
with DDP (3.8 µM) and DMC (5 µM). (B and C) A549/DDP cells were
treated with high concentrations of DDP (10 and 20 µM) and DMC (5
µM). (D) MRC-5 cells were treated with different concentrations of
DMC for 48 h. MRC-5 cells were also treated with single DMC or DDP
(0, 2.5, 5, 10, 20, 40 and 80 µM) or the combination of DMC (5, 10
and 20 µM) and different concentrations of DDP (0, 2.5, 5, 10, 20,
40 and 80 µM) for 48 h. *P<0.05, **P<0.01. DMC,
demethoxycurcumin; DDP, cisplatin; OD, optical density. |
 | Table I.Synergistic roles of DMC and DDP in
A549/DDP cells. |
Table I.
Synergistic roles of DMC and DDP in
A549/DDP cells.
| A, 5 µM DMC |
|---|
|
|---|
|
| Cell survival
effects, % |
|
|---|
|
|
|
|
|---|
| DDP, µM | DMC | DDP | DMC + DDP | CDIa |
|---|
| 3.8 | 94.71±0.49 | 99.40±0.14 | 80.94±2.01 | 0.86 |
| 10 | 94.71±0.49 | 95.52±0.09 | 62.77±0.14 | 0.69 |
| 20 | 94.71±0.49 | 90.14±0.05 | 53.40±0.23 | 0.63 |
|
| B, 10 µM
DMC |
|
|
| Cell survival
effects, % |
|
|
|
|
|
| DDP, µM | DMC | DDP | DMC +
DDP | CDIa |
|
| 3.8 | 87.16±0.49 | 99.40±0.14 | 73.31±1.19 | 0.85 |
| 10 | 87.16±0.49 | 95.52±0.09 | 56.11±0.34 | 0.67 |
| 20 | 87.16±0.49 | 90.14±0.05 | 41.34±0.08 | 0.52 |
|
| C, 20 µM
DMC |
|
|
| Cell survival
effects, % |
|
|
|
|
|
| DDP, µM | DMC | DDP | DMC +
DDP | CDIa |
|
| 3.8 | 59.6±0.11 | 99.40±0.14 | 53.32±1.19 | 0.94 |
| 10 | 59.6±0.11 | 95.52±0.09 | 36.19±0.34 | 0.64 |
| 20 | 59.6±0.11 | 90.14±0.05 | 27.33±0.08 | 0.50 |
Combination drug treatment promotes
A549/DDP cell apoptosis by activating the caspase-3 signaling
pathway
The MTT assay demonstrated that the drug combination
significantly attenuated the proliferation of the A549/DDP cells.
Furthermore, following treatment with the drug combination for 48
h, the results from the TUNEL assay revealed that the apoptosis
rate of the A549/DDP cells in the combined drug group was
significantly elevated compared with control and monotherapy groups
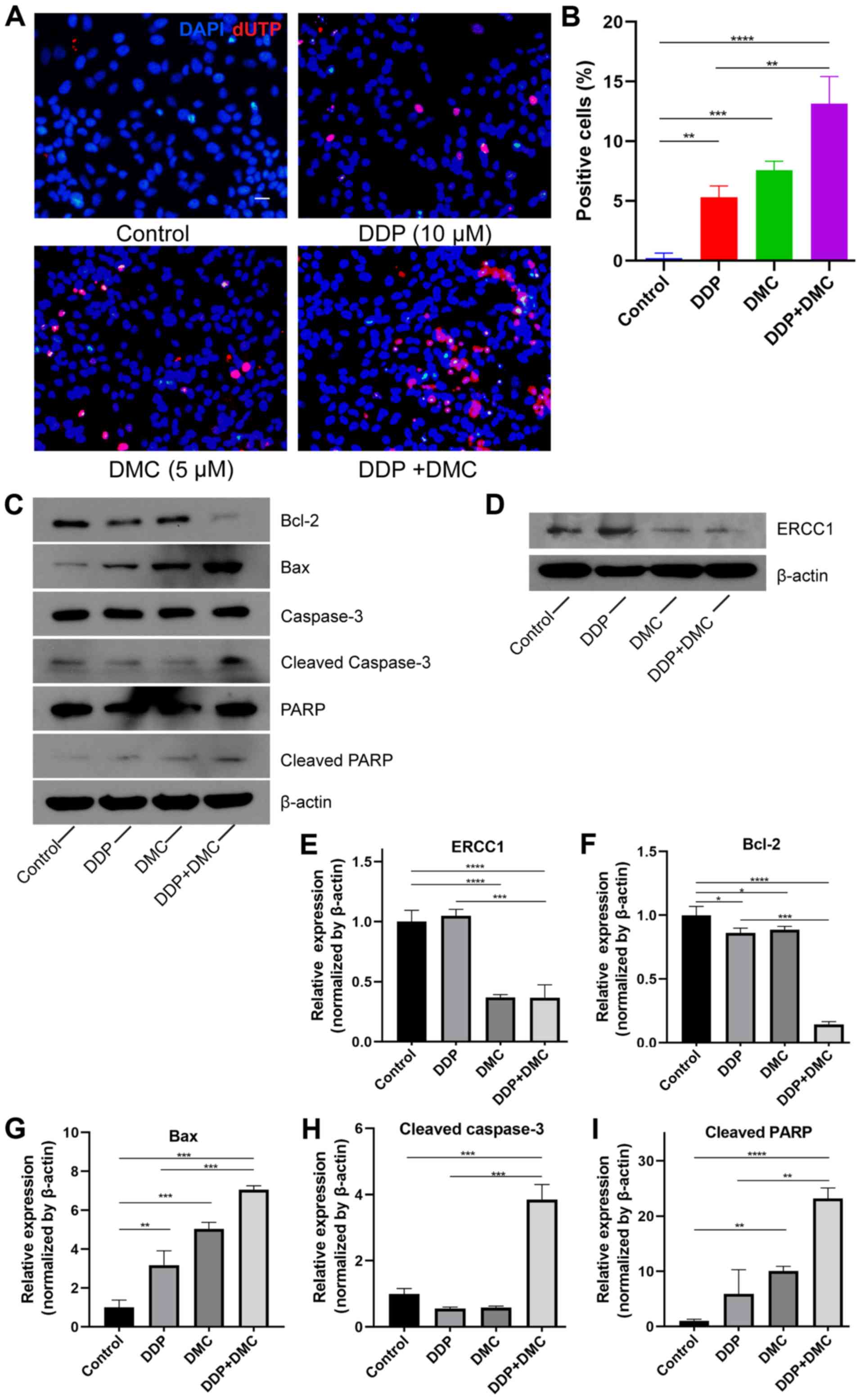
(DDP or DMC) (Fig. 3A). The
apoptosis rates in the single drug groups were 5.30±0.95 and
7.57±0.76% in DDP and DMC groups, respectively, whereas it reached
13.13±2.29% in the combined drug group. In addition, DMC increased
the sensitivity of A549 cells to DDP (P<0.01; Fig. 3B). To investigate the mechanism
underlying A549/DDP cell apoptosis, the expression of
apoptosis-related proteins was detected by western blotting. ERCC1
expression was significantly decreased in the DMC treatment group
(Fig. 3D and E). Bax and Bcl-2
proteins belong to the Bcl-2 family of genes, whereas Bcl-2 is
involved in the inhibition of apoptosis. By contrast, Bax not only
antagonizes the inhibitory effect of Bcl-2 but also promotes
apoptosis (26,27). CMN co-treatment enhanced the effects
on apoptosis in A549/DDP cells. The results demonstrated that Bcl-2
and Bax expression was decreased and increased, respectively, in
the combination groups (Fig. 3C and
F-G). In addition, expression of cleaved caspase-3 was
increased in the combination group compared with control and single
drug groups (Fig. 3C and H). Poly
ADP-ribose polymerase (PARP), a cleavage substrate of caspase, is a
major contributor to apoptosis and cleaved PARP is considered an
important indicator of apoptosis and an activator of caspase 3
cleavage (28). The results
demonstrated that cleaved PARP expression was increased in the
combined drug group, indicating that the caspase-mediated apoptosis
pathway was activated (Fig.
3C-I).
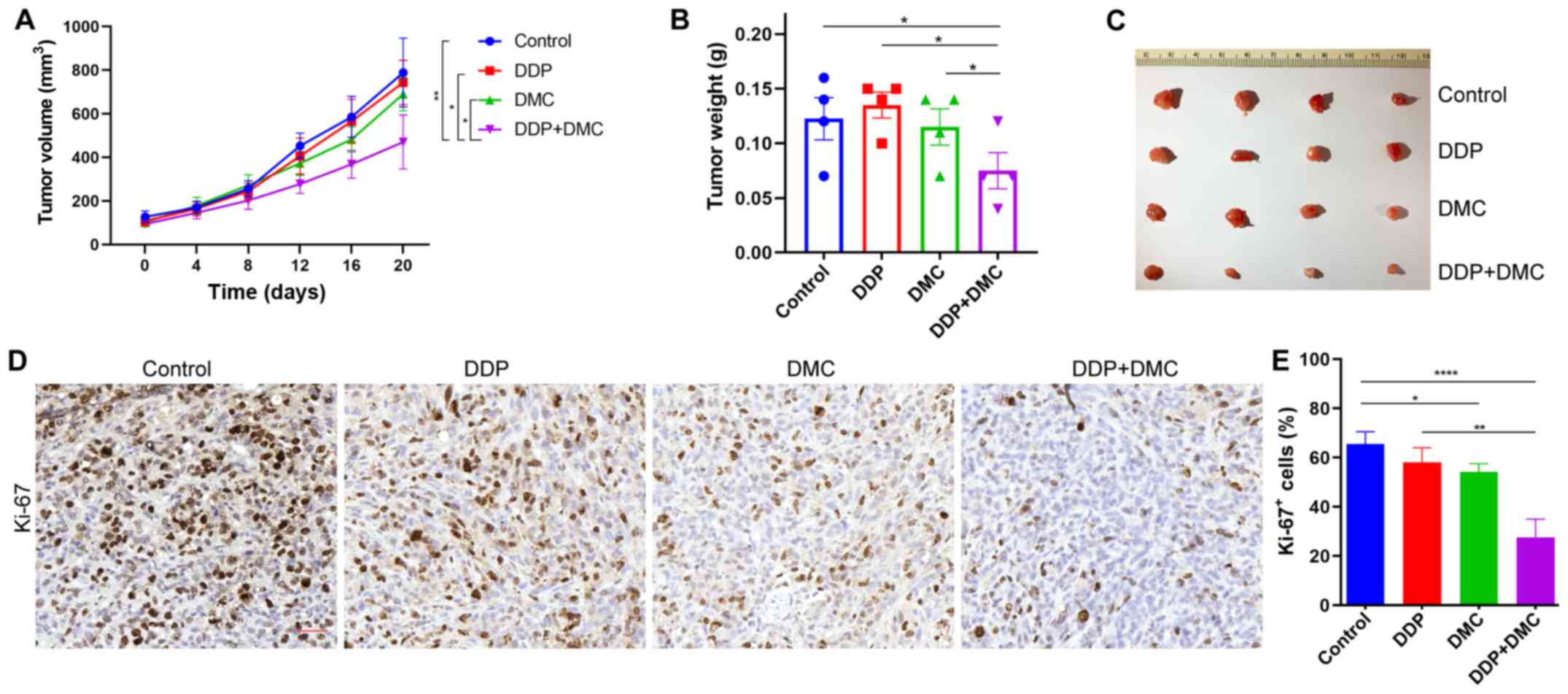
Combination drug treatment inhibits
the proliferation of A549/DDP cells in vivo
The present study investigated whether the drug
combination could exert the same effects on inhibiting
proliferation in vivo. To do so, a subcutaneous ectopic
tumor formation model was established by injecting A549/DDP cells
into nude mice, which were then treated with DMSO (Control group),
single drug (DDP or DMC) and a combination treatment. The results
revealed that the tumor size in the combination group was
significantly lower compared with that in the single drug groups
(Fig. 4A). Following tumor growth in
nude mice for 20 days, tumors were removed and weighed. Consistent
with the tumor size measurements, tumor weight in the combination
group was also lower than that in the other groups (Fig. 4B and C). In addition,
immunohistochemical staining indicated that the Ki-67 proliferation
index was significantly decreased in the combined drug group
compared with that in the other groups (Fig. 4D and E). These findings confirmed the
in vitro effects of DDP treatment combined with DMC.
Discussion
DDP is considered as a classic chemotherapeutic drug
for patients with NSCLC; however, prolonged chemotherapy gradually
increases the risk of resistance and side effects may occur,
leading therefore to reduced treatment efficacy and sharp
deteriorations in the health of patients (29,30).
NSCLC treatment strategies involving the combination of DDP with
other chemotherapy drugs, including taxanes, pemetrexed and
gemcitabine, may significantly reduce drug resistance (31). There is increasing attention for the
combined use of drugs as a way to reduce resistance to single drugs
(32).
Although DMC and BDMC exhibit structural
similarities to CMN, it is difficult to isolate both drugs from
Curcuma longa, making their study also difficult (33). A previous study reported that CMN,
DMC and BDMC were efficiently isolated from Curcuma longa
with a purity >98% (34).
Compared with CMN and BDMC, DMC has the most potent inhibitory
effect on the migration and balloon injury-induced neointimal
formation of vascular smooth muscle cells (35). Furthermore, DMC shows the greatest
inhibitory effect among all curcuminoids, as demonstrated by
rhodamine 123 efflux and calcineurin-AM accumulation assays
(36). Numerous studies have
revealed that curcuminoids could restore drug sensitivity in
drug-resistant cancer cells (37,38).
Furthermore, in colorectal cancer cell lines with acquired
resistance to oxaliplatin (OXA), treatment with CMN in combination
with OXA is more potent and results in reversion of OXA resistance
(39). In addition, DMC in
combination with DDP significantly improves post-target resistance
to DDP in NSCLC cells (40). In the
present study, the DDP-resistant NSCLC A549/DDP cell line was first
induced. The results demonstrated that treatment with DDP or DMC
alone at low concentrations (5 µM) could not inhibit A549/DDP cell
proliferation. However, DDP (10 and 20 µM) in combination with DMC
enhanced the sensitivity of A549/DDP cells to DDP. Nevertheless,
this effect was not robust when cells were treated with 3.8 µM of
DDP, which was the IC50 value of DDP in non-resistant
A549 cells. These results suggested that DMC decreased DDP
resistance of A549/DDP cells; however, DDP resistance was not
completely restored. Unfortunately, only one cell line was used in
the present study, which is a limitation. Further investigation
using additional cell lines would confirm the results from the
present study.
Weakening the DNA damage repair capacity is one of
the important mechanisms of DDP resistance in cancer cells
(41). ERCC1 exerts an essential
function at the 5′ site of impaired DNA, whereas hyperexpression of
ERCC1 in cancer cells is associated with the removal of DDP adducts
from genomic DNA and drug resistance (25). ERCC1 is considered as a key
therapeutic target for lung cancer treatment. It has been
demonstrated that targeting ERCC1 can recover treatment sensitivity
to platinum-based drugs (25).
Numerous studies have reported the mechanisms underlying the
effects of DMC in combination with DDP treatment in enhanced DDP
sensitivity. A study demonstrated that combined administration of
CMN and DDP can increase DDP cytotoxicity in cancer cells by
downregulating the expression of thymidine phosphorylase (TP) and
ERCC1, as well as inactivating the ERK1/2 signaling pathway
(42). Another study demonstrated
that enhanced DDP cytotoxicity, when the drug is administrated in
combination with DMC, is regulated via the PI3K-Akt-Snail pathway
and induced by downregulating the expression of TP and ERCC1
(43). Consistent with the previous
study, the present study demonstrated that ERCC1 expression was
significantly decreased in the combination group compared with that
in the DMC treatment group. This finding suggested that ERCC1
upregulation may increase A549/DDP cell sensitivity to DDP.
Furthermore, combination treatment enhanced the inhibitory effect
of DDP on A549/DDP cells and promoted apoptosis. Finally, the
results obtained by comparing tumor size and weight among different
treatment groups suggested that the combined drug group exhibited
marked therapeutic effects in vivo compared with the
monotherapy groups (DMC or DDP alone), indicating, therefore, that
DMC may function in vivo.
The results from the present study indicated that
DDP in combination with DMC may restore DDP sensitivity in
DDP-resistant NSCLC cells. Therefore, co-treatment of DMC with
chemotherapeutic drugs may be considered as an attractive approach
to overcome drug resistance in patients with NSCLC.
Acknowledgements
Not applicable.
Funding
This study was supported by Zhejiang Province
Traditional Chinese Medicine Science and Technology Program (grant
no. 2016ZA030).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
ZQ and YC designed and performed the experiments. YC
performed the in vitro experiments and wrote the manuscript.
CH performed the in vivo experiments. XC analyzed data and
modified the paper. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All animal procedures were approved by the
Institutional Animal Care and Use Committee of Nanjing Medical
University (approval no. IACUC-1907009).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nasim F, Sabath BF and Eapen GA: Lung
cancer. Med Clin North Am. 103:463–473. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peters S, Camidge DR, Shaw AT, Gadgeel S,
Ahn JS, Kim DW, Ou SI, Pérol M, Dziadziuszko R, Rosell R, et al:
Alectinib versus crizotinib in untreated ALK-positive
non-small-cell lung cancer. N Engl J Med. 377:829–838. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rinaldi M, Cauchi C and Gridelli C: First
line chemotherapy in advanced or metastatic NSCLC. Ann Oncol. 17
(Suppl 5):v64S–v67S. 2006. View Article : Google Scholar
|
|
4
|
Ke B, Wei T, Huang Y, Gong Y, Wu G, Liu J,
Chen X and Shi L: Interleukin-7 resensitizes non-small-cell lung
cancer to cisplatin via inhibition of ABCG2. Mediators Inflamm.
2019:72414182019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Galluzzi L, Senovilla L, Vitale I, Michels
J, Martins I, Kepp O, Castedo M and Kroemer G: Molecular mechanisms
of cisplatin resistance. Oncogene. 31:1869–1883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amable L: Cisplatin resistance and
opportunities for precision medicine. Pharmacol Res. 106:27–36.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Janyou A, Changtam C, Suksamrarn A,
Tocharus C and Tocharus J: Suppression effects of
O-demethyldemethoxycurcumin on thapsigargin triggered on
endoplasmic reticulum stress in SK-N-SH cells. Neurotoxicology.
50:92–100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Simental-Mendia LE, Pirro M, Gotto AM Jr,
Banach M, Atkin SL, Majeed M and Sahebkar A: Lipid-modifying
activity of curcuminoids: A systematic review and meta-analysis of
randomized controlled trials. Crit Rev Food Sci Nutr. 59:1178–1187.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Benediktsdottir BE, Baldursson O,
Gudjonsson T, Tonnesen HH and Masson M: Curcumin,
bisdemethoxycurcumin and dimethoxycurcumin complexed with
cyclodextrins have structure specific effect on the paracellular
integrity of lung epithelia in vitro. Biochem Biophys Rep.
4:405–410. 2015.PubMed/NCBI
|
|
10
|
Yuan F, Dong H, Gong J, Wang D, Hu M,
Huang W, Fang K, Qin X, Qiu X, Yang X and Lu F: A systematic review
and meta-analysis of randomized controlled trials on the effects of
turmeric and curcuminoids on blood lipids in adults with metabolic
diseases. Adv Nutr. 10:791–802. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Imran M, Ullah A, Saeed F, Nadeem M,
Arshad MU and Suleria HAR: Cucurmin, anticancer, & antitumor
perspectives: A comprehensive review. Crit Rev Food Sci Nutr.
58:1271–1293. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Al Ayoub Y, Gopalan RC, Najafzadeh M,
Mohammad MA, Anderson D, Paradkar A and Assi KH: Development and
evaluation of nanoemulsion and microsuspension formulations of
curcuminoids for lung delivery with a novel approach to
understanding the aerosol performance of nanoparticles. Int J
Pharm. 557:254–263. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Adiwidjaja J, McLachlan AJ and Boddy AV:
Curcumin as a clinically-promising anti-cancer agent:
Pharmacokinetics and drug interactions. Expert Opin Drug Metab
Toxicol. 13:953–972. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shanmugam MK, Rane G, Kanchi MM, Arfuso F,
Chinnathambi A, Zayed ME, Alharbi SA, Tan BK, Kumar AP and Sethi G:
The multifaceted role of curcumin in cancer prevention and
treatment. Molecules. 20:2728–2769. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hassanalilou T, Ghavamzadeh S and Khalili
L: Curcumin and gastric cancer: A review on mechanisms of action. J
Gastrointest Cancer. 50:185–192. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mendonca LM, Machado Cda S, Teixeira CC,
Freitas LA, Bianchi ML and Antunes LM: Comparative study of
curcumin and curcumin formulated in a solid dispersion: Evaluation
of their antigenotoxic effects. Genet Mol Biol. 38:490–498. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Quitschke WW: Differential solubility of
curcuminoids in serum and albumin solutions: Implications for
analytical and therapeutic applications. BMC Biotechnol. 8:842008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen G, Chen Y, Yang N, Zhu X, Sun L and
Li G: Interaction between curcumin and mimetic biomembrane. Sci
China Life Sci. 55:527–532. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hatamipour M, Ramezani M, Tabassi SAS,
Johnston TP and Sahebkar A: Demethoxycurcumin: A naturally
occurring curcumin analogue for treating non-cancerous diseases. J
Cell Physiol. 234:19320–19330. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee JW, Hong HM, Kwon DD, Pae HO and Jeong
HJ: Dimethoxycurcumin, a structural analogue of curcumin, induces
apoptosis in human renal carcinoma caki cells through the
production of reactive oxygen species, the release of cytochrome C,
and the activation of caspase-3. Korean J Urol. 51:870–878. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ni X, Zhang A, Zhao Z, Shen Y and Wang S:
Demethoxycurcumin inhibits cell proliferation, migration and
invasion in prostate cancer cells. Oncol Rep. 28:85–90.
2012.PubMed/NCBI
|
|
22
|
Ko YC, Lien JC, Liu HC, Hsu SC, Lin HY,
Chueh FS, Ji BC, Yang MD, Hsu WH and Chung JG:
Demethoxycurcumin-induced DNA damage decreases DNA
Repair-associated protein expression levels in NCI-H460 human lung
cancer cells. Anticancer Res. 35:2691–2698. 2015.PubMed/NCBI
|
|
23
|
Ko YC, Lien JC, Liu HC, Hsu SC, Ji BC,
Yang MD, Hsu WH and Chung JG: Demethoxycurcumin induces the
apoptosis of human lung cancer NCI-H460 cells through the
mitochondrial-dependent pathway. Oncol Rep. 33:2429–2437. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang HB and Chen BH: Inhibition of lung
cancer cells A549 and H460 by curcuminoid extracts and
nanoemulsions prepared from Curcuma longa linnaeus. Int J
Nanomedicine. 10:5059–5080. 2015.PubMed/NCBI
|
|
25
|
McNeil EM and Melton DW: DNA repair
endonuclease ERCC1-XPF as a novel therapeutic target to overcome
chemoresistance in cancer therapy. Nucleic Acids Res.
40:9990–10004. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Adams JM and Cory S: The Bcl-2 protein
family: Arbiters of cell survival. Science. 281:1322–1326. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ashkenazi A, Fairbrother WJ, Leverson JD
and Souers AJ: From basic apoptosis discoveries to advanced
selective BCL-2 family inhibitors. Nat Rev Drug Discov. 16:273–284.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rheaume E, Cohen LY, Uhlmann F, Lazure C,
Alam A, Hurwitz J, Sékaly RP and Denis F: The large subunit of
replication factor C is a substrate for caspase-3 in vitro and is
cleaved by a caspase-3-like protease during Fas-mediated apoptosis.
EMBO J. 16:6346–6354. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rotolo F, Dunant A, Le Chevalier T, Pignon
JP and Arriagada R; IALT Collaborative Group, : Adjuvant
cisplatin-based chemotherapy in nonsmall-cell lung cancer: New
insights into the effect on failure type via a multistate approach.
Ann Oncol. 25:2162–2166. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ricci S, Antonuzzo A, Galli L, Tibaldi C,
Bertuccelli M, Lopes Pegna A, Petruzzelli S, Bonifazi V, Orlandini
C and Franco Conte P: A randomized study comparing two different
schedules of administration of cisplatin in combination with
gemcitabine in advanced nonsmall cell lung carcinoma. Cancer.
89:1714–1719. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moldvay J, Rokszin G, Abonyi-Toth Z,
Katona L and Kovacs G: Analysis of drug therapy of lung cancer in
Hungary. Magy Onkol. 57:33–38. 2013.(In Hungarian). PubMed/NCBI
|
|
32
|
Bayat Mokhtari R, Homayouni TS, Baluch N,
Morgatskaya E, Kumar S, Das B and Yeger H: Combination therapy in
combating cancer. Oncotarget. 8:38022–38043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hung CM, Su YH, Lin HY, Lin JN, Liu LC, Ho
CT and Way TD: Demethoxycurcumin modulates prostate cancer cell
proliferation via AMPK-induced down-regulation of HSP70 and EGFR. J
Agric Food Chem. 60:8427–8434. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sheu MJ, Lin HY, Yang YH, Chou CJ, Chien
YC, Wu TS and Wu CH: Demethoxycurcumin, a major active curcuminoid
from Curcuma longa, suppresses balloon injury induced
vascular smooth muscle cell migration and neointima formation: An
in vitro and in vivo study. Mol Nutr Food Res. 57:1586–1597. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Teng YN, Hsieh YW, Hung CC and Lin HY:
Demethoxycurcumin modulates human P-glycoprotein function via
uncompetitive inhibition of ATPase hydrolysis activity. J Agric
Food Chem. 63:847–855. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ruiz de Porras V, Bystrup S,
Martinez-Cardus A, Pluvinet R, Sumoy L, Howells L, James MI, Iwuji
C, Manzano JL, Layos L, et al: Curcumin mediates
oxaliplatin-acquired resistance reversion in colorectal cancer cell
lines through modulation of CXC-Chemokine/NF-KB signalling pathway.
Sci Rep. 6:246752016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao S, Pi C, Ye Y, Zhao L and Wei Y:
Recent advances of analogues of curcumin for treatment of cancer.
Eur J Med Chem. 180:524–535. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Changtam C, Hongmanee P and Suksamrarn A:
Isoxazole analogs of curcuminoids with highly potent
multidrug-resistant antimycobacterial activity. Eur J Med Chem.
45:4446–4457. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen YY, Lin YJ, Huang WT, Hung CC, Lin
HY, Tu YC, Liu DM, Lan SJ and Sheu MJ: Demethoxycurcumin-Loaded
chitosan nanoparticle downregulates DNA repair pathway to improve
cisplatin-induced apoptosis in non-small cell lung cancer.
Molecules. 23:32172018. View Article : Google Scholar
|
|
40
|
Zhu H, Luo H, Zhang W, Shen Z, Hu X and
Zhu X: Molecular mechanisms of cisplatin resistance in cervical
cancer. Drug Des Devel Ther. 10:1885–1895. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vikhanskaya F, Marchini S, Marabese M,
Galliera E and Broggini M: P73a overexpression is associated with
resistance to treatment with DNA-damaging agents in a human ovarian
cancer cell line. Cancer Res. 61:935–938. 2001.PubMed/NCBI
|
|
42
|
Tsai MS, Weng SH, Kuo YH, Chiu YF and Lin
YW: Synergistic effect of curcumin and cisplatin via
down-regulation of thymidine phosphorylase and excision repair
cross-complementary 1 (ERCC1). Mol Pharmacol. 80:136–146. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lin CY, Hung CC, Wang CCN, Lin HY, Huang
SH and Sheu MJ: Demethoxycurcumin sensitizes the response of
non-small cell lung cancer to cisplatin through downregulation of
TP and ERCC1-related pathways. Phytomedicine. 53:28–36. 2019.
View Article : Google Scholar : PubMed/NCBI
|