Introduction
Osteosarcoma (OS) is a malignant tumor that
originates in bones, especially in children and adolescents. In
addition, OS accounts for about 5% of all childhood tumors
(1). Due to the high malignancy of
OS and poor prognosis, patients may develop lung metastases within
a few months or weeks. The 3- to 5-year survival rate of OS
patients with amputation is usually between 5 and 20% (2). The key factors affecting the prognosis
of OS are early diagnosis, complete tumor resection, chemotherapy
and radiotherapy before and after surgery (3). In addition, it is also related to the
tissue type, size and local lymph node status of the tumor cells
(4). Although existing treatment
methods have greatly improved the survival rate of OS patients, new
and effective treatment methods are still needed.
MicroRNAs (miRNAs/miRs) are highly conserved
endogenous RNAs, which play important regulatory roles in human
cancers (5). It has been found that
miRNAs are involved in biological activity through a complex
regulatory network. This complex regulatory network can regulate
multiple genes involved in cellular activities through an miRNA,
and can also finely regulate a gene through the interaction of
multiple miRNAs (6). In particular,
it has been found that miRNAs can exhibit regulatory effect on bone
cancers (7). For example, miR-214
was found to function as an oncogene in human OS by targeting TRAF3
(8). In contrast, miR-876-5p
inhibited cell viability and metastasis in OS by targeting c-Met
(9). Recently, the different roles
of miR-615 in human cancers have attracted our attention.
Upregulation of miR-615 has been found in gastric cancer. Moreover,
overexpression of miR-615 was found to promote cell proliferation
and migration and inhibit apoptosis in gastric cancer (10). However, downregulation of miR-615 was
found in non-small cell lung cancer and renal cell carcinoma
(11,12). In addition, miR-615 was found to play
an inhibitory role in esophageal squamous cell carcinoma by
targeting IGF2 (13). These findings
indicate that miR-615 has a tissue-specific function in different
types of cancer. At present, the specific role of miR-615 in OS is
unclear and needs to be investigated.
As an important regulator, hexokinase 2 (HK2) has
been found to be involved in various human cancers, such as
laryngeal carcinoma and liver cancer (14,15). In
detail, upregulation of HK2 was detected in hepatocellular
carcinoma and breast cancer (16,17).
Functionally, knockdown of HK2 was found to inhibit the growth of
lung carcinoma A549 cells (18). In
addition, it was found that upregulation of HK2 promoted the
proliferation of ovarian cancer cells (19). Lu et al demonstrated that
miR-603 inhibited the malignancy of ovarian cancer cells by
targeting HK2 (20). However, the
relationship between HK2 and miR-615 has not been reported in
previous studies. In addition, the phosphoinositide 3-kinase
(PI3K)/protein kinase B (AKT)/HK2 axis has been proposed to
regulate the development of breast cancer (21). It has been suggested that the
PI3K/AKT pathway is involved in the regulation of tumorigenesis,
such as renal cancer and esophageal squamous cell carcinoma
(22,23). Xu et al demonstrated that
miR-149 inhibited cell growth by regulating the PI3K/AKT pathway in
human OS (24). Therefore, the
effect of miR-615 on HK2 expression and the PI3K/AKT pathway was
also explored in OS. More importantly, the effect of miR-615 on OS
cell viability and metastasis was investigated in this study. The
present study facilitates the understanding of the pathogenesis of
OS.
Materials and methods
Clinical tissues
A total of 92 paired OS tissues were collected from
OS patients (60 males and 32 females, mean age 21 years, range from
14 to 33 years) who had undergone resection between January 2017
and January 2019 at the Shandong Provincial Third Hospital.
Moreover, All OS patients underwent surgery, and none of them
received preoperative radiotherapy or chemotherapy. All of the
patients provided informed consents. Approval for this research was
acquired from the Institutional Ethics Committee of Shandong
Provincial Third Hospital (2016SPT-44).
Cell culture
Human osteoblast hFOB1.19 cells (CRL-11372) and OS
cell line HOS (CRL-1543) were purchased from the American Type
Culture Collection (ATCC). The cells were incubated in RPMI-1640
medium, supplemented with 10% fetal bovine serum (FBS) and 100 U/ml
penicillin-streptomycin (all from HyClone; GE Healthcare). The
cells were cultured at 37°C in a humidified incubator containing 5%
CO2.
Cell transfection
miR-615 mimics (5′-UCCGAGCCUGGGUCUCCCUCUU-3′),
mimics-NC (5′-UUCUCGAACGUGUCACGUUUU-3′), miR-615 inhibitor
(5′-ACCGAGUCAGGGAUACCCACAA-3′), inhibitor-NC
(5′-CAGUACUUUUGUGUAGUACAA-3′) and the HK2 overexpression plasmid
were obtained from GenePharma. These sequences and plasmid were
transfected into HOS cells using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), respectively. For cell function
assay, cells were collected 12 h after transfection. For RT-qPCR
and western blot analysis, cells were collected 24 and 48 h after
transfection. For the dual luciferase assay, cells were collected
48 h after transfection.
RNA isolation and RT-qPCR
Total RNA isolation was performed using Trizol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The miScript
Reverse Transcription kit (Qiagen) was then used to obtain cDNA
solution. RT-qPCR assay was performed using the miScript
SYBR®-Green PCR kit (Qiagen) based on the manufacturer's
instructions. In brief, 20 µl mixtures were heated at 95°C for 3
min for enzyme activation, then the 20 µl reaction mixture was
incubated as follows: 95°C for 3 sec and 61°C for 20 sec for 40
cycles. U6 or GAPDH was used as a control for miR-615 or HK2. The
expression levels of miR-615 and HK2 were quantified using the
2−∆∆Cq method (25). The
forward primer for miR-615 was 5′-CTGCCTTTCACCTTGGAGAC-3′, and the
reverse primer was 5′-CGTTTCCTGGGGATGAGATA-3′. The internal control
GAPDH was forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. The primers for HK2 were
5′-CTTCTTCACGGAGCTCAACC-3 (forward) and 5′-AAGCCCTTTCTCCATCTCCT-3′
(reverse). The internal control was U6 (forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′).
Transwell assay
For the invasion assay, Matrigel (BD Biosciences)
was diluted 1:4 with serum-free DMEM and used to coat the Transwell
inserts (pore size, 8-µm; EMD Millipore) to form a matrix barrier.
For the migration assay, transfected cells were suspended in
FBS-free DMEM containing 0.1% bovine serum albumin (BSA) (Bioworld
Technology, Inc.). After 30 min, HOS cell suspension
(3×103 cells/well) was added to the Transwell upper
chamber, and RPMI-1640 medium (10% FBS) was added to a 24-well
plate in the lower chamber. After 24 h, the cells that had migrated
or invaded through the membrane were fixed with 95% ethyl alcohol
for 15 min at room temperature and stained with 0.1% crystal violet
for 10 min at room temperature. Observation and photographing were
performed using a light microscope (magnification, ×200).
MTT assay
First, the transfected HOS cells (4×103
cells/well) were prepared in a 96-well plate. Then, HOS cells were
incubated for 24, 48, 72 or 96 h in fresh medium, respectively.
After that, 10 µl of MTT solution was added to incubate the cells
for 4 h. Next, the supernatant fractions were discarded, and 150 µl
dimethyl sulfoxide (DMSO) was added to each well to dissolve the
crystals. The absorbance at 490 nm was examined using a
spectrophotometric plate reader (Olympus Corp.).
Dual luciferase reporter assay
First, wild-type WT-HK2-3′UTR or mutant
MUT-HK2-3′UTR was inserted into pmirGLO luciferase reporter vector
(Promega Corp.). Next, a total of 4×105 HOS cells were
seeded per well into 6-well plates and co-transfected with either
50 nM miR-615-3p mimics or mimics-NC and 2 µg plasmid vector using
Lipofectamine® 2000, according to the manufacturer's
protocol. The cells were lysed and assayed for luciferase activity
at 48 h post-transfection using a Dual-Luciferase Assay kit (cat.
no. E1910, Promega Corp.). The firefly luciferase was used as a
reference for normalization.
Western blot analysis
Protein samples were obtained using RIPA lysis
buffer (Beyotime Institute of Biotechnology). Next, proteins were
separated by 10% SDS-PAGE. A total of 30 µg protein samples were
transferred to the PVDF membrane and were blocked with 5% skim
milk. The protein samples were incubated with E-cadherin (rabbit
monoclonal; dilution, 1:1,000; cat. no. ab1416; Abcam), N-cadherin
(rabbit polyclonal; dilution, 1:1,000; cat. no. ab18203; Abcam),
vimentin (rabbit monoclonal; dilution, 1:1,000; cat. no. ab217673;
Abcam), PI3K (rabbit monoclonal; dilution, 1:1,000; cat. no.
ab32089; Abcam), phosphorylated (p-)PI3K (rabbit monoclonal;
dilution, 1:1,000; cat. no. ab154598; Abcam), AKT (rabbit
polyclonal; dilution, 1:1,000; cat. no. ab8805; Abcam), p-AKT
(rabbit monoclonal; dilution, 1:1,000; cat. no. ab81283; Abcam) and
GAPDH (rabbit monoclonal; dilution, 1:1,000; cat. no. ab181602;
Abcam) primary antibodies overnight at 4°C. After that, horseradish
peroxidase-conjugated secondary antibodies (dilution, 1:5,000; cat.
no. ab7090; Abcam) were added and the protein samples were
incubated for 1 h. Protein bands were assessed using an ECL kit
(Beyotime Institute of Biotechnology).
Statistical analysis
The data were analyzed by SPSS 17.0 (SPSS, Inc.) or
Graphpad Prism 6 (GraphPad Software, Inc.). Data are shown as mean
± SD. Differences between groups were analyzed using Student's
t-test or one-way analysis of variance with Tukey's post hoc test.
The relationship between miR-615 expression and the
clinic-pathological characteristics in OS patients was analyzed by
Chi-square test. The univariate Kaplan-Meier method with log-rank
test was used to calculate the overall survival rate and survival
difference. P<0.05 was considered as indicative of statistical
significance.
Results
Downregulation of miR-615 is related
to poor clinical outcomes in OS patients
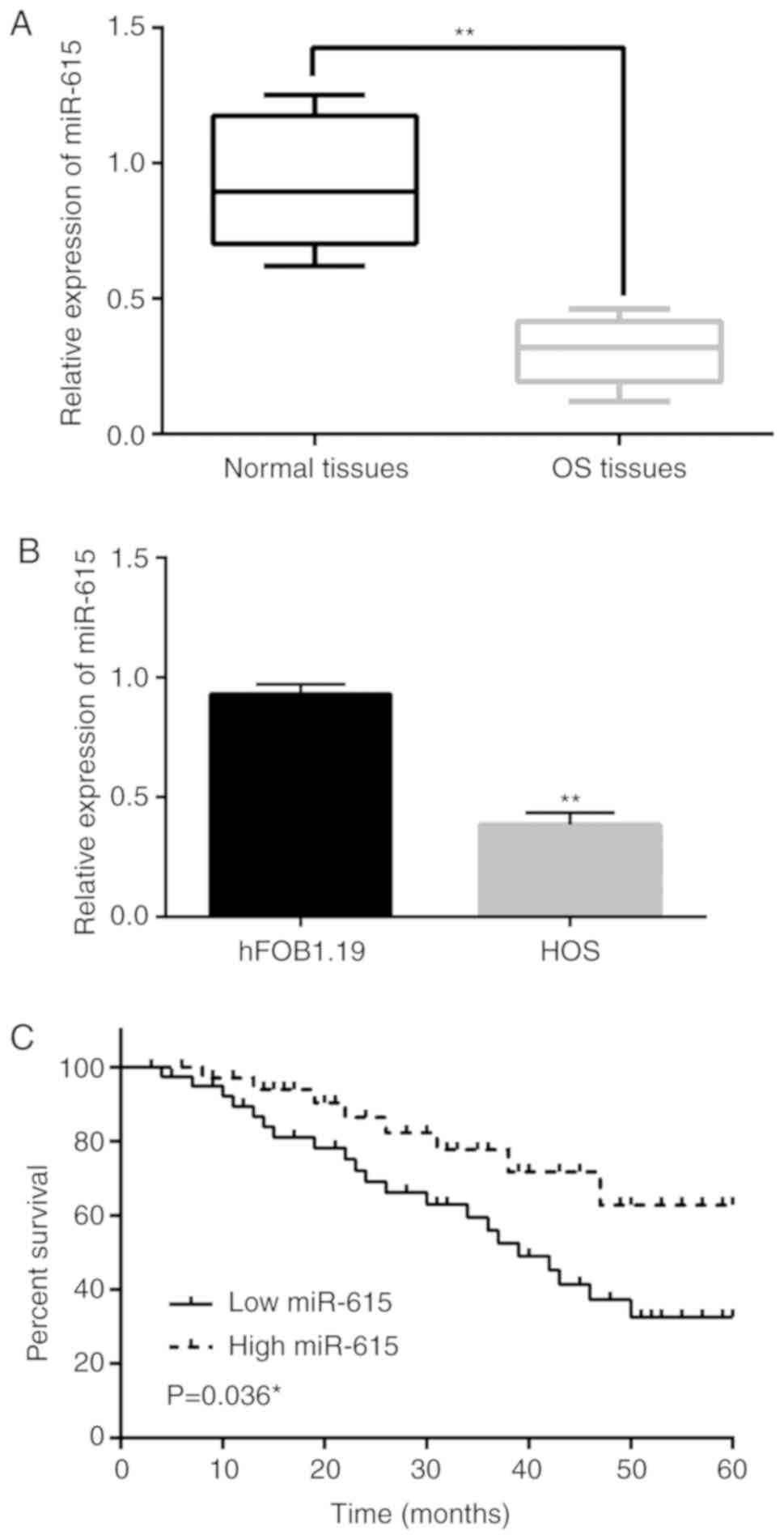
The expression of miR-615 was detected in OS tissues
and cells by RT-qPCR. It was found that miR-615 expression in OS
tissues was downregulated compared to that noted in the normal
tissues (P<0.01; Fig. 1A).
Similarly, the expression of miR-615 in HOS cells was lower than
that in the hFOB1.19 cells (P<0.01; Fig. 1B). Next, we found that the low
expression of miR-615 was associated with TNM stage and lymph node
metastasis in OS patients (P<0.05; Table I). In addition, low miR-615
expression was associated with reduced overall survival of the OS
patients (P=0.036; Fig 1C). These
results indicate that miR-615 may be involved in the tumorigenesis
of OS.
 | Table I.Association between miR-615
expression and the clinicopathological characteristics of the OS
patients. |
Table I.
Association between miR-615
expression and the clinicopathological characteristics of the OS
patients.
|
|
| miR-615 |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Cases | High | Low | P-value |
|---|
| Age (years) |
|
|
| 0.83 |
|
≥18 | 56 | 20 | 36 |
|
|
<18 | 36 | 11 | 25 |
|
| Sex |
|
|
| 0.27 |
|
Male | 60 | 25 | 35 |
|
|
Female | 32 | 6 | 26 |
|
| Tumor size
(cm) |
|
|
| 0.08 |
|
<5 | 63 | 22 | 41 |
|
| ≥5 | 29 | 9 | 20 |
|
| TNM stage |
|
|
| 0.01a |
|
I–II | 71 | 26 | 45 |
|
|
III–IV | 21 | 5 | 16 |
|
| Lymph node
metastasis |
|
|
| 0.01a |
| No | 75 | 26 | 49 |
|
|
Yes | 17 | 5 | 12 |
|
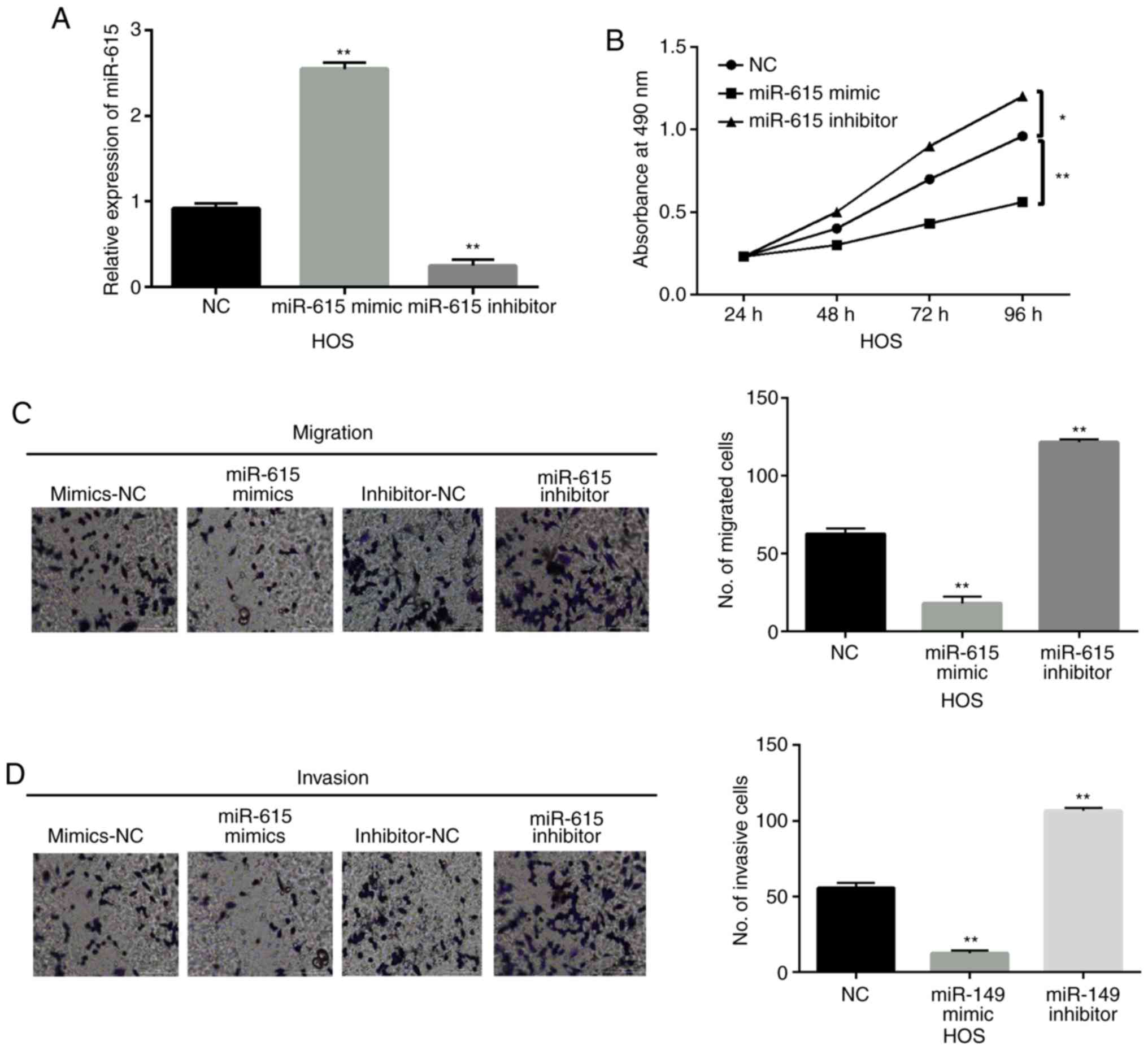
miR-615 inhibits cell viability and
metastasis in OS
Next, miR-615 mimics or inhibitor was transfected
into HOS cells to explore the function of miR-615 in OS. RT-qPCR
showed that the expression of miR-615 was increased by its mimics
and reduced by its inhibitor in HOS cells (P<0.01; Fig. 2A). Functionally, overexpression of
miR-615 inhibited cell proliferation in the HOS cells. In contrast,
downregulation of miR-615 promoted the proliferation of HOS cells
when compared to the NC group (P<0.05 or P<0.01; Fig. 2B). In addition, miR-615 mimics
suppressed cell migration, while miR-615 inhibitor promoted cell
migration in HOS cells when compared to the NC group (P<0.01;
Fig. 2C). Consistently, cell
invasion was also inhibited by miR-615 mimics and promoted by
miR-615 inhibitor in HOS cells when compared to the NC group
(P<0.01; Fig. 2D). These results
indicate that miR-615 functions as a tumor suppressor in the
pathogenesis of OS.
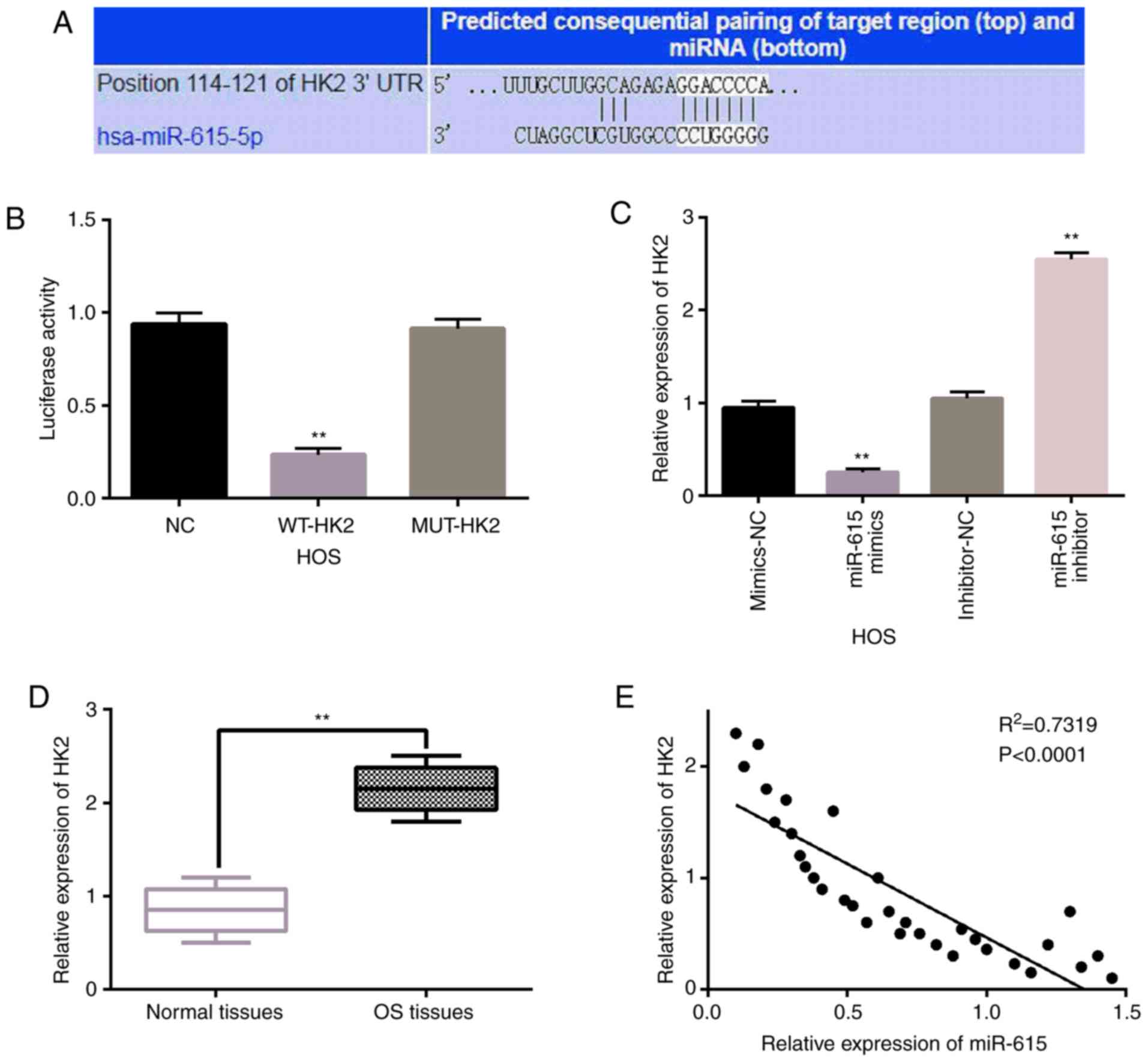
miR-615 directly targets HK2
In addition, TargetScan database (http://www.targetscan.org) indicated that miR-615 has
a binding site that binds to the 3′-UTR (3′-untranslated region) of
HK2 (Fig. 3A). Next, this prediction
was verified by luciferase reporter assay. We found that miR-615
mimics reduced the luciferase activity of WT-HK2 (P<0.01;
Fig. 3B). However, the luciferase
activity of Mut-HK2 was not affected by miR-615 mimics in the HOS
cells. Then, HK2 expression levels were observed in HOS cells with
miR-615 mimics or inhibitor. We found that HK2 expression was
significantly reduced by miR-615 mimics and was significantly
promoted by miR-615 inhibitor in HOS cells (P<0.01; Fig. 3C). Subsequently, the expression of
HK2 was examined in OS tissues to explore the dysregulation of HK2
in the progression of OS. Compared to normal tissues, HK2
expression was significantly upregulated in the OS tissues
(P<0.01; Fig. 3D). In addition,
miR-615 was negatively correlated with HK2 expression in the OS
tissues (P<0.0001, R2=0.7319; Fig. 3E). Taken together, miR-615 directly
targets HK2 and negatively regulates its expression in OS.
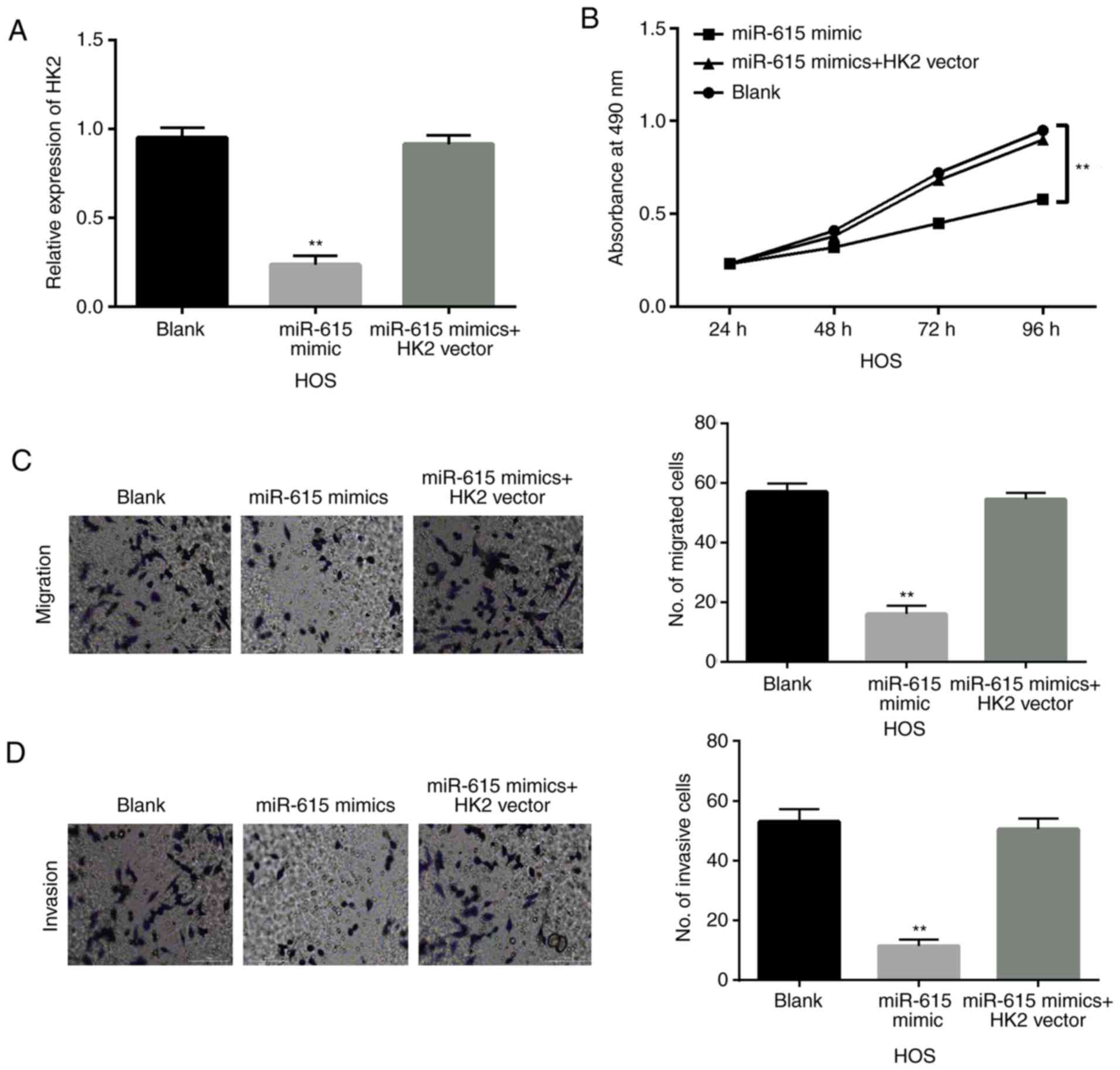
miR-615 is involved in OS progression
through targeting HK2
To explore the interaction between miR-615 and HK2
in OS, HK2 overexpression vector was transfected into HOS cells
with miR-615 mimics. First, the decreased expression of HK2 induced
by miR-615 mimics was recovered by upregulation of HK2 in HOS cells
(Fig. 4A). Similarly, overexpression
of HK2 weakened the inhibitory effect of miR-615 on cell
proliferation in HOS cells (P<0.01; Fig. 4B). Consistently, upregulation of HK2
also weakened the inhibitory effect of miR-615 on cell migration
and invasion in HOS cells (P<0.01; Fig. 4C and D). These findings indicate that
upregulation of HK2 impairs the tumor-suppressive effect of miR-615
in OS.
miR-615 blocks EMT and participates in
the PI3K/AKT pathway in OS
Finally, the effect of miR-615 on EMT and the
PI3K/AKT pathway in HOS cells was investigated to elucidate the
molecular mechanisms of miR-615 in OS. The above results showed
that miR-615 regulates cell migration and invasion in OS. Thus, we
hypothesized that miR-615 may regulate cell metastasis by mediating
EMT. N-cadherin, E-cadherin and vimentin are EMT-associated
proteins. We investigated whether miR-615 regulates EMT by
detecting the effect of miR-615 on N-cadherin, E-cadherin and
vimentin expression. First, overexpression of miR-615 was found to
suppress N-cadherin and vimentin expression in the HOS cells. In
contrast, downregulation of miR-615 promoted N-cadherin and
vimentin expression (Fig. 5). In
addition, E-cadherin expression was promoted by miR-615
overexpression and reduced by miR-615 downregulation in HOS cells
(Fig. 5). In addition, the
expression levels of PI3K, p-PI3K, AKT and p-AKT involved in the
PI3K/AKT pathway were detected in HOS cells with miR-615 mimics or
inhibitor. The expression levels of phosphorylated (p-)PI3K and
p-AKT were suppressed by miR-615 mimics and promoted by miR-615
inhibitor in HOS cells (Fig. 5).
However, PI3K and AKT expression levels were not affected by
miR-615 mimics or inhibitor in HOS cells (Fig. 5). Based on these results, miR-615 can
block EMT and inactivate the PI3K/AKT pathway in OS.
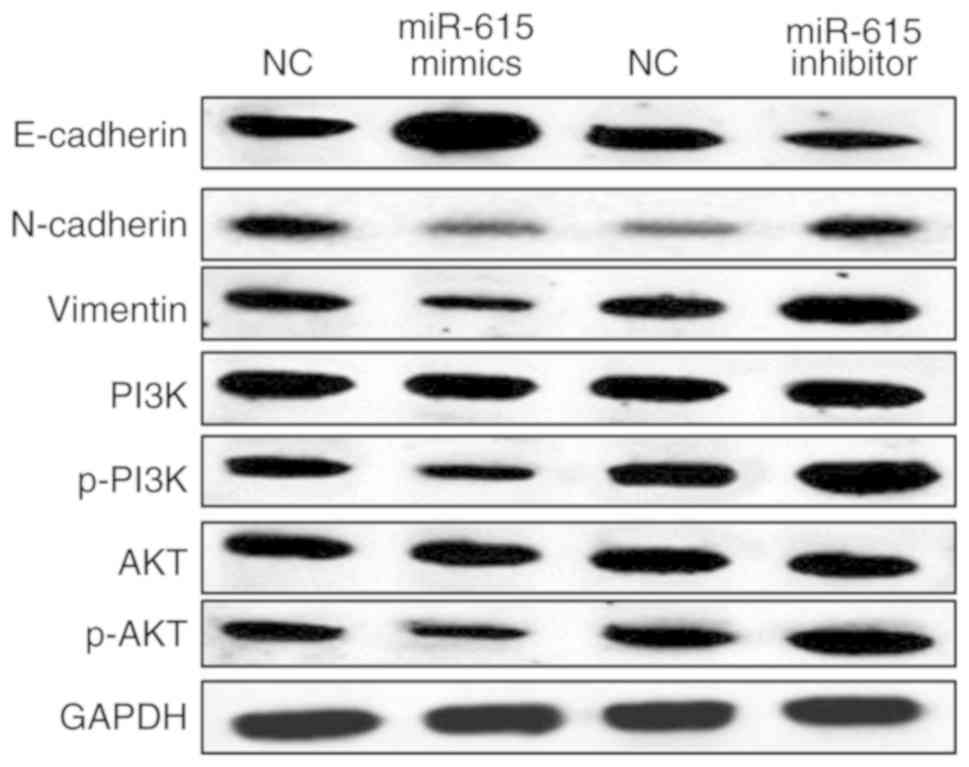 | Figure 5.miR-615 blocks EMT and participates
in the PI3K/AKT pathway in OS. The protein expression levels of
E-cadherin, N-cadherin, vimentin, PI3K, p-PI3K, AKT and p-AKT were
detected in HOS cells by western blot analysis following
transfection with the miR-615 mimics or inhibitor. EMT,
epithelial-mesenchymal transition; PI3K, phosphoinositide 3-kinase;
AKT, protein kinase B; OS, osteosarcoma; p-, phosphorylated. |
Discussion
Many miRNAs have been found to participate in the
pathogenesis of osteosarcoma (OS) by acting as tumor suppressors or
promoters. For example, miR-493-5p was found to inhibit OS cell
proliferation and metastasis by targeting Kruppel-like factor 5
(KLF5) (26). Similarly to the above
study, miR-615 was found to act as a tumor suppressor in OS in the
present study. Specifically, miR-615 expression was reduced in OS
tissues and cells. In addition, the downregulation of miR-615 was
related to the poor clinical outcomes in OS patients. Functionally,
it was found that the overexpression of miR-615 inhibited OS cell
viability and metastasis. In addition, miR-615 also blocked EMT in
OS. EMT refers to the transformation of epithelial cells to
mesenchymal cells, which provides cells with the ability to migrate
and invade. N-cadherin, E-cadherin and vimentin are proteins
associated with EMT. It is well recognized that EMT is activated by
the abnormal expression of N-cadherin, E-cadherin and vimentin. In
this study, we found that miR-615 inactivated EMT in OS cells by
upregulating E-cadherin and downregulating N-cadherin and vimentin.
In the present study, we also found that miR-615 blocked the
PI3K/AKT pathway in OS by inhibiting the expression of p-PI3K and
p-AKT. However, the more specific regulatory mechanisms of the
miR-615/PI3K/AKT pathway are complex and require further study.
Furthermore, miR-615 directly targets hexokinase 2 (HK2), and HK2
expression was upregulated in the OS tissues. More importantly, the
upregulation of HK2 weakened the antitumor effect of miR-615 in OS.
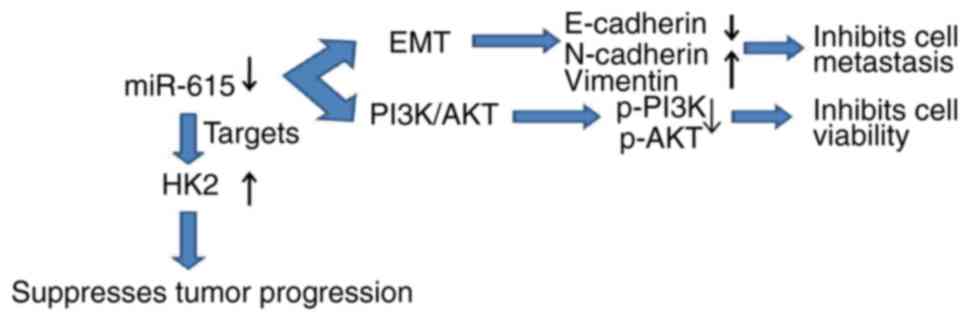
Taken together, the simple regulatory mechanism of miR-615 in OS is
shown in a schematic diagram (Fig.
6).
Consistent with our results, downregulation of
miR-615 was also observed in non-small cell lung (NSCLC) cancer and
pancreatic ductal adenocarcinoma (27,28). In
addition, it has been proposed that the low expression of miR-615
is negatively related to the clinicopathological parameter and poor
prognosis in patients with glioblastoma (29), which is similar to our results.
Functionally, miR-615 has been reported to suppress cell
proliferation and invasion in prostate cancer by directly targeting
cyclin D2 (30). Moreover,
miR-615-3p was found to suppress tumor growth and metastasis in
NSCLC by inhibiting IGF2 (31).
miR-615 was also found to restrain the proliferation of breast
cancer cells by downregulation of AKT2 (32). Consistent with previous studies, we
also found that miR-615 inhibited cell proliferation, invasion and
migration in OS. However, there are still some limitations in the
present study. Further in-depth study should be conducted at the
cellular level, such as cell cycle distribution and apoptosis
experiments. Moreover, miR-615 regulated the progression of OS by
blocking EMT and inactivating the PI3K/AKT pathway, which has not
been reported in previous studies. To make our results more
convincing, we will design experiments to explore whether the
PI3K/AKT pathway can affect miR-615 function in OS.
In the present study, we found that miR-615
inhibited the development of OS by targeting HK2. As a tumor
promoter, HK2 was also confirmed to be a direct target of miR-218
in glioma (33). Here, a negative
correlation between miR-615 and HK2 expression was also identified
in OS tissues. Similarly, it has been demonstrated that
overexpression of miR-9, miR-98, and miR-199 is correlated with the
downregulation of HK2 in colorectal cancer (34). More importantly, upregulation of HK2
impaired the tumor-suppressive effect of miR-615 in OS. The same
interaction between HK2 and other miRNAs, such as miR-125b and
miR-143, have also been identified in human cancers (35,36). In
OS, upregulation of HK2 has been reported to attenuate the
inhibition of cell growth and motility induced by miR-497 (37). In addition, it was found that miR-143
acted as a tumor suppressor in human prostate cancer by targeting
HK2 (38). In the present study,
miR-615 also inhibited the progression of OS by targeting HK2. All
these findings prove the accuracy of our results concerning the
relationship between miR-615 and HK2 in OS. Although we analyzed
the role of miR-615 in the progression of OS, its detailed
regulatory network in OS still needs further exploration.
In conclusion, our study reports the inhibitory
effect of miR-615 on OS cell viability and metastasis. In addition,
miR-615 acts as a tumor suppressor in OS by targeting HK2.
Meanwhile, miR-615 blocks EMT and inactivates the PI3K/AKT pathway
in OS. These findings suggest that miR-615 may be a new target for
the treatment of OS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
LS and JM conceived and designed the study. PW, ZZ,
and KZ performed the experiments. LS wrote the paper. ZX and SL
reviewed and edited the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All of the patients in the present study provided
informed consent. Approval for this research was acquired from the
Institutional Ethics Committee of Shandong Provincial Third
Hospital (2016SPT-44).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Clark JC, Dass CR and Choong PF: A review
of clinical and molecular prognostic factors in osteosarcoma. J
Cancer Res Clin Oncol. 134:281–297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shao Y, Geng Y, Gu W, Huang J, Pei H and
Jiang J: Prognostic role of tissue and circulating microRNA-200c in
malignant tumors: A systematic review and meta-analysis. Cell
Physiol Biochem. 35:1188–1200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bielack SS, Hecker-Nolting S, Blattmann C
and Kager L: Advances in the management of osteosarcoma. F1000Res.
5:27672016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dalmay T: Mechanism of miRNA-mediated
repression of mRNA translation. Essays Biochem. 54:29–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
van Kouwenhove M, Kedde M and Agami R:
MicroRNA regulation by RNA-binding proteins and its implications
for cancer. Nat Rev Cancer. 11:644–656. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nugent M: microRNA and bone cancer. Adv
Exp Med Biol. 889:201–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rehei AL, Zhang L, Fu YX, Mu WB, Yang DS,
Liu Y, Zhou SJ and Younusi A: MicroRNA-214 functions as an oncogene
in human osteosarcoma by targeting TRAF3. Eur Rev Med Pharmacol
Sci. 22:5156–5164. 2018.PubMed/NCBI
|
|
9
|
Xie W, Xiao J, Wang T, Zhang D and Li Z:
MicroRNA-876-5p inhibits cell proliferation, migration and invasion
by targeting c-Met in osteosarcoma. J Cell Mol Med. 23:3293–3301.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang J, Liu L, Sun Y, Xue Y, Qu J, Pan S,
Li H, Qu H, Wang J and Zhang J: miR-615-3p promotes proliferation
and migration and inhibits apoptosis through its potential target
CELF2 in gastric cancer. Biomed Pharmacother. 101:406–413. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen L, Nan A, Zhang N, Jia Y, Li X, Ling
Y, Dai J, Zhang S, Yang Q, Yi Y and Jiang Y: Circular RNA 100146
functions as an oncogene through direct binding to miR-361-3p and
miR-615-5p in non-small cell lung cancer. Mol Cancer. 18:132019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Q, Wu G, Zhang Z, Tang Q, Zheng W,
Chen X, Chen F, Li Q and Che X: Long non-coding RNA HOTTIP promotes
renal cell carcinoma progression through the regulation of the
miR-615/IGF-2 pathway. Int J Oncol. 53:2278–2288. 2018.PubMed/NCBI
|
|
13
|
Yang B, Xie R, Wu SN, Gao CC, Yang XZ and
Zhou JF: MicroRNA-615-5p targets insulin-like growth factor 2 and
exerts tumor-suppressing functions in human esophageal squamous
cell carcinoma. Oncol Rep. 39:255–263. 2018.PubMed/NCBI
|
|
14
|
Zhong JT and Zhou SH: Warburg effect,
hexokinase-II, and radioresistance of laryngeal carcinoma.
Oncotarget. 8:14133–14146. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiao L, Zhang HL, Li DD, Yang KL, Tang J,
Li X, Ji J, Yu Y, Wu RY, Ravichandran S, et al: Regulation of
glycolytic metabolism by autophagy in liver cancer involves
selective autophagic degradation of HK2 (hexokinase 2). Autophagy.
14:671–684. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Z, Huang S, Wang H, Wu J, Chen D,
Peng B and Zhou Q: High expression of hexokinase domain containing
1 is associated with poor prognosis and aggressive phenotype in
hepatocarcinoma. Biochem Biophys Res Commun. 474:673–679. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei L, Zhou Y, Dai Q, Qiao C, Zhao L, Hui
H, Lu N and Guo QL: Oroxylin A induces dissociation of hexokinase
II from the mitochondria and inhibits glycolysis by SIRT3-mediated
deacetylation of cyclophilin D in breast carcinoma. Cell Death Dis.
4:e6012013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xi F and Ye J: Inhibition of lung
carcinoma A549 cell growth by knockdown of hexokinase 2 in situ and
in vivo. Oncol Res. 23:53–59. 2016. View Article : Google Scholar
|
|
19
|
Mukherjee A, Ma Y, Yuan F, Gong Y, Fang Z,
Mohamed EM, Berrios E, Shao H and Fang X: Lysophosphatidic acid
up-regulates hexokinase II and glycolysis to promote proliferation
of ovarian cancer cells. Neoplasia. 17:723–734. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu J, Wang L, Chen W, Wang Y, Zhen S, Chen
H, Cheng J, Zhou Y, Li X and Zhao L: miR-603 targeted hexokinase-2
to inhibit the malignancy of ovarian cancer cells. Arch Biochem
Biophys. 661:1–9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang T, Zhu X, Wu H, Jiang K, Zhao G,
Shaukat A, Deng G and Qiu C: Targeting the ROS/PI3K/AKT/HIF-1α/HK2
axis of breast cancer cells: Combined administration of Polydatin
and 2-Deoxy-d-glucose. J Cell Mol Med. 23:3711–3723. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu JH, Yang S, Nan CJ, Zhou CC, Lu DQ, Li
S and Mu HQ: MiR-182 affects renal cancer cell proliferation,
apoptosis, and invasion by regulating PI3K/AKT/mTOR signaling
pathway. Eur Rev Med Pharmacol Sci. 22:351–357. 2018.PubMed/NCBI
|
|
23
|
Guanen Q, Junjie S, Baolin W, Chaoyang W,
Yajuan Y, Jing L, Junpeng L, Gaili N, Zhongping W and Jun W:
MiR-214 promotes cell meastasis and inhibites apoptosis of
esophageal squamous cell carcinoma via PI3K/AKT/mTOR signaling
pathway. Biomed Pharmacother. 105:350–361. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu RD, Feng F, Yu XS, Liu ZD and Lao LF:
miR-149-5p inhibits cell growth by regulating TWEAK/Fn14/PI3K/AKT
pathway and predicts favorable survival in human osteosarcoma. Int
J Immunopathol Pharmacol. 32:20587384187866562018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Z, Luo G, Yu C, Yu G, Jiang R and
Shi X: MicroRNA-493-5p inhibits proliferation and metastasis of
osteosarcoma cells by targeting Kruppel-like factor 5. J Cell
Physiol. 234:13525–13533. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dong Y, Huo X, Sun R, Liu Z, Huang M and
Yang S: lncRNA Gm15290 promotes cell proliferation and invasion in
lung cancer through directly interacting with and suppressing the
tumor suppressor miR-615-5p. Biosci Rep. 38:BSR201811502018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao W, Gu Y, Li Z, Cai H, Peng Q, Tu M,
Kondo Y, Shinjo K, Zhu Y, Zhang J, et al: miR-615-5p is
epigenetically inactivated and functions as a tumor suppressor in
pancreatic ductal adenocarcinoma. Oncogene. 34:1629–1640. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ji Y, Sun Q, Zhang J and Hu H: MiR-615
inhibits cell proliferation, migration and invasion by targeting
EGFR in human glioblastoma. Biochem Biophys Res Commun.
499:719–726. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang F, Zhao H, Du Z and Jiang H: miR-615
inhibits prostate cancer cell proliferation and invasion by
directly targeting cyclin D2. Oncol Res. 27:293–299. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu J, Jia Y, Jia L, Li T, Yang L and
Zhang G: MicroRNA 615-3p inhibits the tumor growth and metastasis
of NSCLC via inhibiting IGF2. Oncol Res. 27:269–279. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bai Y, Li J, Li J, Liu Y and Zhang B:
MiR-615 inhibited cell proliferation and cell cycle of human breast
cancer cells by suppressing of AKT2 expression. Int J Clin Exp Med.
8:3801–3808. 2015.PubMed/NCBI
|
|
33
|
Liu H, Liu N, Cheng Y, Jin W, Zhang P,
Wang X, Yang H, Xu X, Wang Z and Tu Y: Hexokinase 2 (HK2), the
tumor promoter in glioma, is downregulated by miR-218/Bmi1 pathway.
PLoS One. 12:e01893532017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Snezhkina AV, Krasnov GS, Zhikrivetskaya
SO, Karpova IY, Fedorova MS, Nyushko KM, Belyakov MM, Gnuchev NV,
Sidorov DV, Alekseev BY, et al: Overexpression of microRNAs miR-9,
−98, and −199 correlates with the downregulation of HK2 expression
in colorectal cancer. Mol Biol (Mosk). 52:220–230. 2018.(In
Russian). View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li W, Hao J, Zhang L, Cheng Z, Deng X and
Shu G: Astragalin reduces hexokinase 2 through increasing miR-125b
to inhibit the proliferation of hepatocellular carcinoma cells in
vitro and in vivo. J Agric Food Chem. 65:5961–5972. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gregersen LH, Jacobsen A, Frankel LB, Wen
J, Krogh A and Lund AH: MicroRNA-143 down-regulates hexokinase 2 in
colon cancer cells. BMC Cancer. 12:2322012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song J, Wu X, Liu F, Li M, Sun Y, Wang Y,
Wang C, Zhu K, Jia X, Wang B and Ma X: Long non-coding RNA PVT1
promotes glycolysis and tumor progression by regulating miR-497/HK2
axis in osteosarcoma. Biochem Biophys Res Commun. 490:217–224.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou P, Chen WG and Li XW: MicroRNA-143
acts as a tumor suppressor by targeting hexokinase 2 in human
prostate cancer. Am J Cancer Res. 5:2056–2063. 2015.PubMed/NCBI
|