Introduction
According to the WHO data released in May 2019, the
number of new liver cancer cases was 841080 worldwide in 2018
ranked sixth, and the number of deaths was 781631, the death rate
ranked third among all known types of cancer. In Asia, liver cancer
ranks fourth in morbidity and third in mortality (1). The main treatment methods of primary
liver cancer include surgical resection, local ablation,
transhepatic arterial chemotherapy and embolization (TACE),
molecular targeting and immunotherapy under exploration (2). However, the treatment outcomes remain
still unsatisfactory. As an important indicator for the diagnosis
of hepatocellular carcinoma (HCC), compared to healthy subjects,
the serum α-fetoprotein (AFP) is increased to varying degrees in
≤70% of HCC cases. However, it has been demonstrated that AFP
levels do not predict the tumor stage or prognosis (3). The clinical indicators commonly used to
predict the prognosis of HCC include tumor size, degree of
cirrhosis, tumor differentiation and microvascular invasion
(4,5). Due to the differences in the
progression of liver cancer among individuals, there is a growing
need for prognostic biomarkers to accurately stratify patients for
appropriate risk-adaptive treatment.
DNA topoisomerase is an enzyme that catalyzes the
transformation of DNA topologies; it is present in the nucleus of
living organisms and not only serves a catalytic role in the
breaking and binding of DNA strands, but also affects the
three-dimensional structure of DNA (6). DNA topoisomerase type I (TOP1) breaks
the single strand, whereas DNA topoisomerase II (TOP2) cuts off the
double chain (7). Although the two
TOPs are highly homologous in structure, their distribution in the
organism is significantly different, and they are not produced by
the same transportation process (8).
TOP2 is divided into two subtypes, of which the 1,531 amino acid
170 kDa subtype encoded by the TOP2A gene is topoisomerase IIα, and
the 1,621 amino acid 180 kDa subtype encoded by the TOP2B gene is
topoisomerase IIβ (9). TOP2A serves
an important role in DNA synthesis and transcription, as well as
chromosome segregation during mitosis (10). The location of the human TOP2A gene
is on chromosome 17, and its expression is associated with cell
proliferation and the cell cycle; its production and degradation
are regulated by the cell cycle, and it decreases rapidly after
completing mitosis (11,12). Previous studies have reported that
the accumulation of the TOP2A protein occurs due to the stable
upregulation of TOP2A transcription (13,14).
In addition to its physiological function, TOP2A has
been reported to be a sensitive and specific marker of active
proliferating cells in the late S, G and M phases of the cell
cycle, indicating its importance in cancer research (15). Increased cell proliferation is a
hallmark of malignant tumors, and the expression of TOP2A in
proliferating cells is significantly increased; therefore, its
expression levels reflect the tumor proliferation level to a
certain extent and indicates invasive behavior and poor prognosis
(16). In colorectal (17), liver (18), esophageal (19) and gastric cancer (20), the expression levels of TOP2A are
higher compared with those in the adjacent normal tissues. TOP2A
can be used as a potential biomarker to predict the prognosis of
patients with malignant tumors and screen out high-risk cases in
order to enable individualized treatment (21). In a study on triple-negative breast
cancer, a significant association was observed between the TOP2A
gene and chromosome 17 polysomy (CEP17) status, and TOP2A has a
certain predictive effect on the therapeutic effect of
anthracycline drugs (22). Wittmann
et al (23) confirmed that
the high expression of TOP2A was associated with tumor metastasis
and death outcome by analyzing 102 cases of nephroblastoma. A
large-scale retrospective study has demonstrated that TOP2A high
expression is associated with poor differentiation and neural
invasion of esophageal cancer, and is also an independent risk
factor affecting the prognosis of esophageal cancer (24). De Resende et al (25) have reported that high expression of
TOP2A protein is associated with severe lymphatic vascular invasion
and a low relapse-free survival rate in prostate cancer. In
addition, in breast cancer, TOP2A expression is associated with
Ki67 expression, mitotic count, tumor grade and nipple
infiltration, and high expression of TOP2A adversely affects
prognosis (26). High expression of
TOP2A in tumors, such as pancreatic cancer, prostate cancer and
breast cancer is also associated with regional lymph node
metastasis and metastasis to other organs, and these behaviors lead
to a poor prognosis (27–29). In studies on endometrial (30) and adrenal gland (31) cancer, high expression of TOP2A has
been demonstrated to indicate a poor prognosis. However, to the
best of our knowledge, no studies on TOP2A expression and prognosis
in primary liver cancer are currently available.
The present study aimed to investigate the
association between TOP2A expression and the clinicopathological
characteristics of patients with liver cancer, and to conduct
survival analysis on the prognosis of the entire liver cancer
cohort and the Asian cohort in The Cancer Genome Atlas (TCGA)
database to explore the feasibility of TOP2A as a prognostic
indicator.
Materials and methods
Patients and samples
The study protocol and acquisition of tissue
specimens were approved by the Clinical Ethics Committee of The
Affiliated Tumor Hospital of Nantong University (approval no.
2018-024). The patients provided informed consent for participation
and publication of the present study. The inclusion criteria were
patients who were clinically diagnosed with HCC and were about to
undergo surgical resection, regardless of tumor size, sex and age.
All specimens were verified as HCC by pathological analysis and
non-HCC patients will be excluded. A total of 15 HCC and paired
paracancerous liver tissue specimens were collected during
hepatectomy at The Affiliated Tumor Hospital of Nantong University
between March and August 2018. Among the 15 cases, 7 were males and
8 were females, age range, 47–77 years old with a mean age of 63
years. The liver tissues within 2 cm from the edge of the tumor was
obtained as paracancerous tissues. The specimens were immediately
stored in liquid nitrogen until further experiments. These same
specimens have been used in a previous study (32).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA preparation
Total RNA was extracted from 15 pairs of HCC and
paired paracancerous tissues by TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.).
Synthesis of cDNA chain 1
Total RNA was reverse transcribed into cDNA, and
Beacon Designer 8.14 (http://www.premierbiosoft.com/molecular_beacons/index.html)
was used to design the RT-PCR primers. Total RNA was extracted from
tumor or paracanerous tissue using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific Inc). The RNA was
subsequently treated with RNase-free DNase I (Roche Applied
Science, Inc). Reverse transcription was performed using the
BcaBest® RNA PCR kit (Takara Biotechnology, Co., Ltd)
with a temperature protocol of 30°C for 5 min, followed by 65°C for
30 min and 98°C for 5 min. The quantitative polymerase chain
reaction was conducted using the StepOnePlus® Real-Time
PCR System (Applied Biosystems; Thermo Fisher Scientific Inc.). The
primer sequences were as follows: TOP2A forward,
5′-CATTGAAGACGCTTCGTTATGG-3′ and reverse,
5′-CAGAAGAGAGGGCCAGTTGTG-3′; and β-actin forward,
5′-ATAGCACAGCCTGGATAGCAACGTAC-3′ and reverse,
5′-CACCTTCTACAATGAGCTGCGTGTG-3′. The qPCR mix comprised the
following: 10 µl SYBR® Green I (BioRuler, Inc.), 0.8 µl
(10 µmol/l) qPCR upstream and downstream primers, 2 µl cDNA, 0.4 µl
50X ROX reference dye and deionized water to 20 µl. The reaction
conditions were as follows: 95°C for 1 min, followed by 40 cycles
of 95°C for 30 sec and 60°C for 40 sec. All experiments were
repeated three times. The relative quantitative was used by
2−ΔΔCq method (33). The
results were analyzed by paired Student's t-test.
Gene expression data and clinical data
from TCGA database
Gene expression and clinical data from patients with
HCC were obtained from TCGA-liver hepatocellular carcinoma (LIHC)
dataset (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
A total of 371 cases and 424 samples of gene expression data were
downloaded. Among them, 50 tumor tissue samples were paired with
paracancerous tissues, and the rest of the 374 samples were tumor
tissue. A total of 377 cases of clinical data files were obtained,
371 cases retained after matching with tissue specimen data when
analyzing the prognostic factors. Among the 371 cases, 252 were
from male and 119 were from female patients, with a mean age of
59.44 years (age range, 16–90 years). Among the 371 cases, one case
provided 2 tumor tissue specimens, another case provided 3 tumor
tissue specimens, and the remaining 369 cases each provided one
tumor tissue specimen. The total number of specimens was 374. Of
the 371 cases, there was one case provided 3 tumor samples, and
another case provided 2 tumor samples, so the total number of tumor
samples was 374. Among these 374 tumor specimens, some clinical
information was missing. Seven cases lacked T stage, 17 lacked
clinical stage and 2 lacked histological grade. These cases were
deleted accordingly during analysis.
Differential TOP2A expression
analysis
Perl language (www.perl.org)
was used to organize the downloaded data, and R (version 3.6.0)
(34) was used to analyze the
differences in TOP2A expression between paracancerous and tumor
tissues. ‘Limma’ (35) and
‘beeswarm’ (36) packages were used
to analyze the differences in the expression of TOP2A between
paracancerous and tumor tissues, and Perl was used to pair the
tumor tissues with paracancerous tissues for analysis.
Associations between TOP2A expression,
clinicopathological characteristics and prognosis
To study the associations between TOP2A expression
and pathological grade, clinical and T stage, logistic regression
analysis was performed using R. Univariate and multivariate
analyses were conducted for age, sex, pathological grade, clinical
staging, T stage and TOP2A expression levels using the ‘survival’
package, and Kaplan-Meier analysis was conducted to compare the
survival of patients stratified into high and low TOP2A expression
groups.
Statistical analysis
The quantitative data are presented as the mean ±
SD, and the qualitative data are presented as counts or
percentages. R (version 3.6.0) was used for statistical analysis.
The median TOP2A expression was used as the cut-off value to
determine the high and low expression groups. The exact median
expression value was assigned to the low expression group. Paired
t-test was used to analyze the differences in TOP2A mRNA expression
in 15 pairs of fresh tumors and paracancerous tissues. When
analyzing TOP2A expression data downloaded from TCGA, all 374 tumor
samples were compared with 50 paracancerous tissues using unpaired
Student's t-test, and 50 pairs of tumors and paracancerous tissues
from the same patients were compared using paired Student's t-test.
Kruskal-Wallis tests were performed to determine the associations
between TOP2A expression and clinicopathological characteristics.
Cox multivariate regression was used to ascertain the expression of
TOP2A in tumor and adjacent tissues, and Renyi test was used for
survival analysis.
Results
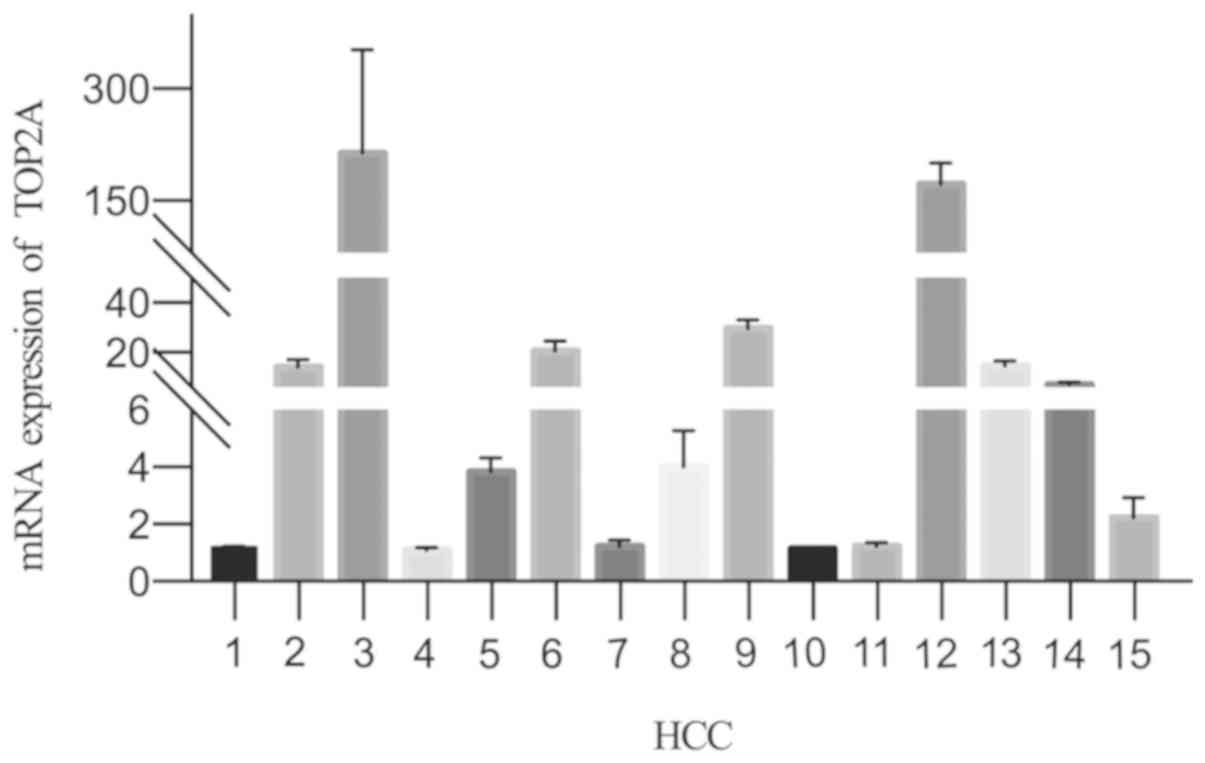
TOP2A mRNA expression in tumor and
paracancerous tissues
To determine the differences in TOP2A expression
between tumor and paracancerous tissues, RNA was extracted from 15
pairs of tumor and paracancerous tissues and analyzed by RT-qPCR.
The mRNA expression levels of TOP2A in tumor tissues was
significantly higher compared with those in paracancerous tissues
normalized to β-actin (P=0.0064; Fig.
1).
TOP2A expression and
clinicopathological features in TCGA-liver hepatocellular carcinoma
(LIHC) dataset
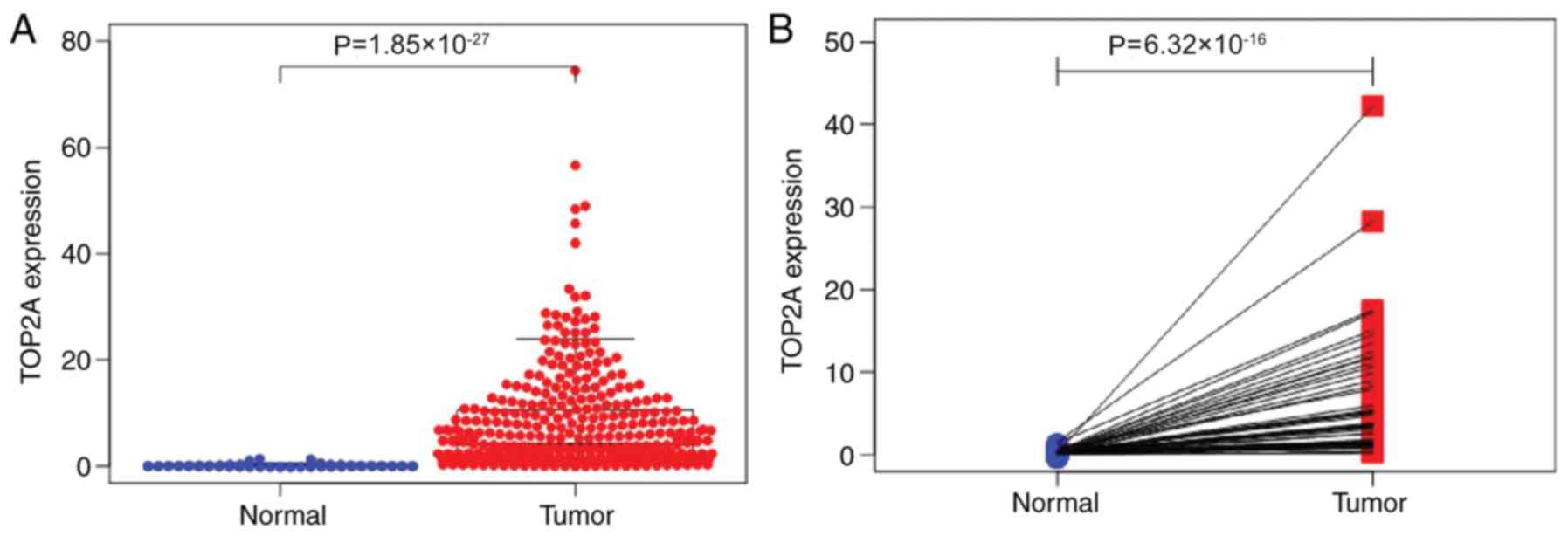
In the TCGA-LIHC dataset, the expression of TOP2A in
374 tumor tissue specimens was higher compared with that in 50
paracancerous tissues (P=1.85×10−27; Fig. 2A). In 50 paired samples of tumor and
paracancerous tissues, the expression levels of TOP2A in tumor
tissues were higher compared with those in the paired paracancerous
tissues (P=6.319×10−16; Fig.
2B).
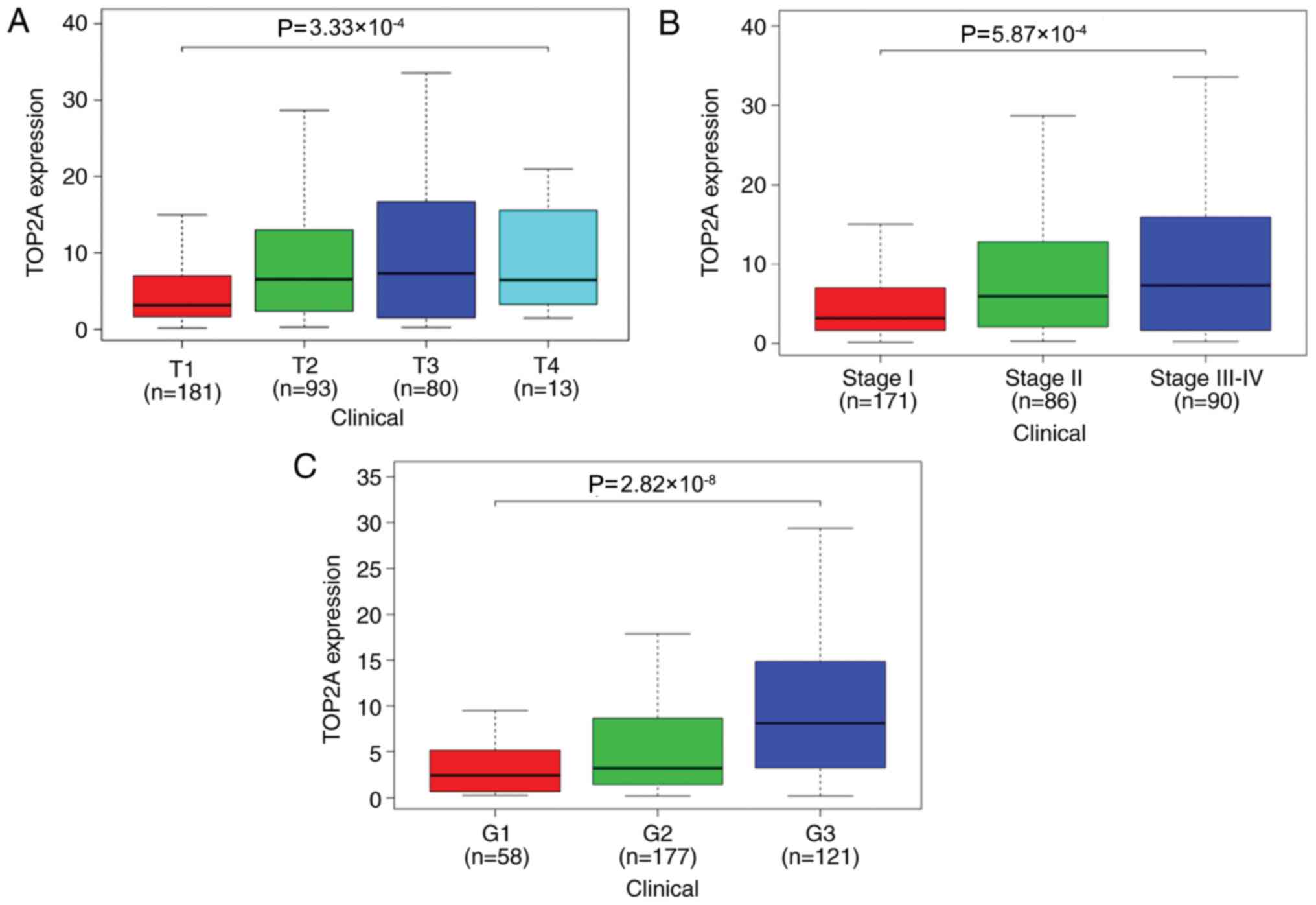
The association between the expression of TOP2A and
T stage, clinical stage and histological grade were analyzed by
logistic regression. The expression levels of TOP2A in stages T3-T4
were higher compared with those in stages T1-T2 (OR, 1.86; 95% CI,
1.16–3.02; P=0.011; Table I). The
expression of TOP2A in clinical stage II tumors was higher compared
those in with stage I (OR, 1.88; 95% CI, 1.11–3.17; P=0.012); the
expression of TOP2A in stage III–IV was higher compared with that
in stage I (OR, 2.71; 95% CI, 1.59–4.69; P=2.90×10−4);
and the expression of TOP2A in histological grade G3-G4 was higher
compared with that in grade G1-G2 (OR, 3.22; 95% CI, 2.07–5.07;
P=3.12×10−7; Table I).
Kruskal-Wallis test identified significant differences in the
expression of TOP2A among different histological grades, clinical
stages and T stages (Fig. 3).
 | Table I.Logistic regression of TOP2A
expression and patient clinicopathological characteristics. |
Table I.
Logistic regression of TOP2A
expression and patient clinicopathological characteristics.
| Characteristic | n | Total, n | Odds ratio (95%
CI) | P-value |
|---|
| T stage, T3-T4 vs.
T1-T2 | 93 vs. 274 | 367 | 1.86
(1.16–3.02) |
1.10×10−2a |
| Clinical stage |
| 347 |
|
|
| Stage
II vs. stage I | 86 vs. 171 | 257 | 1.88
(1.11–3.17) |
1.20×10−2a |
| Stage
III–IV vs. stage I | 90 vs. 86 | 176 | 2.71
(1.59–4.69) |
2.90×10−4a |
| Histological grade,
G3-G4 vs. G1-G2 | 137 vs. 235 | 372 | 3.22
(2.07–5.07) |
3.12×10−7a |
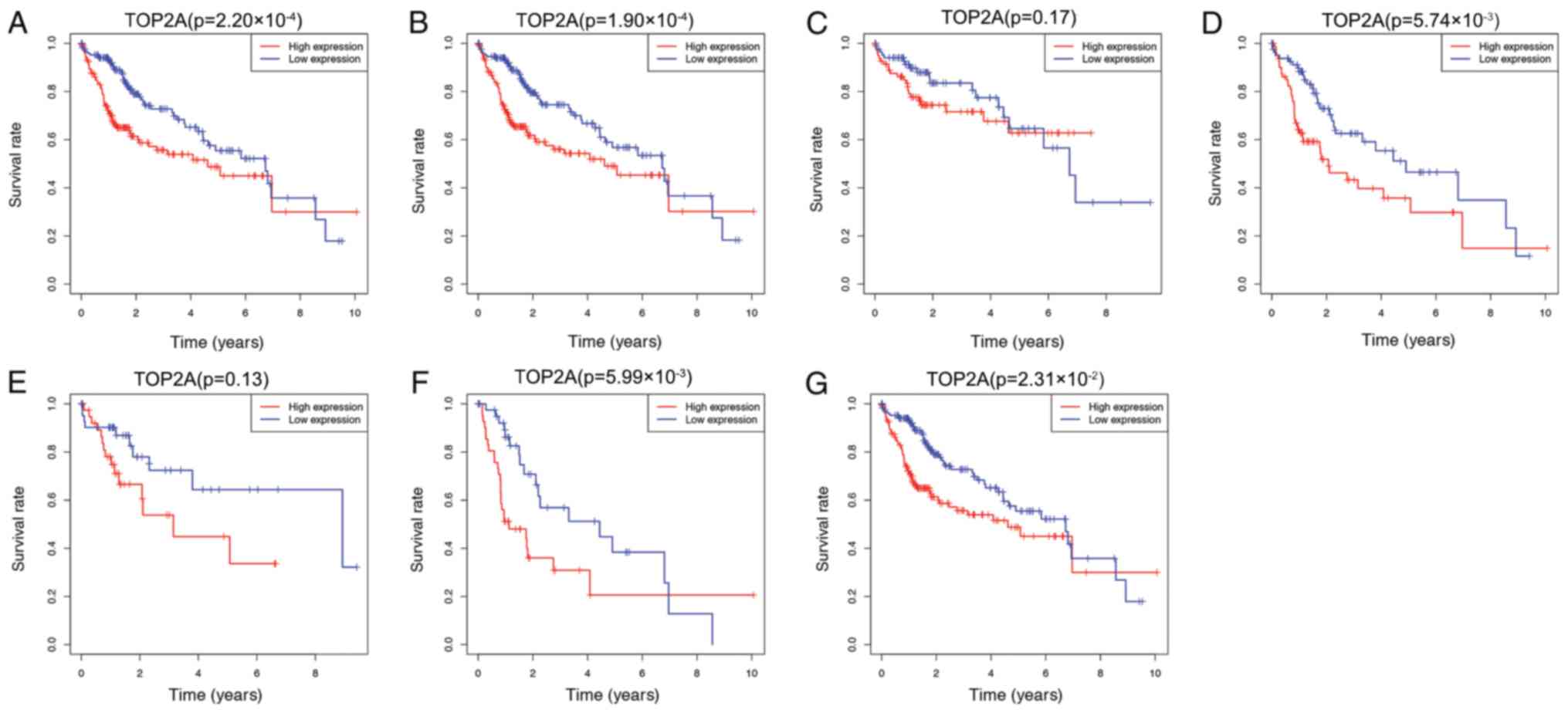
High expression of TOP2A is associated
with a poor prognosis
To determine the association between TOP2A
expression and prognosis, Kaplan-Meier analysis was performed in
data subsets stratified by clinicopathological factors. As
presented in Fig. 4, the overall
survival of patients with low expression of TOP2A was higher
compared those with high expression (P=2.20×10−4;
Fig. 4A). The 5-year survival rate
of the TOP2A high expression group was 48.7%, whereas that of the
low expression group was 55.4%. Among different clinical stages,
the overall survival of the high expression group does not present
with improved survival in any of the presented groups. Clinical
stage I–III (P=1.90×10−4; Fig. 4B), stage II–III
(P=5.74×10−4; Fig. 4D),
stage III (P=5.99×10−3; Fig.
4F) and stage III–IV (P=2.31×10−2; Fig. 4G) exhibited significant differences
between the two groups. No significant differences were observed
between the high and low TOP2A expression groups in the overall
survival of patients at stage I and stage II (P=0.17; Fig. 4C and P=0.13; Fig. 4E), and no independent survival
analysis was performed for patients at stage IV as the number of
patients was low.
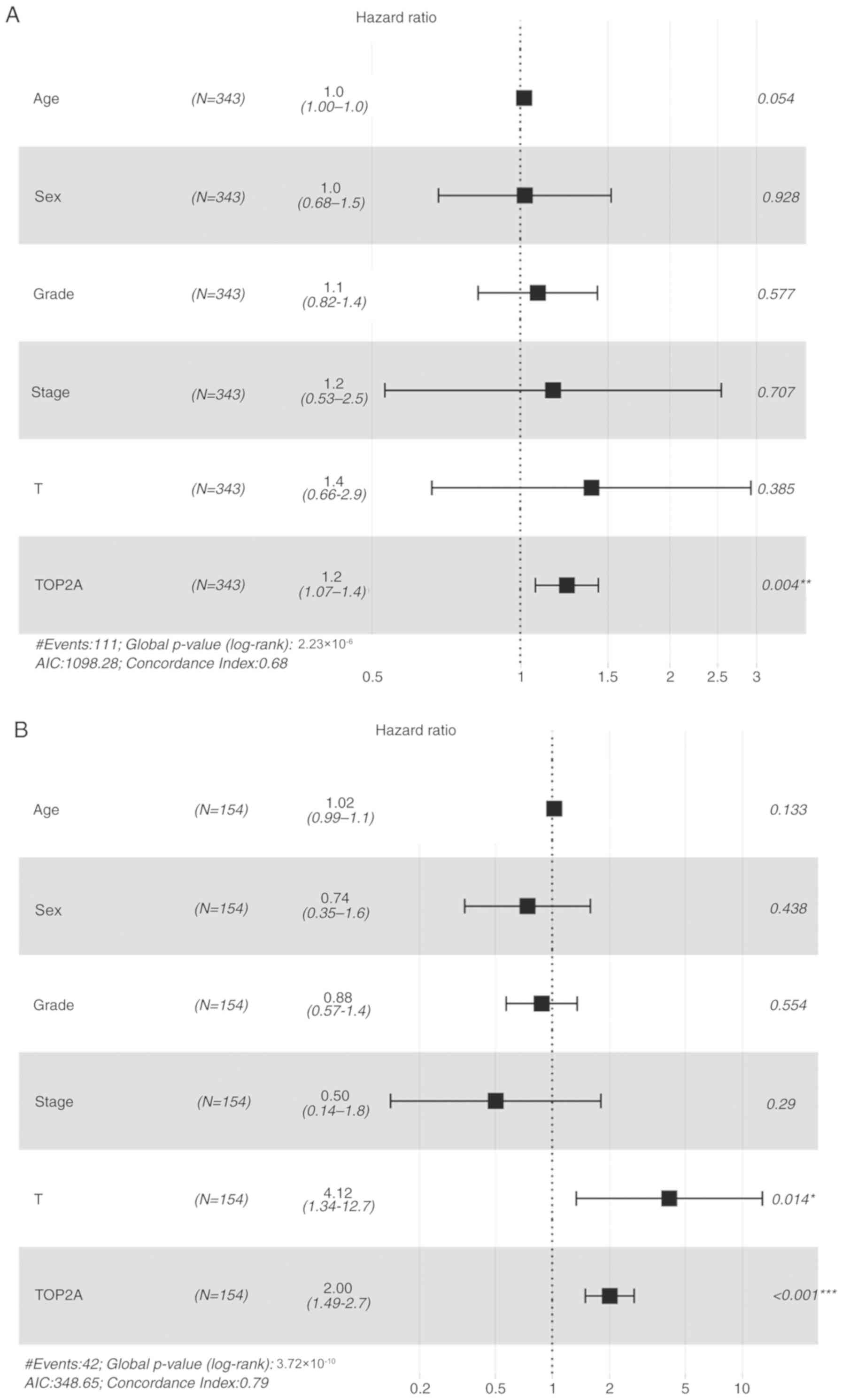
In the multivariate analysis, high expression of
TOP2A was the only independent factor for poor prognosis of HCC
(HR, 1.240; 95% CI, 1.071–1.435; P=2.23×10−6; Fig. 5A).
Effects of TOP2A expression on
survival in different ethnic groups
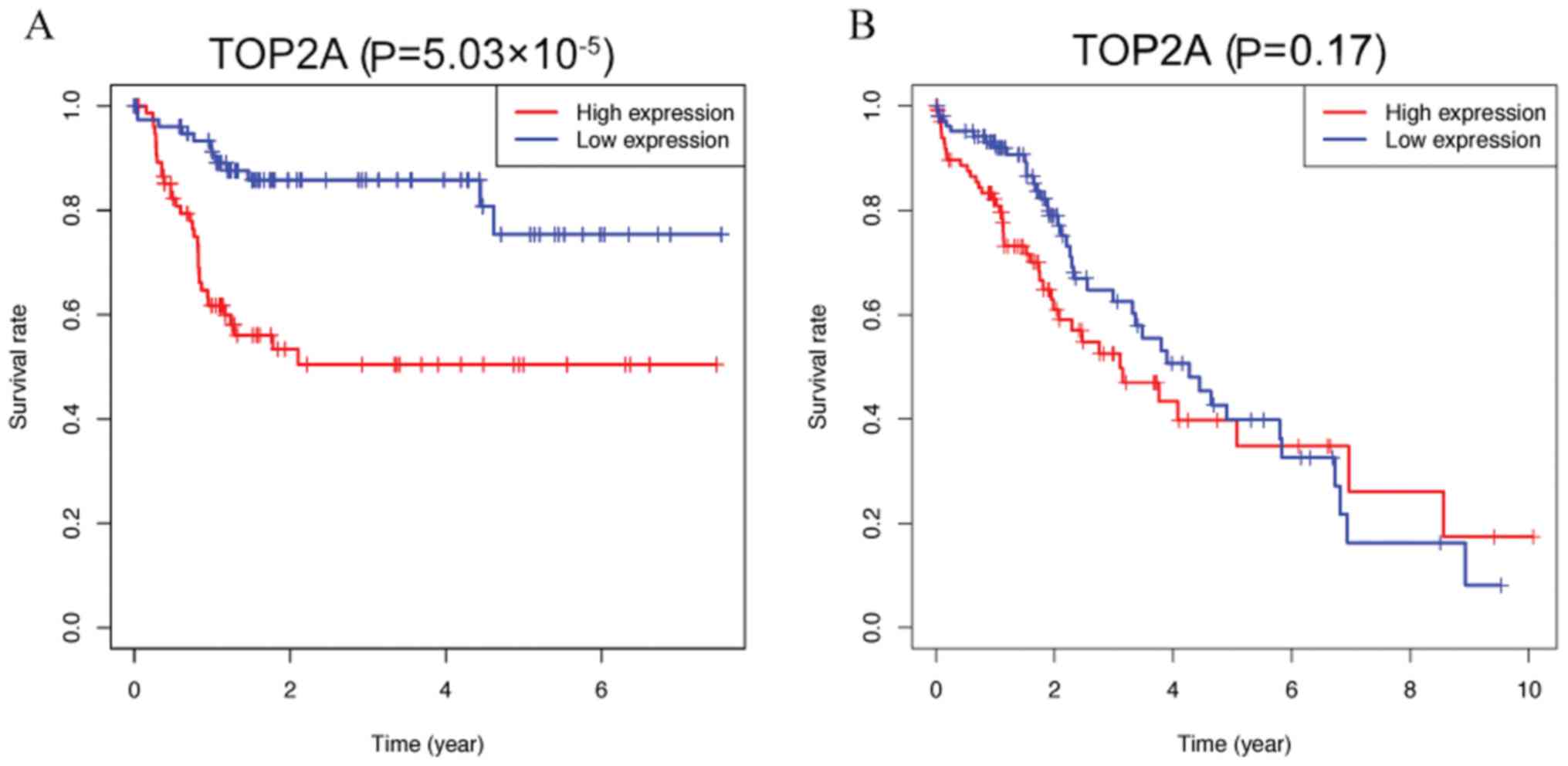
Kaplan-Meier analysis was performed using data from
Asian and non-Asian patients (all ethnic groups) in the dataset to
identify whether differences in ethnicity were associated with the
effects of TOP2A on patient survival in HCC. As presented in
Fig. 6, the overall survival of
Asian patients in the high and low TOP2A expression groups was
significantly different (P=5.03×10−5), whereas among the
non-Asian groups, the overall survival differences due to abnormal
TOP2A expression were not significant (P=0.212). Multivariate
analysis results indicated that high expression of TOP2A was an
independent prognostic factor in the Asian population (HR, 2.005;
95% CI, 1.493–2.692; P=3.74×10−6; Fig. 5B).
Discussion
HCC is one of the most common malignant tumors
worldwide (37). After years of
development, the treatment methods for HCC have gradually evolved
from early surgical resection, liver transplantation, TACE,
chemotherapy and radiotherapy to molecular targeted therapy
combined with immunotherapy (38).
AFP was previously recommended by the guidelines as a serum marker
for monitoring HCC (39). A study by
Agopian et al (40) reported
that among 665 patients with HCC, 31.3% had AFP levels within the
normal range. Although AFP is still used in the clinic, it cannot
be used to monitor patients who present with normal levels of AFP
(41). The European Association for
the Study of the Liver (EASL) recommends that in addition to AFP,
vascular endothelial growth factor and angiopoietin 2 can also be
used as prognostic markers (42). In
addition, efforts to identify new effective prognostic biomarkers
for HCC are ongoing. According to previous reports, high expression
levels of collagen type XXIV alpha 1 chain (43), ubiquilin 2 (44), sushi, von Willebrand factor type A,
EGF and pentraxin domain containing 1 (45), acyl-CoA synthetase long chain family
member 4 (46) and YTH
N6-methyladenosine RNA binding protein 1 (47) are associated with the progression of
HCC. Another study has demonstrated that in addition to the
American Joint Committee on Cancer staging system, a long
non-coding RNA-based risk score is another important factor
affecting the overall survival of patients with HCC (48). However, none of the above methods are
currently widely used in the clinic. Therefore, there is an urgent
need to identify new prognostic markers for HCC.
TOP2A serves an important role in various biological
behaviors of cells, including DNA replication, chromosome
separation, chromatin concentration and gene expression (49). As a DNA replication- and cell
division-regulating enzyme, TOP2A is the main target of several
anticancer drugs, such as doxorubicin, etoposide, and mitoxantrone
(50). High expression levels of
TOP2A in various types of tumors, such as lung adenocarcinoma,
bladder urothelial carcinoma, breast, prostate and colon cancer,
indicates a poor prognosis (51–55),
although this has not been previously reported in HCC. However,
Panvichian et al (56) have
demonstrated that the high expression of TOP2A in HCC is associated
with the high expression of Ki67, and Ki-67 expression has been
found to correlate with tumor growth rate and poor prognosis in HCC
(57), which indicates that TOP2A is
a potential target for the treatment of HCC. In the present study,
TOP2A was selected as a potential prognostic marker of HCC for
further study. Analysis of the differential expression of TOP2A in
50 pairs of tumor and paracancerous tissue specimens in TCGA
database revealed that the expression of TOP2A in tumor tissue was
significantly higher compared with that in paracancerous tissue. In
fresh clinical samples, the mRNA levels in HCC and paired samples
confirmed that TOP2A was significantly highly expressed in tumor
tissues, consistent with a previous study (56), suggesting that TOP2A may be
associated with the occurrence and development of liver cancer.
These results suggested that the high expression levels of TOP2A in
HCC may be associated with tumor growth and metastasis, and that
TOP2A may be used as a biomarker to predict tumor prognosis in
clinical practice.
In the present study, a high T stage, poor
differentiation degree and a high clinical stage were associated
with high expression levels of TOP2A. In the univariate and
multivariate analyses, high expression of TOP2A was identified as
an independent predictor of HCC prognosis. Zhang et al
(53) have reported that TOP2A
promotes colon cancer progression by regulating ERK and AKT. The
ERK and AKT pathways are two key signaling pathways involved in
various tumor processes associated with the malignant proliferation
of cancer (58,59). Therefore, in HCC, TOP2A may also
promote the proliferation of HCC by regulating these two
pathways.
The results of the present study demonstrated that
the cumulative survival rate of patients with high expression of
TOP2A was higher compared with that of patients with low expression
of TOP2A. Due to ethnic differences, the role of TOP2A expression
in predicting tumor prognosis in different ethnic groups was also
different (60). The association
between high expression of TOP2A and poor prognosis was more
significant in the Asian population compared with that the white
population in the present study. The etiology of Asian HCC is
mostly associated with viral hepatitis (37), which may be due to the effects of the
hepatitis virus on DNA replication. Thus, the predictive effect of
TOP2A on HCC may be different between the Asian and non-Asian
populations.
However, there were certain limitations to the
present study. First, the present study determined that high
expression of TOP2A may lead to a poor prognosis of HCC using TCGA;
while TCGA is a useful resource for obtaining research clues, this
result needs to be verified in clinical samples, which will be
performed in future studies. Second, since the present study was
preliminary, the difference in the expression of TOP2A in 15 pairs
of tumors and paracancerous tissues was only verified at the mRNA
level, and these differences will be verified at the protein level
in more cases in the future study. Third, in the survival analysis
between the high and low TOP2A expression groups, a number of
survival curves reversed in the late follow-up period. As
Rocha-Singh (61) has reported,
retrospective and observational studies present inherent potential
moderate and strong bias, which may obscure, overestimate and even
reverse the real effects. The appearance of this reversal may mean
that beyond this time point, the value of TOP2A as a risk factor
for poor prognosis of HCC began to decrease. On the other hand,
cases in the TCGA database are followed up for ≤10 years. With the
extension of survival time, a number of patients may succumb to
other diseases than HCC, which may lead to bias in the results at
the later stage of follow-up. In addition, further studies will be
required in the future to establish the prediction model, in
vitro and in vivo functions, signal transduction
pathways and molecular mechanisms that affect the expression of
TOP2A in HCC.
In conclusion, the results of the present study
demonstrated that high expression of TOP2A in HCC was associated
with an advanced clinical stage, a low tumor differentiation grade
and a high T stage. The expression of TOP2A may be an important
biomarker for predicting the prognosis of HCC, especially in the
Asian population.
Acknowledgements
Not applicable.
Funding
This work was supported by the Nantong Science and
Technology Plan (grant no. MSZ18223), the Nantong Youth Medical
Talent Fund (grant no. Youth 088) and the Youth Fund of Nantong
Health Commission (grant no. WKZL2018045).
Availability of data and materials
The datasets used or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZC and HC were responsible for the conceptualization
of the study, drafting and revision of the manuscript. BS and YZ
performed the experiments and analyzed the data. All the authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol and acquisition of tissue
specimens were approved by the Clinical Ethics Committee of The
Affiliated Tumor Hospital of Nantong University (Nantong, China)
(approval no. 2018-024) and all patients provided signed informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
WHO: Cancer today. https://gco.iarc.fr/today/homeJune 24–2019
|
|
2
|
Daher S, Massarwa M, Benson AA and Khoury
T: Current and future treatment of hepatocellular carcinoma: An
updated comprehensive review. J Clin Transl Hepatol. 6:69–78. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Park SJ, Jang JY, Jeong SW, Cho YK, Lee
SH, Kim SG, Cha SW, Kim YS, Cho YD, Kim HS, et al: Usefulness of
AFP, AFP-L3, and PIVKA-II, and their combinations in diagnosing
hepatocellular carcinoma. Medicine (Baltimore). 96:e58112017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sharma SA, Kowgier M, Hansen BE, Brouwer
WP, Maan R, Wong D, Shah H, Khalili K, Yim C, Heathcote EJ, et al:
Toronto HCC risk index: A validated scoring system to predict
10-year risk of HCC in patients with cirrhosis. J Hepatol. Aug
24–2017.(Epub ahead of print). doi: 10.1016/j.jhep.2017.07.033.
|
|
5
|
Shen J, He L, Li C, Wen T, Chen W, Lu C,
Yan L, Li B and Yang J: Nomograms to predict the individual
survival of patients with solitary hepatocellular carcinoma after
hepatectomy. Gut Liver. 11:684–692. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Visone V, Vettone A, Serpe M, Valenti A,
Perugino G, Rossi M and Ciaramella M: Chromatin structure and
dynamics in hot environments: Architectural proteins and DNA
topoisomerases of thermophilic archaea. Int J Mol Sci.
15:17162–17187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Choi IY, Chung IK and Muller MT:
Eukaryotic topoisomerase II cleavage is independent of duplex DNA
conformation. Biochim Biophys Acta. 1264:209–214. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Austin CA, Lee KC, Swan RL, Khazeem MM,
Manville CM, Cridland P, Treumann A, Porter A, Morris NJ and Cowell
IG: TOP2B: The first thirty years. Int J Mol Sci. 19:27652018.
View Article : Google Scholar
|
|
9
|
Bush NG, Evans-Roberts K and Maxwell A:
DNA topoisomerases. EcoSal Plus. 6:2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Antoniou-Kourounioti M, Mimmack ML, Porter
ACG and Farr CJ: The impact of the C-terminal region on the
interaction of topoisomerase II alpha with mitotic chromatin. Int J
Mol Sci. 20:12382019. View Article : Google Scholar
|
|
11
|
Demoulin B, Hermant M, Castrogiovanni C,
Staudt C and Dumont P: Resveratrol induces DNA damage in colon
cancer cells by poisoning topoisomerase II and activates the ATM
kinase to trigger p53-dependent apoptosis. Toxicol In Vitro.
29:1156–1165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sudan S and Rupasinghe HPV:
Flavonoid-enriched apple fraction AF4 induces cell cycle arrest,
DNA topoisomerase II inhibition, and apoptosis in human liver
cancer HepG2 cells. Nutr Cancer. 66:1237–1246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kania EE, Carvajal-Moreno J, Hernandez VA,
English A, Papa JL, Shkolnikov N, Ozer HG, Yilmaz AS, Yalowich JC
and Elton TS: hsa-miR-9-3p and hsa-miR-9-5p as post-transcriptional
modulators of DNA topoisomerase IIα in human leukemia K562 cells
with acquired resistance to etoposide. Mol Pharmacol. 97:159–170.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanagasabai R, Karmahapatra S, Kientz CA,
Yu Y, Hernandez VA, Kania EE, Yalowich JC and Elton TS: The novel
c-terminal truncated 90-kDa isoform of topoisomerase IIα (TOP2α/90)
is a determinant of etoposide resistance in K562 leukemia cells via
heterodimerization with the TOP2α/170 isoform. Mol Pharmacol.
93:515–525. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
D Arcy N and Gabrielli B: Topoisomerase II
inhibitors and poisons, and the influence of cell cycle
checkpoints. Curr Med Chem. 24:1504–1519. 2017.PubMed/NCBI
|
|
16
|
Wang JC: Cellular roles of DNA
topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol.
3:430–440. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jain CK, Roychoudhury S and Majumder HK:
Selective killing of G2 decatenation checkpoint defective colon
cancer cells by catalytic topoisomerase II inhibitor. Biochim
Biophys Acta. 1853:1195–1204. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deng SP and Guo WL: Identifying key genes
of liver cancer by networking of multiple data sets. IEEE/ACM Trans
Comput Biol Bioinform. 16:792–800. 2019. View Article : Google Scholar
|
|
19
|
Yu Y, Ding S, Liang Y, Zheng Y, Li W, Yang
L, Zheng X and Jiang J: Expression of ERCC1, TYMS, TUBB3, RRM1 and
TOP2A in patients with esophageal squamous cell carcinoma: A
hierarchical clustering analysis. Exp Ther Med. 7:1578–1582. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xing C, Cai Z, Gong J, Zhou J, Xu J and
Guo F: Identification of potential biomarkers involved in gastric
cancer through integrated analysis of non-coding RNA associated
competing endogenous RNAs network. Clin Lab. 64:1661–1669. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu LM, Xiong DD, Lin P, Yang H, Dang YW
and Chen G: DNA topoisomerase 1 and 2A function as oncogenes in
liver cancer and may be direct targets of nitidine chloride. Int J
Oncol. 53:1897–1912. 2018.PubMed/NCBI
|
|
22
|
Eltohamy MI, Badawy OM, El kinaai N, Loay
I, Nassar HR, Allam RM and Ali Sakr M: Topoisomerase II α gene
alteration in triple negative breast cancer and its predictive role
for anthracycline-based chemotherapy (Egyptian NCI Patients). Asian
Pac J Cancer Prev. 19:3581–3589. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wittmann S, Wunder C, Zirn B, Furtwängler
R, Wegert J, Graf N and Gessler M: New prognostic markers revealed
by evaluation of genes correlated with clinical parameters in wilms
tumors. Genes Chromosomes Cancer. 47:386–395. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu XL, Zheng WH, Fu ZX, Li ZP, Xie HX, Li
XX, Jiang LH, Wang Y, Zhu SM and Mao WM: Topo2A as a prognostic
biomarker for patients with resectable esophageal squamous cell
carcinomas. Med Oncol. 32:3962015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
de Resende MF, Vieira S, Chinen LT,
Chiappelli F, da Fonseca FP, Guimarães GC, Soares FA, Neves I,
Pagotty S, Pellionisz PA, et al: Prognostication of prostate cancer
based on TOP2A protein and gene assessment: TOP2A in prostate
cancer. J Transl Med. 11:362013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Şahin S, Gönül II, Çakır A, Seçkin S and
Uluoğlu O: Clinicopathological significance of the proliferation
markers Ki67, RacGAP1, and topoisomerase 2 alpha in breast cancer.
Int J Surg Pathol. 24:607–613. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pei YF, Yin XM and Liu XQ: TOP2A induces
malignant character of pancreatic cancer through activating
β-catenin signaling pathway. Biochim Biophys Acta Mol Basis Dis.
1864:197–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Labbe DP, Sweeney CJ, Brown M, Galbo P,
Rosario S, Wadosky KM, Ku SY, Sjöström M, Alshalalfa M, Erho N, et
al: TOP2A and EZH2 provide early detection of an aggressive
prostate cancer subgroup. Clin Cancer Res. 23:7072–7083. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Villman K, Sjöström J, Heikkilä R,
Hultborn R, Malmström P, Bengtsson NO, Söderberg M, Saksela E and
Blomqvist C: TOP2A and HER2 gene amplification as predictors of
response to anthracycline treatment in breast cancer. Acta Oncol.
45:590–596. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ito F, Furukawa N and Nakai T: Evaluation
of TOP2A as a predictive marker for endometrial cancer with
taxane-containing adjuvant chemotherapy. Int J Gynecol Cancer.
26:325–330. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Roca E, Berruti A, Sbiera S, Rapa I, Oneda
E, Sperone P, Ronchi CL, Ferrari L, Grisanti S, Germano A, et al:
Topoisomerase 2α and thymidylate synthase expression in
adrenocortical cancer. Endocr Relat Cancer. 24:319–327. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cai HY, Zhu XH, Qian F, Shao B, Zhou Y,
Zhang Y and Chen Z: High expression of TOP2A gene predicted poor
prognosis of hepatocellular carcinoma after radical hepatectomy.
Transl Cancer Res. 9:983–992. 2020. View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
R Core Team (2014): R, . A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: http://www.R-project.org/
|
|
35
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ecklund A: beeswarm: The Bee Swarm Plot,
an Alternative to Stripchart. https://CRAN.R-project.org/package=beeswarm
|
|
37
|
Kulik L and El-Serag HB: Epidemiology and
management of hepatocellular carcinoma. Gastroenterology.
156:477–491 e471. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yau T, Hsu C, Kim TY, Choo SP, Kang YK,
Hou MM, Numata K, Yeo W, Chopra A, Ikeda M, et al: Nivolumab in
advanced hepatocellular carcinoma: Sorafenib-experienced Asian
cohort analysis. J Hepatol. 71:543–552. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Di Bisceglie AM and Hoofnagle JH:
Elevations in serum alpha-fetoprotein levels in patients with
chronic hepatitis B. Cancer. 64:2117–2120. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Agopian VG, Harlander-Locke MP, Markovic
D, Zarrinpar A, Kaldas FM, Cheng EY, Yersiz H, Farmer DG, Hiatt JR
and Busuttil RW: Evaluation of patients with hepatocellular
carcinomas that do not produce α-fetoprotein. JAMA Surg. 152:55–64.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Luo P, Wu S, Yu Y, Ming X, Li S, Zuo X and
Tu J: Current status and perspective biomarkers in AFP negative
HCC: Towards screening for and diagnosing hepatocellular carcinoma
at an earlier stage. Pathol Oncol Res. 26:599–603. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
European Association For The Study Of The
Liver; European Organisation For Research and Treatment Of Cancer,
. EASL-EORTC clinical practice guidelines: Management of
hepatocellular carcinoma. J Hepatol. 56:908–943. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yan L, Xu F and Dai C: Overexpression of
COL24A1 in hepatocellular carcinoma predicts poor prognosis: A
study based on multiple databases, clinical samples and cell lines.
Onco Targets Ther. 13:2819–2832. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Luo YD, Yu HQ, Liu XY, Huang D, Dai HS,
Fang L, Zhang YJ, Lai JJ, Jiang Y, Shuai L, et al: Prognostic and
predicted significance of Ubqln2 in patients with hepatocellular
carcinoma. Cancer Med. 9:4083–4094. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen L, Liu D, Yi X, Qi L, Tian X, Sun B,
Dong Q, Han Z, Li Q, Song T, et al: The novel miR-1269b-regulated
protein SVEP1 induces hepatocellular carcinoma proliferation and
metastasis likely through the PI3K/Akt pathway. Cell Death Dis.
11:3202020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen J, Ding C, Chen Y, Hu W, Lu Y, Wu W,
Zhang Y, Yang B, Wu H, Peng C, et al: ACSL4 promotes hepatocellular
carcinoma progression via c-Myc stability mediated by
ERK/FBW7/c-Myc axis. Oncogenesis. 9:422020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhao X, Chen Y, Mao Q, Jiang X, Jiang W,
Chen J, Xu W, Zhong L and Sun X: Overexpression of YTHDF1 is
associated with poor prognosis in patients with hepatocellular
carcinoma. Cancer Biomark. 21:859–868. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li F, Bai L, Li S, Chen Y, Xue X and Yu Z:
Construction and evaluation of a prognosis lncRNA model for
hepatocellular carcinoma. J Cell Biochem. Apr 29–2020.(Epub ahead
of print). doi: https://doi.org/10.1002/jcb.29608.
|
|
49
|
Kou F, Sun H, Wu L, Li B, Zhang B, Wang X
and Yang L: TOP2A promotes lung adenocarcinoma cells' malignant
progression and predicts poor prognosis in lung adenocarcinoma. J
Cancer. 11:2496–2508. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu T, Zhang H, Yi S, Gu L and Zhou M:
Mutual regulation of MDM4 and TOP2A in cancer cell proliferation.
Mol Oncol. 13:1047–1058. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zeng S, Liu A, Dai L, Yu X, Zhang Z, Xiong
Q, Yang J, Liu F, Xu J, Xue Y, et al: Prognostic value of TOP2A in
bladder urothelial carcinoma and potential molecular mechanisms.
BMC Cancer. 19:6042019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Badawy OM and Loay I: FISH analysis of
TOP2A and HER-2 aberrations in female breast carcinoma on archived
material: Egyptian NCI experience. Appl Immunohistochem Mol
Morphol. 27:216–222. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang R, Xu J, Zhao J and Bai JH:
Proliferation and invasion of colon cancer cells are suppressed by
knockdown of TOP2A. J Cell Biochem. 119:7256–7263. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xiong Y, Yuan L, Chen L, Zhu Y, Zhang S,
Liu X, Xiao Y and Wang X: Identifying a novel biomarker TOP2A of
clear cell renal cell carcinoma (ccRCC) associated with smoking by
co-expression network analysis. J Cancer. 9:3912–3922. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nelson WG, Haffner MC and Yegnasubramanian
S: The structure of the nucleus in normal and neoplastic prostate
cells: Untangling the role of type 2 DNA topoisomerases. Am J Clin
Exp Urol. 6:107–113. 2018.PubMed/NCBI
|
|
56
|
Panvichian R, Tantiwetrueangdet A,
Angkathunyakul N and Leelaudomlipi S: TOP2A amplification and
overexpression in hepatocellular carcinoma tissues. Biomed Res Int.
2015:3816022015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ng IO, Na J, Lai EC, Fan ST and Ng M:
Ki-67 antigen expression in hepatocellular carcinoma using
monoclonal antibody MIB1. A comparison with proliferating cell
nuclear antigen. Am J Clin Pathol. 104:313–318. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Steelman LS, Chappell WH, Abrams SL, Kempf
RC, Long J, Laidler P, Mijatovic S, Maksimovic-Ivanic D, Stivala F,
Mazzarino MC, et al: Roles of the Raf/MEK/ERK and
PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity
to therapy-implications for cancer and aging. Aging (Albany NY).
3:192–222. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhao J, Ou B, Han D, Wang P, Zong Y, Zhu
C, Liu D, Zheng M, Sun J, Feng H and Lu A: Tumor-derived CXCL5
promotes human colorectal cancer metastasis through activation of
the ERK/Elk-1/Snail and AKT/GSK3β/β-catenin pathways. Mol Cancer.
16:702017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Echevarria MI, Awasthi S, Cheng CH,
Berglund AE, Rounbehler RJ, Gerke TA, Takhar M, Davicioni E, Klein
EA, Freedland SJ, et al: African American specific gene panel
predictive of poor prostate cancer outcome. J Urol. 202:247–255.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Rocha-Singh KJ: Retrospective real-world
studies of paclitaxel and mortality: Defining the many faces of
bias. JACC Cardiovasc Interv. May 14–2020.(Epub ahead of print).
doi: 10.1016/j.jcin.2020.05.006. View Article : Google Scholar : PubMed/NCBI
|