Introduction
Colorectal cancer (CRC) is one of the most prevalent
types of cancer worldwide. Despite the existence of an established
screening system, CRC remains to be the second leading cause of
cancer-associated mortality in a number of countries. Therefore,
the identification of novel biomarkers with therapeutic potentials
is a subject of intense research (1). Persistent accumulation of genetic and
epigenetic changes in the epithelial cells of the colon and rectum
has been documented to promote the malignant transformation of
normal mucosa into invasive cancer. Among the possible candidates
of proteins associated with CRC are transporters belonging to the
solute carrier (SLC) family. It was previously reported that solute
carrier organic anion transporter family member 4A1 (OATP4A1), a
transporter from the family of OATPs, encoded by the solute carrier
organic anion (SLCO)4A1 gene is overexpressed in CRC
tumors (2,3). Studies in cancer cell lines revealed
further that OATP4A1 mediates the uptake of endogenous compounds
including prostaglandin E2 (PGE2), sulfated and glucuronidated
steroid hormone precursors, bile acids, and drugs such as
benzylpenicillin and isoprostane (4–8). For the
analysis of OATP-associated transport in vitro,
OATP4A1-mediated uptake can be measured using the sodium
fluorescein surrogate substrate (9).
Notably, several endogenous OATP4A1 substrates have been identified
to mediate a significant role in CRC by regulating signaling
pathways that were implicated in the malignant transformation of
the mucosa and tumor progression. The proinflammatory prostaglandin
PGE2 is reportedly taken up from the extracellular space into the
mucosa cells by OATP4A1, which facilitates its enzymatic
inactivation to suppress its pro-inflammatory effects (10). In addition, bile acids are also
substrates of OATP4A1, which are taken up by the mucosa cells for
further metabolism to prevent their accumulation in the colonic
lumen. This observation has been hypothesized to be beneficial as
certain bile acids can promote carcinogenesis in the colon
(11). OATP4A1 can also transport
steroid hormone precursors such as estrone sulfate, which has been
demonstrated to increase cell proliferation following its
metabolization to 17β-estradiol, the most biologically active
estrogen. Knockdown of OATP4A1 expression in colon cancer cells
lines has been identified to significantly disrupt estrone sulfate
uptake (12). However, the role of
estrogen in CRC remains controversial. Previous epidemiological
studies have suggested that estrogen-based hormone replacement
therapy may decrease the risk of CRC, whilst data from in
vitro studies demonstrated that estrogens promote cancer cell
proliferation (13,14).
The association between OATP4A1 expression and
patient prognosis in a number of different types of tumor,
especially that of CRC, and lung cancer, remains unclear (15). In a previous study, OATP4A1 abundance
was reported to associate significantly with a decreased risk of
tumor recurrence within 5 years. In the present study, patients
with early stage CRC were investigated. In particular, high
expression levels of OATP4A1 expression in tumor immune cells were
revealed to associate with a decreased rate of early tumor
recurrence (16). By contrast,
another study by Ban et al (17) previously revealed that patients with
more advanced CRC tumors exhibit increased levels of OATP4A1 in the
tumor cells, have worse prognoses and decreased overall survival
rates. These data suggest that OATP4A1 expression may serve
different roles in the tumor cells and their microenvironment
depending on the CRC stage.
Genetic variants in OATP4A1, which can alter the
structure and function of the transporter, may also affect the
susceptibility to CRC and/or alter the course of the disease. It is
well known that SLCO genes are characterized by a high
genetic variability, with a minor allele frequency of >5
identified for OATP1B1, OATP1B3, and OATP1A2 (18). However, to the best of our knowledge,
there is no information as yet on SLCO4A1 polymorphisms in
patients with CRC compared with those in healthy controls. This is
important as OATP variants have been documented to affect
transporter properties, where single nucleotide polymorphisms
(SNPs) identified in the transporter genes have been demonstrated
to alter the expression and/or cellular localization of the protein
(19). SNPs in the SLCO1B3
gene, which codes for OATP1B3, have been widely studied in CRC
(20). It has been reported that the
overexpression of OATP1B3 in CRC cells decreases the
transcriptional activity of p53 and expression of its downstream
transcription targets, cyclin-dependent kinase inhibitor 1 and
p53-upregulated modulator of apoptosis (21). In particular, the OATP1B3 (SNP
583G>E) variant, which lacks transport function, was not
identified to exert any effect on the transcriptional activity of
p53, suggesting that tumor inhibition is associated with transport
activity (21). By contrast, 2
common SNPs in the SLCO1B1 (rs4149056 and rs2306283) and
SLCO2B1 (rs2306168 and rs12422149) genes were not associated
with CRC risk in patients (22). The
role of the additional SNPs detected in 3 exons of the
SLCO1B3 gene in tumor samples from 30 patients treated for
CRC in a Greek hospital remains unknown (23). In addition, even less is known about
the role of the OATP4A1 variants in cancer physiology.
Although previous studies have reported the
overexpression of OATP4A1 in CRC, to the best of our knowledge, the
possibility that SNPs in the SLCO4A1 gene can serve as a
risk factor for CRC has not been investigated previously.
Therefore, the present study aimed to determine whether the OATP4A1
variants were associated with an increased risk for CRC. Firstly,
NCBI, dbSNP, and Ensembl databases were used to identify the most
frequent non-synonymous mutations in the SLCO4A1 gene.
According to the Ensemble database, it was identified that 2
variants, G209>A (rs34419428, chr20:61288015) and G232>A
(rs1047099, chr20:61288038), were the most frequent mutations
present within the European population. In addition, the CRC
variants and their association with the clinicopathological data,
including sex, Union for International Cancer Control (UICC) stage
0/I vs. II and tumor recurrence following initial surgery within
the follow-up period of 5±0.25 years, were investigated in a sample
of 178 patients with early stage CRC and 65 healthy controls.
Paraffin-embedded CRC tissue samples were also used to determine if
OATP4A1 abundance in the tumor samples is associated with these
OATP4A1 genotypes. The functional activity of these OATP4A1
variants were studied in cells overexpressing them.
Material and methods
Patient samples
The present study was performed following the
Declaration of Helsinki and the Ethical guidelines of the
Institutions summarized in https://www.medunigraz.at/ethikkommission/index_dwnld.html.
Inclusion criteria for the present study were as
follows: Patients with CRC stage 0/I and stage II primary tumors
[classification according to the 7th edition of the TNM
classification of the UICC (24)];
and aged 35–85 years. Patients were not excluded on sex. Follow-up
for the patients was 5±0.5 years following the primary surgical
removal of the tumor.
Archived material from 178 patients of
middle-European ethnicity with early stage CRC (77 females and 101
males; mean age 63.5±12.8 years) were included into the present
study. The guidelines of the Ethical Committee of the City of
Vienna for retrospective studies (25) were applied. All patients had
undergone surgery for primary CRC at the hospital
Donauspital/Sozialmedizinisches Zentrum Ost between January 1992
and December 2009. In the cohort, stage 0/I tumors were present in
67 patients, whereas stage II tumors were present in 111 patients.
Clinical data for a follow-up period of 5±0.25 years were available
for these patients. Relapse of the tumor either at the original
site or at a distant site during follow-up was observed in 43 (25%)
of patients (mean time to relapse 809±164 days). Patients with
motality due to causes unrelated to CRC and patients with
additional cancers were excluded from the present study.
From the surgically removed tumors,
paraffin-embedded tissue blocks were routinely prepared at the
Department of Pathology (Donauspital/Sozialmedizinisches Zentrum
Ost) using neutral phosphate-buffered 10% formalin for fixation of
the tissue for 16–24 h at room temperature. Serial sections (4 µm)
were cut from archived paraffin blocks, which were stored at 4°C at
the Department of Pathology (Donauspital/Sozialmedizinisches
Zentrum Ost). A section from each paraffin block was stained with
Mayer's hematoxilin (Dako; Agilent Technologies GmbH) and eosin
G-Lösung 0.5% (Sigma-Aldrich; Merck KGaA) for 3–5 min at room
temperature following deparaffinization with xylene (three times),
followed by treatment with ethanol gradients (100% ethanol, twice
for 3 min; 95, 70 and 50% ethanol for 3 min, each) (26). Stained sections were then examined
for tissue integrity and for the presence of areas enriched with
cancer, immune, and stroma cells and normal mucosa adjacent to the
tumor using a Zeiss AXIO Observer Light microscope (Zeiss, Jena,
Deutschland) and the TissueFAXS Plus® cytometric system
(TissueGnostics, Vienna, Austria) at a magnification, ×20.
Unstained sections were used for genotyping and
immunohistochemistry.
DNA were obtained from 65 healthy volunteers of
middle-European ethnicity (mean age, 33.5±8.2 years; 32: 33
females: males). DNA was extracted from blood cells provided by the
Hungarian National Blood Transfusion Service and was used for
genotype controls. Permission for using the samples for the study
was obtained from the Medical Research Council of Hungary
(https://ett.aeek.hu/en/secretariat/;
3027/2013). Informed consent was provided by the participants.
DNA isolation from formalin-fixed
paraffin-embedded (FFPE) tissue sections
In the present retrospective study, DNA was isolated
from the FFPE tissue sections, prepared from the archived tissue
blocks, and then the frequency of the 2 OATP4A1 variants rs34419428
(G209>A) and rs1047099 (G232>A) was determined. Tissue
scrapes were prepared using a razor blade from the sections, which
were then collected into 1.5 ml DNA LoBind tubes (Eppendorf). DNA
was isolated using a NucleoSpin® FFPE XS DNA kit
(Macherey-Nagel GmbH & Co., KG) according to the manufacturer's
protocol. Briefly, a dissolver provided in the
NucleoSpin® FFPE XS DNA kit was used to remove the
paraffin from the FFPE tissue scrapes. Then, proteinase digestion
was used to solubilize the fixed tissue and release DNA into
solution. Heat incubation at 90°C for 30 min with the Decrosslink
Buffer D-Link from the kit was then applied to eliminate crosslinks
from the released DNA. Then, following the addition of ethanol, the
lysate was applied to the NucleoSpin® DNA FFPE XS
Column, where the DNA is bound to the silica membrane. Following 2
washing steps, the DNA was finally eluted under low ionic strength
conditions in a small volume (20 µl) of NucleoSpin® FFPE
XS DNA kit Elution Buffer BE.
For each extraction, the DNA yields were quantified
using a Qubit™ 3.0 Fluorometer with the Quant-It kit (Thermo Fisher
Scientific, Inc). The purity of the extracted nucleic acids was
determined using the 260/280 nm absorbance ratios obtained using a
NanoDrop™ spectrophotometer.
Restriction fragment length
polymorphism (RFLP)
To determine the presence of the G209>A
(rs34419428, chr20:61288015; http://www.ncbi.nlm.nih.gov/snp/rs34419428) and
G232>A (rs1047099, chr20:61288038; http://www.ncbi.nlm.nih.gov/snp/rs1047099) variants in
CRC and control samples, the DNA fragments containing both SNPs
were amplified by PCR using the following primers: Forward,
5′-GACAAGCCGCTCACCTTC-3′ and reverse 5′-CACAGGAAGAACAGGATGCC-3′
(Invitrogen; Thermo Fisher Scientific, Inc.). PCR conditions (35
cycles) were as follows: Annealing of the primers at 55°C for 40
sec and extension at 72°C for 1 min. Prior to initiation of the 35
cycles, pre-incubation at 95°C was performed for 5 min. The PCR
fragment was digested with restriction enzymes Eco88I and/or Alw26I
(Fermentas; Thermo Fisher Scientific, Inc.) to detect the presence
of G209>A and G232>A, respectively, and subsequently analyzed
on agarose gels after staining with SYBR Safe DNA Gel Stain
(Invitrogen; Thermo Fisher Scientific, Inc.). Bands were documented
and analyzed using the GelDoc Go Imaging System together with the
Image Lab Touch 2.4 Software (Bio-Rad Laboratories, Inc.).
Haplotype frequencies and
linkage-disequilibrium (LD) analysis
Haplotype frequencies were calculated by integrating
the population-specific variant frequencies (Table SI) with the linkage information from
the corresponding population (27).
Data on the populations were provided by the 1000 Genomes Project
using LDLink (28). To determine the
LD, which gives the nonrandom association of alleles at different
loci, for rs1047099 and rs34419428 in different ethnic groups, the
LDpair analytical tool was used (https://analysistools.nci.nih.gov/LDlink/?var1=rs34419428&var2=rs1047099&pop=TSI&tab=ldpair)
(28).
Immunohistochemistry of FFPE
sections
Immunohistochemical staining of tissue sections was
performed using an affinity-purified anti-OATP4A1 polyclonal rabbit
antibody (cat. no. HPA030669; Atlas Antibodies AB). If not stated
otherwise, all steps were performed at room temperature (22°C). In
the preliminary experiments, serial dilutions of the antibody were
examined to obtain optimal staining for OATP4A1 and minimize
unspecific background staining of the tissue. Following
de-paraffinization of the sections with xylene followed by
treatment with an ethanol gradient (xylene, twice for 3 min;
xylene/ethanol mixture for 3 min; 100% ethanol, twice for 3 min;
95, 70 and 50% ethanol for 3 min, each), endogenous peroxidase
activity was blocked using 0.3% H2O2 in PBS
for 10 min. Repeated washing (three times) with PBS was performed,
followed by a blocking step for unspecific binding sites on the
antigen using the horseradish peroxidase polymer blocking solution
provided in the Thermo Scientific Lab Vision™ UltraVision™ LP
Detection System (Thermo Fisher Scientific, Inc.) for 1 h at room
temperature. The UltraVision™ LP detection system contains a
universal secondary antibody formulation conjugated to an
HRP-labeled polymer that recognizes mouse and rabbit IgG. To detect
OATP4A1, the HPA030669 antibody (Atlas Antibodies AB; 1:50 dilution
in PBS containing 0.5% bovine serum albumin) was applied at 4°C
overnight. Following repeated washing, the UltraVision™ LP antibody
enhancer solution containing goat anti-rabbit IgG was added for 1
h. The UltraVision™ LP Detection System HRP polymer and DAB Plus
Chromogen were then used for the detection of OATP4A1 staining.
Finally, cell nuclei were stained using water-based Mayer's
hematoxylin solution (Dako; Agilent Technologies GmbH) for 1 min
and sections were mounted using dibutylphthalate polystyrene xylene
mounting medium (Sigma-Aldrich; Merck KGaA).
Quantitative image cytometry of the
FFPE sections
The TissueFAXS Plus® cytometric system
(TissueGnostics GmbH) equipped with an AXIO Imager Zeiss 1
automated light microscope (Carl Zeiss AG) and the
HistoQuest® software (version 4.0.4.150; TissueGnostics
GmbH) was used for image acquisition at magnification, ×20, and
quantitative evaluation of OATP4A1-positive cells as described
previously (29). To determine the
proportion of OATP4A1-positive cells, the gray values of the DAB
signal intensity were assessed after correcting for the background
values from the negative control samples, where the primary
antibody was replaced with non-immunogenic IgG. The proportions of
OATP4A1-positive tumor, immune, stroma, and adjacent mucosa cells
were calculated from the total number of OATP4A1-positive tumor,
immune, stroma, and adjacent mucosa cells per area, respectively
(set to 100%).
The blue color of hematoxylin-stained nuclei allows
for an estimation of tissue quality and staining efficacy of the
procedure in the individual samples. From the original cohort,
samples from 6 patients were excluded due to the poor morphological
preservation of the tissue, which did not result in a staining
pattern that was acceptable for the automated analysis by the FAXS
system. Analysis was performed on sections from 172 patients on
defined regions of interest covering the following areas in the
tumor sample: Cancer cell-enriched areas and the surrounding
stroma; infiltrating immune cells; and morphological normal mucosa
cells as described previously (29,30).
Studies on the G209>A (rs34419428) and
G232>A (rs1047099) SLCO4A1 variants in Sf9 insect cells and A431
cancer cell lines
Generation of the Sf9 and A431 cells
overexpressing the OATP4A1 variants
Generation of the plasmid construct for expressing
OATPs in Sf9 cells was performed as described previously (9). The baculovirus transfer vector pAcUW21
(BD Biosciences) was engineered to give the modified plasmid
pAcUW-L2, which is suitable for cloning of all OATPs (9). Full-length cDNA sequences encoding
human OATP4A1 variants were introduced into the pAcUW-L2 vector.
Sequencing of the OATP4A1 construct from the Harvard Plasmid ID
Repository (Harvard Medical School) revealed that the cDNA
contained the 232G>A SNP (V78I, rs1047099). Therefore, the
canonical sequence for wild-type (wt) OATP4A1 (HGNC: 10953; Entrez
Gene: 28231; Ensembl: ENSG00000101187; OMIM: 612436; UniProtKB:
Q96BD0-1) was generated using the QuikChange Mutagenesis kit
(Thermo Fisher Scientific, Inc.). Also, other probes for the
variants R70Q and R70Q/V78I (GenBank accession numbers: variant
rs34419428, NM_016354.4: c.209G>A; NP_057438.4: p.R70Q and
variant rs1047099, NM_016354.4: c.232G>A; NP_057438.4: p.V78I;
Table SI) were generated by PCR
site-directed mutagenesis. The following primers were used: Wt
forward, 5′-AGGTGCGGTACGTCTCGG-3′; Wt reverse,
5′-CCGAGACGTACCGCACCT-3′. R70Q forward, 5′-GCCCAGGGGACCCATGA-3′;
R70Q reverse, 5′-TCATGGGTCCCCTGGGC-3′; R70Q/V78I forward,
5′-GCCCAGGGGACCCATGA-3′; and R70Q/V78I reverse,
5′-TCATGGGTCCCCTGGGC-3′. The PCR reaction was performed using 2.5
U/µl Phusion High-Fidelity DNA Polymerase (Thermo Fisher
Scientific, Inc.). DNA sequences of all the constructs were
verified using the DNA Sanger sequencing method. At least 2 clones
were sequenced from each plasmid.
The human epidermoid carcinoma A431 cell line was
obtained from ATCC and cultured in DMEM supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.). Cells were maintained at
37°C in a humidified atmosphere under 5% CO2. Wt and
variant OATP4A1-overexpressing A431 cells were generated by the
transposase (Thermo Fisher Scientific, Inc.) mediated genomic
insertion method (31) using a
pSB-CMV vector (Promega Corporation). For transfection, the PCR
fragments were cloned between the NotI-HindIII (New England
Biolabs Inc.) sites of the pSB-CMV vector (29,30).
Cell selection was performed with 1 µg/ml puromycin (Sigma-Aldrich;
Merck KGaA). Thereafter, the cells were maintained in DMEM (Gibco,
Thermo Fischer) supplemented with 10% FBS, 2 mM L-glutamine, 100
U/ml penicillin, and 100 µg/ml streptomycin (Sigma-Aldrich; Merck
KGaA) at 37°C with 5% CO2 and 95% humidity (32,33).
Sf9 cells from Spodoptera frugiperda were
obtained from ATCC (Sf9 ATCC® CRL-1711™) and were grown
in a 0.5 liter round-bottom spinner flask in TNM-FH insect medium
(Sigma-Aldrich; Merck KGaA), supplemented with 10% FBS, 100 U/ml
penicillin and 100 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA)
at 27°C. These cells were previously used to study OATP-mediated
transport (34). Recombinant
baculoviruses, carrying the different human SLCO4A1 gene
sequences, were generated using the BD BaculoGold™ Transfection kit
(BD Biosciences), according to the manufacturer's protocols, and
were stored at 4°C.
Western blot analysis
Western blot analysis was used to detect the
expression of the wtOATP4A1, and the polymorphic variants expressed
in Sf9 and A431 cells. All the following isolation steps were done
at 4°C to obtain the proteins. Cells were suspended in 50 mM
Tris/HCL buffer, pH 7.0, containing 50 mM mannitol, 2 mM EGTA, 10
pg/ml leupeptin, 8 pg/ml aprotinin, 0.5 mM phenylmethylsulfonyl
fluoride and 2 mM beta-mercaptoethanol (all from Sigma-Aldrich;
Merck KGaA). Thereafter, cells in this suspension buffer were
homogenized to obtain a homogenate containing cells, organelles and
membranes. However, it contained also some undisrupted cells and
larger cell debris. To remove undisrupted cells and larger debris
from the homogenate, the sample was centrifuged at 500 × g for 10
min, followed by a centrifugation step at 2,000 × g for 10 min to
remove mitochondria and other cell organelles, such as lysosomes.
Finally, a crude membrane fraction was recovered by centrifugation
(60 min at 100,000 × g). This pellet containing the proteins was
resuspended in the homogenization buffer to obtain a protein
concentration of 2 mg/ml. All procedures were carried out at 4°C
and samples were stored at −70°C for further applications. Protein
concentration was determined using modified Lowry method
(Sigma-Aldrich; Merck KGaA).
Proteins (25 µg) were separated by 4–15% SDS-PAGE
and transferred onto PVDF membranes (Bio-Rad Laboratories, Inc.).
After blocking unspecific binding sites on the protein with the
Invitrogen™ membrane blocking solution (Thermo Fisher Scientific,
Inc.) for 1 h at room temperature, membranes were incubated with
the polyclonal anti-OATP4A1 antibody overnight at 4°C (1:1,000).
The antibody was provided by Dr. Bruno Stieger, (Division of
Clinical Pharmacology and Toxicology, Universitätsspital Zürich,
Switzerland) or non-immunogenic IgG (Thermo Scientific™ Lab
Vision™) for the negative control. Following primary antibody
incubation, the membranes were incubated with a horseradish
(HRP)-conjugated anti-rabbit secondary antibody at a dilution of
1:10,000 for 1 h at room temperature (cat. no. AB_2307391; Jackson
ImmunoResearch Europe, Ltd.). Immunoreactive protein bands were
detected using the Pierce™ ECL Western Blotting Substrate (Thermo
Fisher Scientific, Inc.).
Immunofluorescence staining
A431 cells overexpressing mutant or wtOATP4A1
cultured on cover slides were fixed using methanol (stored at
−20°C) for 5 min. Following repeated washing and blocking of
unspecific binding sites with 10% bovine serum albumin
(Sigma-Aldrich; Merck KGaA) for 1 h, the antibody against OATP4A1
(cat. no. HPA030669, Atlas Antibodies AB) was diluted 1:100 in PBS
containing 2% rabbit serum (Vector Laboratories; Maravai
LifeSciences) and applied overnight. Prior to the staining
procedure, optimal antibody concentrations were determined by
titrating serial antibody dilutions. The applied dilution
corresponds to the minimum concentration necessary to produce a
positive signal. Following primary antibody incubation, a secondary
antibody (Alexa Fluor 488-anti-rabbit IgG; cat. no. AB_2338052;
Jackson ImmunoResearch Europe Ltd.), diluted 1:500 in PBS was
applied for 1 h at room temperature. For staining of the nuclei,
DAPI (Roche Diagnostics) was used at a concentration of 0.2 µg/ml
for 10 min at room temperature. In the control experiments, the
primary antibody was replaced by non-immunogenic IgG (Thermo Fisher
Scientific Lab Vision™; Thermo Fisher Scientific, Inc.). No
staining for OATP4A1 was observed. Finally, following repeated
washing, the slides were mounted in Fluoromount™ mounting medium
(Sigma-Aldrich; Merck KGaA) prior to visualization using the LSM
900 Confocal Laser Scanning Microscope (Carl Zeiss AG;
magnification, ×20).
Uptake of sodium-fluorescein in Sf9
and A431 cells overexpressing OATP4A1 and its polymorphic
variants
Transport experiments in Sf9 and A431 cells were
performed using the fluorescent OATP substrate, sodium fluorescein,
as previously described (35).
Briefly, 5×105 cells were incubated with 1 µM sodium
fluorescein (Sigma-Aldrich, Merck KG) at pH 5.5 for 10 min. The
reaction was stopped using 1 ml ice-cold PBS. The cellular
fluorescence was measured at the emission wavelength of 530 nm
following the excitation wavelength of 488 nm using an Attune™
acoustic focusing cytometer (Thermo Fisher Scientific, Inc.).
Position of the two SNPs in a
two-dimensional (2D) structure of OATP4A1
The 2D structure of human OATP4A1 has been
determined using the HMMTOP transmembrane topology prediction
server; 2.0 version (36). The
diagram was drawn according to the information available at:
http://emboss.bioinformatics.nl/cgi-bin/emboss
(version: EMBOSS:6.6.0.0).
Statistical analysis
Statistical analysis was performed using SAS
software (version 9.4; SAS Institute, Inc.), and categorical
variables are presented as counts and percentages.
Clinical data from the 172 patients including sex,
UICC stage 0/I and II, relapse within 5±0.25 years, were used for
the statistical calculations. Statistical comparisons between
groups were performed using χ2 tests, whilst Cramer's V
was used to quantify the associations between categorical
variables. For Cramer's V, values range between +1 for perfect
agreement and −1 for perfect disagreement. A Kruskal-Wallis test
was used to compare the percentages of OATP4A1-positive cells
between tumor (tumor, immune, and stroma cells) and the adjacent
mucosa cells. Within each OATP4A1 variant, multiple comparisons
among the three cell types in the tumor and the tumor-adjacent
mucosa cells were corrected by the Bonferroni-Holm adjustment.
P≤0.05 was considered to indicate a statistically significant
difference.
Results
OATP4A1 allele frequency in patients
with CRC and controls
The frequency of the two most common missense
mutations in the SLCO4A1 gene in the European population,
rs34419428; c. 209G>A, which encodes the polymorphic variant
R70Q, and rs1047099; c. 232G>A, which encodes the R78I variant,
were assessed (Table SI). The
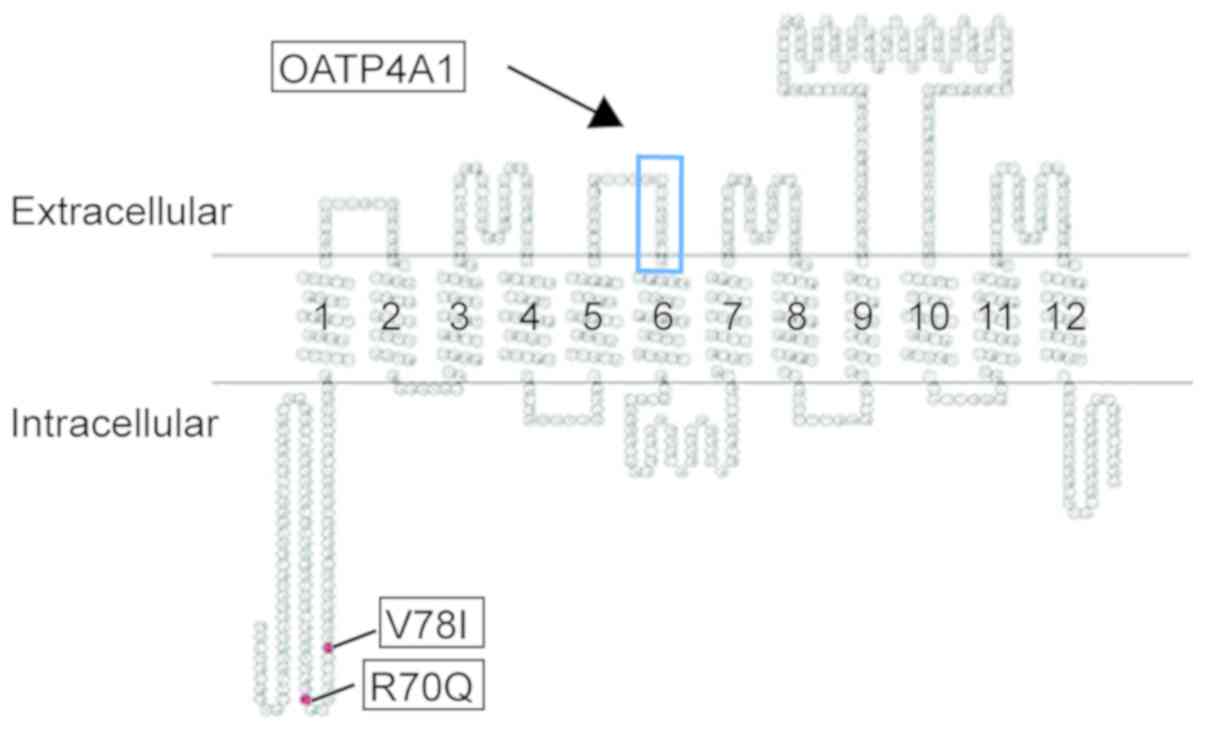
structure of rodent Oatp1 was used as a template to illustrate the
position of the mutant amino acids in the two OATP4A1 variants
(37). According to computer-based
hydropathy analysis, all of the OATP4A1 variants were identified to
share a similar transmembrane domain organization, with 12
predicted transmembrane domains, where the N- and C-terminal ends
are located on the cytoplasmic side of the membrane. The sequence
defining the OATP superfamily is located on the boundary of the
extracellular loop 3 and transmembrane domain 6. The two SNPs
investigated in the present study were identified to be in the
first intracellular loop of the cytoplasmic domain (Fig. 1).
Genotyping of the 2 variants was conducted by RFLP
using DNA from 178 patients with CRC and 65 healthy controls. A
frequency of 0.038 was observed for the R90Q allele and 0.292 for
the V78I allele in patients with CRC, whilst similar frequencies of
0.051 and 0.286, were observed for the R90Q and the V78I alleles,
respectively, in the control group (Table I). Statistical analysis revealed that
there was no significant difference between the genotype frequency
in the control and CRC groups (P=0.56 and P=0.52 for the R90Q and
the V78I alleles, respectively).
 | Table I.Wild-type OATP4A1 and the V78I and
R90Q minor alleles in patients with colorectal cancer and in
healthy individuals as controls. |
Table I.
Wild-type OATP4A1 and the V78I and
R90Q minor alleles in patients with colorectal cancer and in
healthy individuals as controls.
|
| Controls
(n=65) | Patients with CRC
(n=187) |
|---|
|
|
|
|
|---|
| Individuals | R70Q | V78I | R70Q | V78I |
|---|
| Minor alleles, n
(%) | 5 (3.8) | 38 (29.2) | 19 (5.1) | 107 (28.6) |
| Minor allele,
frequency | 0.038 | 0.292 | 0.051 | 0.286 |
| Wt individuals, n
(%) | 60 (92.3) | 31 (47.7) | 168 (89.8) | 97 (51.9) |
| Heterozygous
individuals, n (%) | 5 (7.7) | 30 (46.1) | 19 (10.2) | 73 (39.0) |
| Homozygous
individuals, n (%) | 0 (0.0) | 4 (6.2) | 0 (0.0) | 17 (9.1) |
| Non-wt individuals,
n (%) | 5 (7.7) | 34 (52.3) | 19 (10.2) | 90 (48.1) |
| Chromosomes, n | 130 | 130 | 374 | 374 |
The linkage equilibrium between the two alleles in
the mixed European population was next determined by the LDpair
analytical tool using data for the R90Q and the V78I allele
frequency from the 1000 genome project (Table SI). The statistical analysis for the
random association of the two alleles in the different populations
revealed a value of D′=1, suggesting a high level of allelic
association. The r2 value of 0.12 and the high
χ2 value of 120 (P<0.0001) further suggested that
these two alleles are associated with each other (Table SII).
All individuals in the CRC and control groups were
identified to carry either hetero- or homozygous mutations for the
V78I allele, but were always heterozygous for the R70Q allele
(Table I). No significant difference
was observed between the patients with CRC and controls in the
distribution of these two minor alleles. In the CRC group, 7.7% of
patients were heterozygous for the R90Q allele and 46.2% for the
V78I allele. Similarly, in the control group, 10.2% of individuals
were heterozygous for the R70Q allele and 39.0% for the V78I
allele. There was also no difference between the CRC and the
control group regarding the proportion of V78I homozygous
individuals. 9.1% individuals were homozygous in the CRC and 6.2%
in control groups, respectively (Table
I).
OATP4A1 variants and patient
characteristics
The association between the 2 polymorphic variants
of OATP4A1 in patients with CRC and their respective
clinicopathological characteristics [female (n=74) and male (n=98);
UICC stage 0/I (n=61) and UICC stage II (n=111); relapse within the
follow-up period of 5±0.25 years (n=43) and no relapse (n=129)] was
analyzed using the Cramer's V and χ2 test. From these
172 patients, FFPE sections were available for the quantitative
analysis of OATP4A1 expression. No association between the
parameter was observed. Data are summarized in Table II.
 | Table II.Association between variant OATP4A1
and clinicopathological data from patients with colorectal
carcinoma. |
Table II.
Association between variant OATP4A1
and clinicopathological data from patients with colorectal
carcinoma.
|
| Clinicopathological
data |
|---|
|
|
|
|---|
|
| Sex, n
(female:male, 74:98) | UICC Stage, n
0/I:61/II:111 | Relapse, n
yes:43/no:129 |
|---|
|
|
|
|
|
|---|
| OATP4A1 | Cramer's V | P-value | Cramer's V | P-value | Cramer's V | P-value |
|---|
| R70Q | −0.09 | 0.26 | −0.02 | 0.75 | −0.11 | 0.15 |
| V78I | 0.07 | 0.63 | 0.13 | 0.21 | 0.17 | 0.08 |
| Haplotype | 0.09 | 0.47 | 0.07 | 0.65 | 0.11 | 0.33 |
| V78I_dom | −0.06 | 0.40 | −0.07 | 0.35 | −0.07 | 0.38 |
| V78I_rec | 0.01 | 0.87 | 0.08 | 0.28 | 0.12 | 0.10 |
In addition, this analysis was performed to study
whether haplotypes are associated with these parameters. The 3
haplotypes were examined as a collective (Tables SI and SII). Type 1 consists of wtR70 and wtV78
with nucleotides G and G in the two alleles at amino acid position
70 and 78, respectively; type 2 consists of wtR70 and mutant V78I
resulting from the nucleotides being G and A, respectively; and
type 3 has the R70Q and V78I alleles encoded by the A and A
nucleotide variations, respectively.
The type 4 haplotype with R70Q and wtV78
(nucleotides A and G in the respective allele) was not present in
the samples examined in the present study, consistent with data
from the 1000 Genomes project on the haplotype distribution in the
Middle European population. Further data on the distribution of the
haplotypes among different ethnic groups are provided in detail in
the Tables SI and SII. It was observed that Type 4 is present
only in ethnic groups from Africa.
The association between haplotypes and
clinicopathological data is summarized in Table II. Values obtained for Cramer's V
ranged between-0.09 and 0.09, which indicated that there was no
significant correlation between the clinicopathological parameters
and the haplotypes. However, independence could not be rejected
according to the results from the χ2 test
(P=0.08–0.87).
OATP4A1 variants in CRC tumor
sections
To determine if the percentage of OATP4A1-positive
cells in the CRC tumors is associated with the polymorphic variants
in the CRC patients, the FFPE tumor sections were stained for
OATP4A1. Controls were stained with non-immunogenic IgG. On the
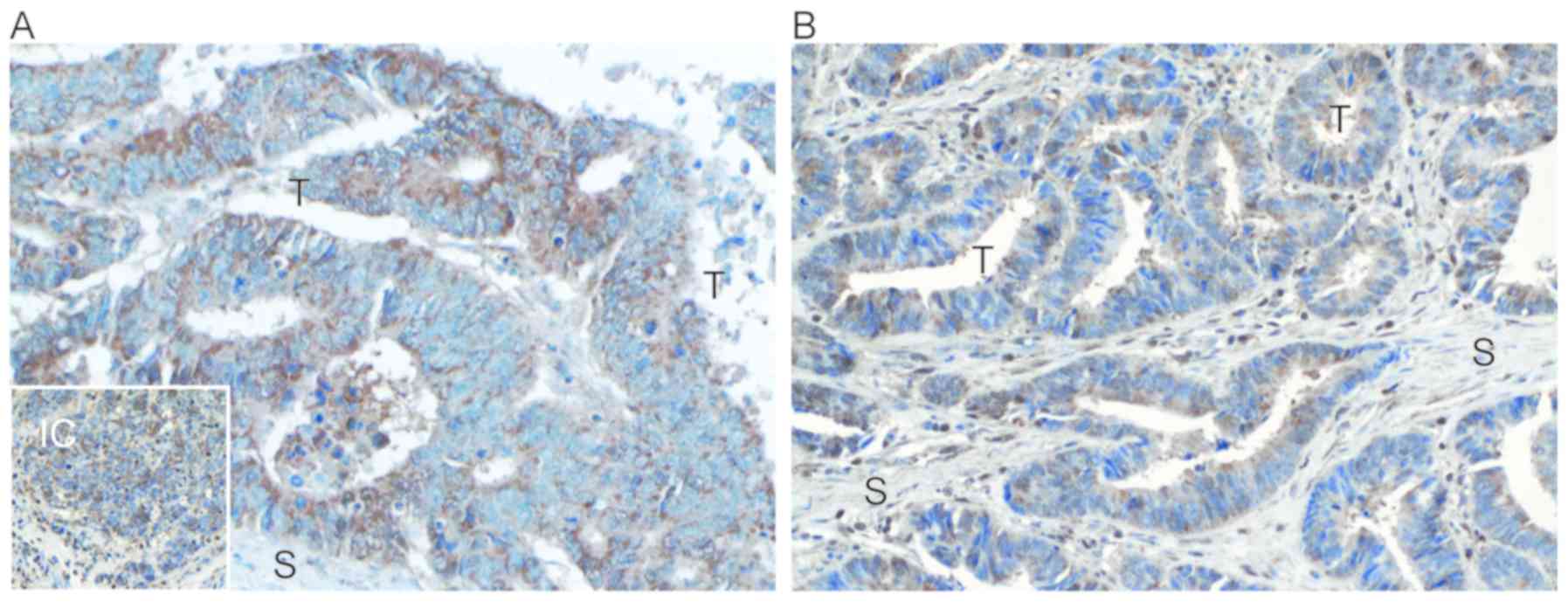
tissue sections, positive staining for OATP4A1 was observed in the
cytosol and on the membrane of tumor cells, while the membrane vs.
cytosol localization was not identified to be significantly
different among patients carrying the wtOATP4A1 or those harboring
variants (Fig. 2). In adjacent
mucosa cells, positive staining for OATP4A1 was almost always
(>95%) observed to be located on the membrane.
An automated quantitative microscopic image analysis
system was used to determine the number of OATP4A1-positive cells
in areas enriched in cancer, immune, and stroma cell and the mucosa
adjacent to the tumor (Fig. 2A and
B). Following correction for the unspecific background staining
of the samples, the percentage of positive cells per area was then
calculated. The portion of OATP4A1-positive cells in all areas was
identified to be independent of whether the patients carried the
wtOATP4A1 or the polymorphic variants (Table III).
 | Table III.Statistical analysis of differences
in the proportion of OATP4A1-positive cells in tumor sections
derived from patients with CRC with variant OATP4A1. |
Table III.
Statistical analysis of differences
in the proportion of OATP4A1-positive cells in tumor sections
derived from patients with CRC with variant OATP4A1.
| OATP4A1
variant | OATP4A1-positive
cellsa | P-value | P-value
(corrb) |
|---|
| R70Q | Immune cells | 0.628 | 1.000 |
|
| Tumor cells | 0.046 | 0.185 |
|
| Adjacent
mucosa | 0.344 | 1.000 |
|
| Stroma cells | 0.548 | 1.000 |
| V78I | Immune cells | 0.619 | 1.000 |
|
| Tumor cells | 0.161 | 0,644 |
|
| Adjacent
mucosa | 0.759 | 1.000 |
|
| Stroma cells | 0.256 | 0.767 |
| Haplotype | Immune cells | 0.402 | 0.804 |
|
| Tumor cells | 0.096 | 0.385 |
|
| Adjacent
mucosa | 0.440 | 0.804 |
|
| Stroma cells | 0.267 | 0.802 |
| V78I_dominant | Immune cells | 0.329 | 0.659 |
|
| Tumor cells | 0.129 | 0.417 |
|
| Adjacent
mucosa | 0.624 | 0.659 |
|
| Stroma cells | 0.104 | 0.417 |
| V78I_recessive | Immune cells | 0.786 | 1.000 |
|
| Tumor cells | 0.586 | 1.000 |
|
| Adjacent
mucosa | 0.687 | 1.000 |
|
| Stroma cells | 0.395 | 1.000 |
Studies in Sf9 and A431 cells
overexpressing OATP4A1 variants
To determine whether the SNPs resulted in changes in
the expression levels, localization, or function of OATP4A1, Sf9
(S. frugiperda) cells and A431 cell lines overexpressing
either wtOATP4A1 or its R70Q, V78I or R70Q/V78I variants were
established. Sf9 cells provide a well-known eukaryotic expression
system that is able to produce large amounts of active recombinant
proteins such as transporters, and is useful for investigating the
biochemical and pharmacological properties of the proteins
expressed (34). These cells have
also been successfully used in the pharmaceutical industry for
studying drug uptake and excretion pharmacologically. However,
unlike transfected human cell lines, Sf9 cells cannot glycosylate
proteins. Therefore, the human skin tumor A430 cell line was also
transfected and expresses sufficient wt and variant OATP4A1
required for transportation studies (9,35).
The expression of wtOATP4A1 or their variants were
first analyzed using western blot analysis and immunofluorescence
staining (Fig. 3). According to the
western blot analysis results, the anti-OATP4A1 antibody produced a
single immunoreactive band at ~70 kDa for both wt and the variant
OATP4A1. The subcellular localization of the transporter was next
studied in A431 cells transfected with wtOATP4A1 or the variants.
Following immunofluorescence staining using Alexa Fluor®
488-labeled antibodies, green fluorescence, representing labelled
OATP4A1 proteins, was visible on the cell membranes of the cells
transfected with wt or OATP4A1 variants (Fig. 3).
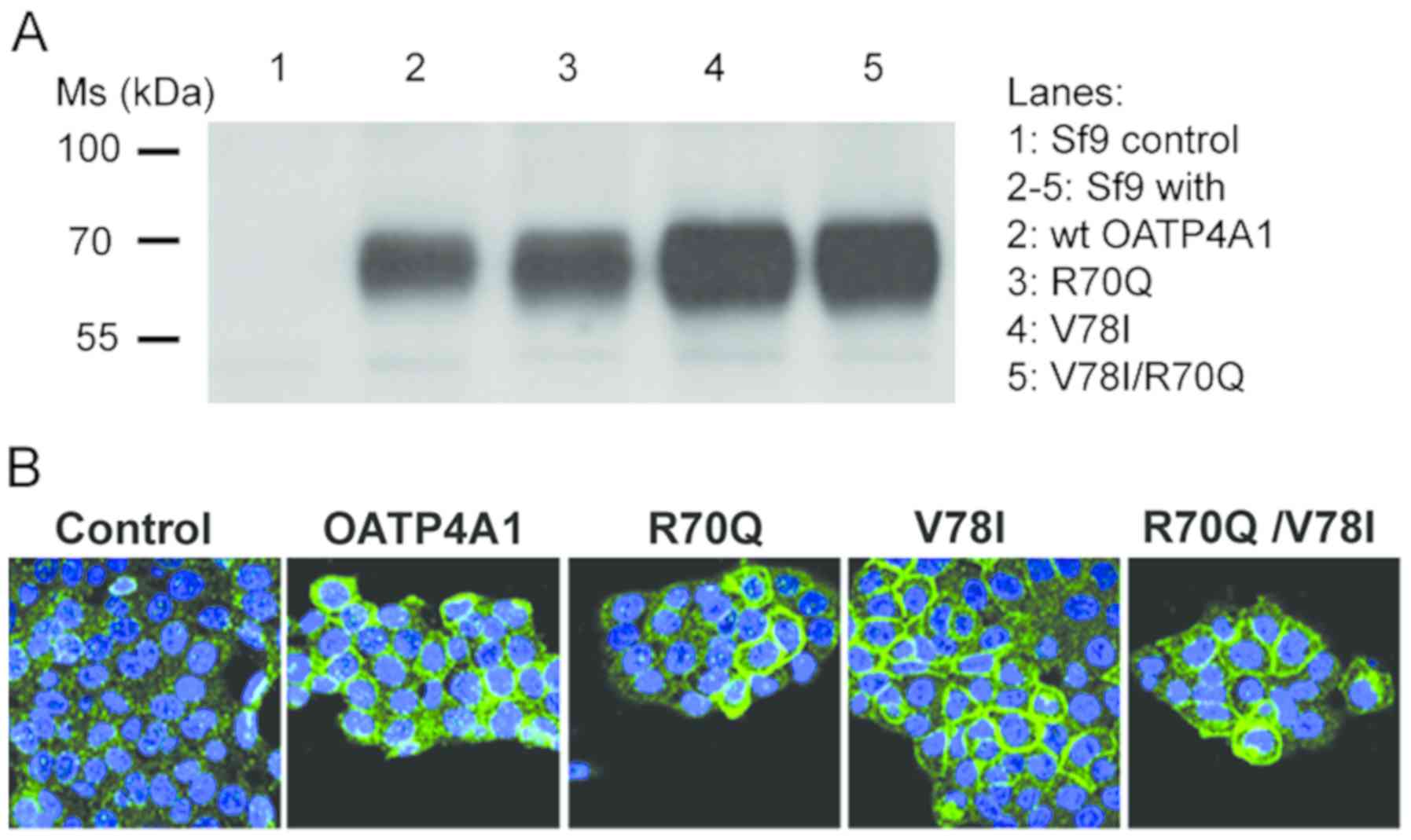 | Figure 3.OATP4A1 (wt, R70Q, V78I, and
R70Q/V78I variant) overexpression in Sf9 insect and A431 cancer
cells. (A) Western blot of Wt and variant OATP4A1 in Sf9 insect
cells probed with a polyclonal anti-OATP4A1 antibody. There were 10
µg whole Sf9 cell lysates were analyzed. Lane 1, sF9 cells with the
empty plasmid (control); lanes 2–5, sF9 cells overexpressing either
wt OATP4A1, (shown in lane 2); or variant OATP4A1 (lanes 3–5).
Variant R70Q is shown in lane 3, V78I in lane 4 and V78I/R70Q in
lane 5. (B) Confocal images of A431 cells transfected with the
empty vector (control) and cells overexpressing wt OATP4A1 and the
R70Q, V78I or R70Q/V78I variants. Magnification, ×20. Nuclei were
stained blue using DAPI. wt, wild-type; OATP4A1, solute carrier
organic anion transporter family member 4A1; CRC, colorectal
cancer; Ms, molecular mass. |
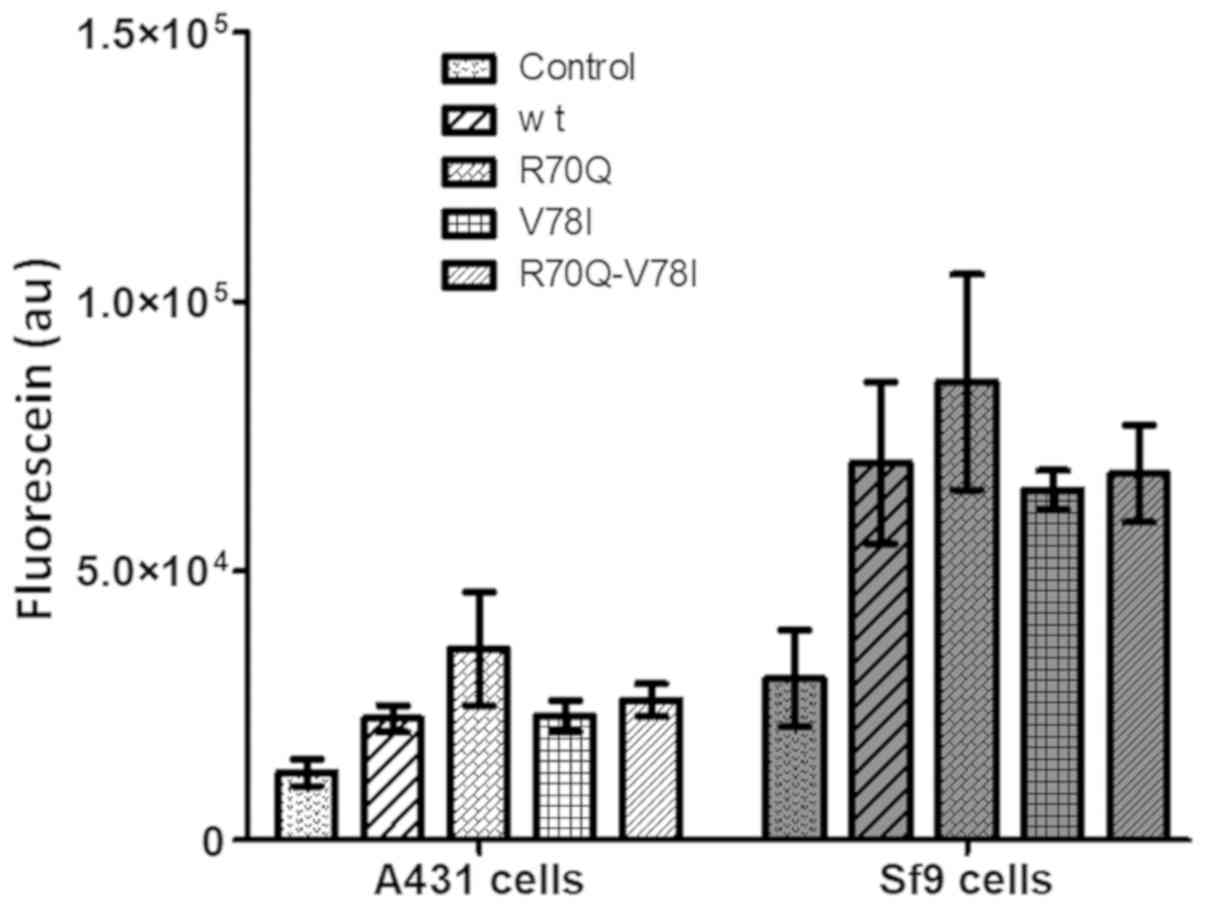
Following the validation of the expression of the
variants, their capability of transporting the sodium fluorescein,
a model substrate used for studying OATP-mediated transport, was
determined (32). An increased
accumulation (2–3-fold) of the fluorescent dye was observed in Sf9
and A431 cells transfected with the wt or OATP4A1 variants compared
with that in cells transfected with the empty plasmid. Importantly,
no significant difference was identified between the transport
function of wtOATP4A1 and that of the variants (Fig. 4).
Discussion
OATPs have been previously identified to exhibit
significant genetic variability (18), and polymorphisms in these
transporters may affect cancer risk and progression, drug responses
and the outcome of subsequent therapeutic efficacy (38,39). To
date, the most commonly studied polymorphisms in the SLCO
genes are those encoding for the OATP1 family (19,40).
SNPs present in OATP1B3 and OATP1B1 have been identified to exert
severe side effects due to the altered uptake kinetics of
anticancer drugs such as that of irinotecan, which is frequently
used for treating CRC (41,42).
Although a previous study observed that OATP4A1 was
overexpressed in CRC, to the best of our knowledge, no data are
available regarding the effect of SNPs in genes encoding for
OATP4A1 on the outcome or progression of CRC. Studies on OATP4A1
expression levels and its role in the altered pharmacokinetic
activity of drugs are limited. Benzylpenicillin and an isoprostane
metabolite have been previously identified as OATP4A1 substrates
in vitro (43). Notably,
methotrexate (MTX), which exerts antiproliferative and cytotoxic
effects in a number of cell types such as colon cancer cells, was
proposed to be OATP4A1 substrate. It was investigated whether the
presence of the polymorphic OATP4A1 variants would alter the
outcome of a specific therapy against rheumatoid arthritis
(44). MTX is an agent that is
frequently used in cancer chemotherapy and an established treatment
strategy for autoimmune and inflammatory diseases such as
rheumatoid arthritis. In a study in patients with arthritis, a
C>T intron variant (rs2236553) with a frequency of 0.34 in the
European population was identified to affect the efficacy of MTX
treatment and promote drug toxicity (44). By contrast, a second study did not
confirm these results (45). In
fact, it suggested that MTX was unlikely to be a substrate for
OATP4A1. The cellular accumulation of MTX may be caused by the
action of other OATPs, including OATP1B1 and OATP1B3 (45).
The expression levels of OATP4A1 have been reported
to be important for CRC predisposition and tumor progression
(16,17). Therefore, the present study aimed to
elucidate whether variants in OATP4A1 may predispose individuals to
CRC. A total of 2 frequent nonsynonymous polymorphisms in OATP4A1
(rs34419428 and rs1047099) were investigated in 178 patients with
CRC and 65 healthy controls of Austrian and Hungarian origin. It
was identified that <50% of all individuals were heterozygous
for the c.232G>A allele (p.V78I) and 7–10% for the c.209G>A
(p.R70Q) allele. These results on the allelic frequency in the CRC
patients and controls were consistent with data involving a mixed
European population, which were previously published in the 1000
Genomes Project (46). Notably, all
individuals harboring the 209A (R70Q) allele also had the minor
allele c.232G>A. As both SNPs are located in the same exon, they
can be co-amplified by one PCR reaction. Using RFLP-PCR, it was
shown that the R70Q minor allele (rs34419428) is associated with
the V78I minor allele (rs1047099) in the genome of patients with
CRC and control individuals in the present cohort.
In the present study, no association was identified
between the sex, UICC stage and tumor recurrence of patients with
early stage CRC and the 2 OATP4A1 polymorphic variants. The
distribution of the 2 alleles differs among different ethnic
groups, but none of the polymorphic genotypes in the present cohort
were associated with the CRC risk and the risk a tumor recurrence
within 5 years. The same conclusion was also derived from
immunohistochemical staining using FFPE tissue sections from the
CRC tumors in the present study. The percentage of OATP4A1-positive
cells present in the tumor, immune, and stroma cells and the
adjacent mucosa cells were not found to be associated with the
OATP4A1 variant present in the patients.
In addition, the effect of SNPs in OATP4A1 on the
functional properties of the OATP4A1 transporter was analyzed. SNPs
in SLCO genes encoding for the different OATPs were
described to alter the subcellular localization of the proteins,
causing a shift of the transporter from the plasma membrane to the
cytosol (47,48). Therefore, these results suggested
that SNPs in SLCO genes may also alter the transport
functionality of the OATPs. For example, a nonsynonymous SNP in
OATP1B1 was previously identified to change its phosphorylation
status, subcellular localization, and the ability to transport its
substrate estradiol glucuronide (49).
In the present study, the intracellular localization
of the OATP4A1 variants and their functional properties as
transporters were investigated following transfection of wtOATP4A1
and its variants in the human cancer A431 cell line and Sf9 insect
cells. As predicted from the amino acid sequence of OATP4A1,
immunoreactive bands at ~70 kDa were observed in the western blot
images generated from Sf9 cell homogenates. It was also
demonstrated that the two SNPs did not alter the membrane
localization of the protein or its function as a transporter for
the fluorescent dye sodium fluorescein. Therefore, it can be
hypothesized that other OATP substrates including steroid hormones,
prostaglandins and metabolic products can also be transported in an
analogous manner by wt and variant OATP4A1. CRC recurrence and its
outcome in patients are unlikely to be affected by these
polymorphisms.
The location of the two SNPs in the N-terminal
cytoplasmic domains, which is also the first intracellular loop of
the protein, is distant to the region described to be important for
the transport of OATP. Based on the comparative analysis of OATPs
in multiple species, OATP-mediated transport is considered to occur
through a central, positively charged pore, which is known as being
a typical example of the rocker-switch type of transporters
(4). Previous studies showed that
transmembrane domain 10 is important for initial substrate
recognition and its subsequent translocation across the membrane in
OATP1B1 and OATP1B3 (50). Other
transmembrane domains, such as the extracellular loop 6, have also
been demonstrated to be important for OATP transport function, as
was demonstrated in these 2 families of OATP1 proteins (51).
In conclusion, the results from the present study
demonstrated that the 2 most frequent polymorphisms in the OATP4A1
gene were not associated with an increased predisposition for CRC,
nor did they alter the functional properties of the transporter,
suggesting that the two OATP4A1 variants are unlikely to be risk
factors for CRC development and early recurrence. In the future,
larger studies investigating other variants of OATP4A1 should be
conducted to facilitate the improvement in our understanding of the
association between these transporters and personalized CRC risk
factors.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Erika Bajna
(Institute of Pathophysiology and Allergy Research, Center for
Pathophysiology, Infectiology and Immunology, Medical University of
Vienna, Vienna, Austria) for her technical assistance.
Funding
This study was funded by a grant from the Austrian
Science Fund (FWF) awarded to WJ (grant no. I 3417-B31), the
Anniversary Fund of the Oesterreichische Nationalbank (Austrian
National Bank), project Nr. 17278 awarded to DM, and a grant from
the Hungarian Research, Development and Innovation Office awarded
to CÖ-L (grant no. OTKA FK 128751, CÖL). The work of CÖ-L was also
supported by the János Bolyai Research Scholarship of the Hungarian
Academy of Sciences (2015–2020).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors contributions
TT, CÖL and WJ participated in research design and
supervision of the project. MS and ON conducted the experiments,
and VBA and CA contributed to data analysis. AT and HA performed
genetic analysis. AR and CA provided research samples and clinical
data. AG, AT, DM and HA performed data analysis. VBA, TT, WJ and
CÖL wrote the manuscript with substantial intellectual
contributions from all authors. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was performed following the
Declaration of Helsinki and the Ethical guidelines of the
Institutions summarized in https://www.medunigraz.at/ethikkommission/index_dwnld.html.
The guidelines of the Ethical Committee of the City of Vienna for
retrospective studies (25) (§15a
Abs. 3a des Wiener Krankenanstaltengesetzes) were applied. Informed
consent was provided by the participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Dyba T, Randi G, Bettio M, Gavin A, Visser O and Bray F: Cancer
incidence and mortality patterns in Europe: Estimates for 40
countries and 25 major cancers in 2018. Eur J Cancer. 103:356–387.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kleberg K, Jensen GM, Christensen DP,
Lundh M, Grunnet LG, Knuhtsen S, Poulsen SS, Hansen MB and Bindslev
N: Transporter function and cyclic AMP turnover in normal colonic
mucosa from patients with and without colorectal neoplasia. BMC
Gastroenterol. 12:782012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rawłuszko-Wieczorek AA, Horst N, Horbacka
K, Bandura AS, Świderska M, Krokowicz P and Jagodziński PP: Effect
of DNA methylation profile on OATP3A1 and OATP4A1 transcript levels
in colorectal cancer. Biomed Pharmacother. 74:233–242. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stieger B and Hagenbuch B: Organic
anion-transporting polypeptides. Curr Top Membr. 73:205–232. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hagenbuch B and Stieger B: The SLCO
(former SLC21) superfamily of transporters. Mol Aspects Med.
34:396–412. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Svoboda M, Riha J, Wlcek K, Jaeger W and
Thalhammer T: Organic anion transporting polypeptides (OATPs):
Regulation of expression and function. Curr Drug Metab. 12:139–153.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Obaidat A, Roth M and Hagenbuch B: The
expression and function of organic anion transporting polypeptides
in normal tissues and in cancer. Annu Rev Pharmacol Toxicol.
52:135–151. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
König J: Uptake transporters of the human
OATP family: Molecular characteristics, substrates, their role in
drug-drug interactions, and functional consequences of
polymorphisms. Handb Exp Pharmacol. 1–28. 2011.doi:
10.1007/978-3-642-14541-4_1. View Article : Google Scholar
|
|
9
|
Patik I, Kovacsics D, Német O, Gera M,
Várady G, Stieger B, Hagenbuch B, Szakács G and Özvegy-Laczka C:
Functional expression of the 11 human organic anion transporting
polypeptides in insect cells reveals that sodium fluorescein is a
general OATP substrate. Biochem Pharmacol. 98:649–658. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schuster VL: Prostaglandin transport.
Prostaglandins Other Lipid Mediat. 68-69:633–647. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nguyen TT, Ung TT, Kim NH and Jung YD:
Role of bile acids in colon carcinogenesis. World J Clin Cases.
6:577–588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gilligan LC, Gondal A, Tang V, Hussain MT,
Arvaniti A, Hewitt AM and Foster PA: Estrone sulfate transport and
steroid sulfatase activity in colorectal cancer: Implications for
hormone replacement therapy. Front Pharmacol. 8:1032017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen C, Gong X, Yang X, Shang X, Du Q,
Liao Q, Xie R, Chen Y and Xu J: The roles of estrogen and estrogen
receptors in gastrointestinal disease. Oncol Lett. 18:5673–5680.
2019.PubMed/NCBI
|
|
14
|
Lavasani S, Chlebowski RT, Prentice RL,
Kato I, Wactawski-Wende J, Johnson KC, Young A, Rodabough R,
Hubbell FA, Mahinbakht A and Simon MS: Estrogen and colorectal
cancer incidence and mortality. Cancer. 121:3261–3271. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Buxhofer-Ausch V, Secky L, Wlcek K,
Svoboda M, Kounnis V, Briasoulis E, Tzakos AG, Jaeger W and
Thalhammer T: Tumor-specific expression of organic
anion-transporting polypeptides: Transporters as novel targets for
cancer therapy. J Drug Deliv. 2013:8635392013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Buxhofer-Ausch V, Sheikh M, Ausch C,
Zotter S, Bauer H, Mollik M, Reiner A, Gleiss A, Jäger W, Sebesta
C, et al: Abundance of the Organic Anion-transporting Polypeptide
OATP4A1 in Early-stage colorectal cancer patients: Association with
disease relapse. Appl Immunohistochem Mol Morphol. 27:185–194.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ban MJ, Ji SH, Lee CK, Bae SB, Kim HJ, Ahn
TS, Lee MS, Baek MJ and Jeong D: Solute carrier organic anion
transporter family member 4A1 (SLCO4A1) as a prognosis marker of
colorectal cancer. J Cancer Res Clin Oncol. 143:1437–1447. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee HH and Ho RH: Interindividual and
interethnic variability in drug disposition: Polymorphisms in
organic anion transporting polypeptide 1B1 (OATP1B1; SLCO1B1). Br J
Clin Pharmacol. 83:1176–1184. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kalliokoski A and Niemi M: Impact of OATP
transporters on pharmacokinetics. Br J Pharmacol. 158:693–705.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang B and Lauschke VM: Genetic
variability and population diversity of the human SLCO (OATP)
transporter family. Pharmacol Res. 139:550–559. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee W, Belkhiri A, Lockhart AC, Merchant
N, Glaeser H, Harris EI, Washington MK, Brunt EM, Zaika A, Kim RB
and El-Rifai W: Overexpression of OATP1B3 confers apoptotic
resistance in colon cancer. Cancer Res. 68:10315–10323. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Falkowski S, Woillard JB, Postil D,
Tubiana-Mathieu N, Terrebonne E, Pariente A, Smith D, Guimbaud R,
Thalamas C, Rouguieg-Malki K, et al: Common variants in
glucuronidation enzymes and membrane transporters as potential risk
factors for colorectal cancer: A case control study. BMC Cancer.
17:9012017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Evangeli L, Ioannis S, Valentinos K,
Antigony M, Elli I, Eleftheria H, Vasiliki G and Evangelos B:
SLCO1B3 screening in colorectal cancer patients using
High-Resolution Melting Analysis method and immunohistochemistry.
Tumour Biol. 39:10104283176911762017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
UICC (International Union Against Cancer).
TNM Classification of Malignant Tumors. Sobin LH, Gospodarowicz MK
and Wittekind CH: 7th. Blackwell, Oxford: pp. 252010
|
|
25
|
https://www.ris.bka.gv.at/eli/lgbl/WI/1987/23/P15a/LWI40012343Aug
5–2020
|
|
26
|
Fischer AH, Jacobson KA, Rose J and Zeller
R: Hematoxylin and eosin staining of tissue and cell sections. CSH
Protoc. May 1–2008.(Epub ahead of print). doi:
10.1101/pdb.prot4986.
|
|
27
|
Slatkin M: Linkage
disequilibrium-understanding the evolutionary past and mapping the
medical future. Nat Rev Genet. 9:477–485. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Machiela MJ and Chanock SJ: LDlink: A
web-based application for exploring population-specific haplotype
structure and linking correlated alleles of possible functional
variants. Bioinformatics. 31:3555–3557. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rogojanu R, Thalhammer T, Thiem U, Heindl
A, Mesteri I, Seewald A, Jäger W, Smochina C, Ellinger I and Bises
G: Quantitative image analysis of epithelial and stromal area in
histological sections of colorectal cancer: An emerging diagnostic
tool. BioMed Res Int. 2015:5690712015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Haisan A, Rogojanu R, Croitoru C, Jitaru
D, Tarniceriu C, Danciu M and Carasevici E: Digital microscopy
assessment of angiogenesis in different breast cancer compartments.
BioMed Res Int. 2013:2869022013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kolacsek O, Krízsik V, Schamberger A,
Erdei Z, Apáti A, Várady G, Mátés L, Izsvák Z, Ivics Z, Sarkadi B
and Orbán TI: Reliable transgene-independent method for determining
sleeping beauty transposon copy numbers. Mob DNA. 2:52011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gál C, Hegedüs C, Szakács G, Váradi A,
Sarkadi B and Özvegy-Laczka C: Mutations of the central tyrosines
of putative cholesterol recognition amino acid consensus (CRAC)
sequences modify folding, activity, and sterol-sensing of the human
ABCG2 multidrug transporter. Biochim Biophys Acta. 1848:477–487.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ozvegy C, Váradi A and Sarkadi B:
Characterization of drug transport, ATP hydrolysis, and nucleotide
trapping by the human ABCG2 multidrug transporter. Modulation of
substrate specificity by a point mutation. J Biol Chem.
277:47980–47990. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tschantz WR, Pfeifer ND, Meade CL, Wang L,
Lanzetti A, Kamath AV, Berlioz-Seux F and Hashim MF: Expression,
purification and characterization of the human membrane transporter
protein OATP2B1 from Sf9 insect cells. Protein Expr Purif.
57:163–171. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Patik I, Székely V, Német O, Szepesi Á,
Kucsma N, Várady G, Szakács G, Bakos É and Özvegy-Laczka C:
Identification of novel cell-impermeant fluorescent substrates for
testing the function and drug interaction of Organic
Anion-transporting polypeptides, OATP1B1/1B3 and 2B1. Sci Rep.
8:26302018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tusnády GE and Simon I: The HMMTOP
transmembrane topology prediction server. Bioinformatics.
17:849–850. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hagenbuch B and Meier PJ: The superfamily
of organic anion transporting polypeptides. Biochim Biophys Acta.
1609:1–18. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Thakkar N, Lockhart AC and Lee W: Role of
organic anion-transporting polypeptides (OATPs) in cancer therapy.
AAPS J. 17:535–545. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kovacsics D, Patik I and Özvegy-Laczka C:
The role of organic anion transporting polypeptides in drug
absorption, distribution, excretion and drug-drug interactions.
Expert Opin Drug Metab Toxicol. 13:409–424. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sissung TM, Goey AK, Ley AM, Strope JD and
Figg WD: Pharmacogenetics of membrane transporters: A review of
current approaches. Methods Mol Biol. 1175:91–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Teft WA, Welch S, Lenehan J, Parfitt J,
Choi YH, Winquist E and Kim RB: OATP1B1 and tumour OATP1B3 modulate
exposure, toxicity, and survival after Irinotecan-based
chemotherapy. Br J Cancer. 112:857–865. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rhodes KE, Zhang W, Yang D, Press OA,
Gordon M, Vallböhmer D, Schultheis AM, Lurje G, Ladner RD, Fazzone
W, et al: ABCB1, SLCO1B1 and UGT1A1 gene polymorphisms are
associated with toxicity in metastatic colorectal cancer patients
treated with first-line irinotecan. Drug Metab Lett. 1:23–30. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Roth M, Obaidat A and Hagenbuch B: OATPs,
OATs and OCTs: The organic anion and cation transporters of the
SLCO and SLC22A gene superfamilies. Br J Pharmacol. 165:1260–1287.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Aslibekyan S, Brown EE, Reynolds RJ,
Redden DT, Morgan S, Baggott JE, Sha J, Moreland LW, O'Dell JR,
Curtis JR, et al: Genetic variants associated with methotrexate
efficacy and toxicity in early rheumatoid arthritis: Results from
the treatment of early aggressive rheumatoid arthritis trial.
Pharmacogenomics J. 14:48–53. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Eektimmerman F, Swen JJ, Böhringer S,
Aslibekyan S, Allaart CF and Guchelaar HJ: SLC04A1, SLC22A2 and
SLC28A2 variants not related to methotrexate efficacy or toxicity
in rheumatoid arthritis patients. Pharmacogenomics. 19:613–619.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fairley S, Lowy-Gallego E, Perry E and
Flicek P: The International Genome sample resource (IGSR)
collection of open human genomic variation resources. Nucleic Acids
Res. 48:D941–D947. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Durmus S, Lozano-Mena G, van Esch A,
Wagenaar E, van Tellingen O and Schinkel AH: Preclinical mouse
models to study human OATP1B1- and OATP1B3-mediated drug-drug
interactions in vivo. Mol Pharm. 12:4259–4269. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Alam K, Crowe A, Wang X, Zhang P, Ding K,
Li L and Yue W: Regulation of organic anion transporting
polypeptides (OATP) 1B1- and OATP1B3-mediated transport: An updated
review in the context of OATP-mediated drug-drug interactions. Int
J Mol Sci. 19:E8552018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Crowe A, Zheng W, Miller J, Pahwa S, Alam
K, Fung KM, Rubin E, Yin F, Ding K and Yue W: Characterization of
plasma membrane localization and phosphorylation status of organic
anion transporting polypeptide (OATP) 1B1 c.521 T>C
nonsynonymous single-nucleotide polymorphism. Pharm Res.
36:1012019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gui C and Hagenbuch B: Amino acid residues
in transmembrane domain 10 of organic anion transporting
polypeptide 1B3 are critical for cholecystokinin octapeptide
transport. Biochemistry. 47:9090–9097. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Meier-Abt F, Mokrab Y and Mizuguchi K:
Organic anion transporting polypeptides of the OATP/SLCO
superfamily: Identification of new members in nonmammalian species,
comparative modeling and a potential transport mode. J Membr Biol.
208:213–227. 2005. View Article : Google Scholar : PubMed/NCBI
|