Introduction
Colorectal cancer (CRC) is one of the most commonly
diagnosed malignant tumors worldwide (1,2). The
latest statistics indicate that CRC has the third highest incidence
among all malignant tumors (3).
Moreover, CRC has the second highest fatality rate among all
malignant tumors, and the morbidity and mortality rates of CRC are
continuing to rise (2). Due to the
lack of effective treatments for CRC, more research efforts need to
be focused on determining the molecular mechanisms of its
oncogenesis. With such research, improved antitumor agents and
immunotherapies aimed at novel specific molecular targets could be
developed (4).
Ubiquitin-conjugating enzyme E2T (UBE2T) is a member
of the ubiquitin-conjugating (E2) enzyme family that plays an
important role in the ubiquitin-proteasome pathway. The proteasome
pathway participates in a number of crucial cell activities, such
as the repair of DNA damage, differentiation, apoptosis,
angiogenesis, inflammation and various cancer-associated processes
(5). Together with E3 ubiquitin
ligase, the E2 enzyme family has been reported to serve roles in
various types of cancer, such as breast cancer (6). As a component of the Fanconi anemia
(FA) pathway, UBE2T participates in DNA damage and repair pathways.
These pathways are recognized to be crucial in the tumorigenesis of
lung, breast, prostate, gastric and nasopharyngeal cancer (7–9).
Furthermore, it has been revealed that UBE2T gene silencing can
lead to FA and that this silencing can increase the sensitivity of
tumor cells to cross-linking agents used to target the DNA
damage-repair response (7).
Therefore, UBE2T may have an important role in the maintenance of
genome integrity. However, the role of UBE2T in CRC remains to be
elucidated. Therefore, the aim of the present study was to examine
UBE2T expression in patients with CRC and evaluate its prognostic
significance by identifying associations between its expression and
various clinicopathological parameters of CRC, and to verify the
role of UBE2T in CRC using The Cancer Genome Atlas (TCGA)
database.
Materials and methods
Human tissue samples and cell
lines
For the purpose of this study, a total of 50 paired
surgically resected specimens of fresh colorectal adenocarcinoma
and adjacent normal tissues (>5 cm away from the tumor edge)
were collected from patients between April 2012 and April 2013 in
Shaanxi Provincial People's Hospital (Shaanxi, China). After
surgical resection, the tissues were immediately frozen in liquid
nitrogen and then transferred to a −80°C refrigerator. Samples were
collected from 29 male and 21 female patients, and the diagnoses of
all patients were confirmed by pathological examination. Only
patients who had not received any radiotherapy or chemotherapy
prior to surgery and who had a complete clinical data record were
included in this study. All patients were followed up for ≥5 years
by telephone. All specimens were classified according to the 8th
edition Tumor Node Metastasis (TNM) staging system enacted by The
International Union Against Cancer and American Joint Committee on
Cancer (10). The human normal colon
mucosal cell line NCM460 and human colorectal adenocarcinoma cell
lines SW480, SW620, Caco2, HCT116, HT-29, LoVo and RKO were
purchased from the American Type Culture Collection and maintained
in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin/streptomycin (Thermo Fisher Scientific) at
37°C under 5% CO2 in Xi'an Jiaotong University First
Affiliated Hospital Urology Research Center Laboratory.
Bioinformatics analysis
Gene expression data and corresponding clinical data
were obtained from TCGA database (https://portal.gdc.cancer.gov). The parameters were
set as follows: Primary Site: Colon, rectum; Program: TCGA;
Project: TCGA-COAD, TCGA-READ; Data Category: Transcriptome
profiling; Workflow Type: HTSeq-FPKM. In total, 612 samples (568
tumor samples and 44 normal samples) were obtained by this search,
and all the samples were downloaded in June 2020. The gene
expression data of UBE2T were extracted to perform both non-paired
and paired differential expression analyses between tumor and
normal tissues. Patients were divided into the UBE2T-high and
UBE2T-low groups based on the median of UBE2T expression level.
Logistic regression analysis, as well as univariate and
multivariate Cox regression analyses were performed to investigate
the associations between UBE2T expression and the clinical
characteristics of the patients. Survival analysis was conducted to
explore whether UBE2T expression influences the prognosis of
patients with CRC. Gene Set Enrichment Analysis (GSEA) was
performed to identify the Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathways according to their association with UBE2T
expression using GSEA software v3.0 (Broad Institute). The C2
(c2.cp.kegg.v7.1.symbols.gmt) sub-collection was chosen as the
reference gene set and gene set permutations were performed 1,000
times for the analysis. A false-discovery rate (FDR) q value
<0.05 was considered to indicate a statistically significant
result.
Immunohistochemistry (IHC)
For IHC analysis, fresh CRC and normal adjacent
tissue were fixed in 4% paraformaldehyde solution at room
temperature for 48 h. Next, tissues were embedded in paraffin, and
4-µm tissue slices were mounted on slides, baked, dewaxed with
xylene and hydrated by a gradient ethanol series with 5-min washes
at room temperature as follows: 100, 100, 95, 95, 80, 70 and 50%
ethanol, followed by two washes with ddH2O. The slides
were then placed in citric acid buffer for antigen retrieval,
heated under high-pressure at 120°C for 5 min and then allowed to
cool to room temperature. Endogenous peroxidase activity was
blocked by immersing the samples in 3%
H2O2-methanol for 30 min at room temperature.
To eliminate non-specific staining, slides were incubated with 5%
bovine serum albumin (Beijing Solarbio Science & Technology
Co., Ltd.) at room temperature for 30 min. Next, slides were
incubated with the primary rabbit polyclonal anti-UBE2T antibody
(cat. no. 10105-2-AP; 1:1,000; ProteinTech Group, Inc.) at 4°C
overnight. The next day, slides were incubated with the secondary
biotinylated antibody (cat. no. GK600510; original liquid; Gene
Tech Co., Ltd.) for 1 h at room temperature. Finally, slides were
stained with 3,3′-diaminobenzidine tetrahydrochloride at room
temperature for 25 sec and counterstained with hematoxylin at room
temperature for 5 min. The negative control was prepared by
omitting the primary antibody. The slides were observed by light
microscopy under ×200 magnification.
Semi-quantitative analysis of IHC
results
IHC staining was evaluated independently by two
senior pathologists, who were blinded to the clinical data. The
slides with disagreements were evaluated by a third pathologist,
and the final results were subject to two matching opinions. The
immunoreactive score was calculated based on the percentage of
stained cells as well as the staining intensity. The intensity of
staining was classified as follows: i) 0, no staining; ii) 1, weak
staining; iii) 2, medium staining; iv) 3, strong staining. The area
of staining was graded as 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%)
or 4 (76–100%). Finally, patients were divided into two groups: Low
UBE2T expression group (≤3) and high UBE2T expression group (>3)
according to the sum of the intensity and area scores.
Reverse transcription-quantitative PCR
(RT-qPCR)
To determine mRNA expression levels in the cell
lines, total RNA from the cell cultures was isolated using
TRIzol® reagent (Thermo Fisher Scientific, Inc.),
reverse transcribed to cDNA using the PrimeScript™ RT reagent kit
(Takara Biotechnology Co., Ltd.) for 15 min at 60°C and 5 sec at
85°C, and stored at 4°C. qPCR was performed on a CFX96 Real-Time
PCR Detection system (Bio-Rad Laboratories, Inc.) with SYBR-Green
PCR Master Mix (Takara Biotechnology Co., Ltd.). The primer
sequences used were as follows: UBE2T forward
5′-AGCCACCCCCAGGCATCACA-3′ and reverse,
5′-TCTCCAAGCACCTTTTGGTGGCA-3′; and GAPDH forward,
5′-ACCCAGAAGACTGTGGATGG-3′ and reverse, 5′-TGCTGTAGCCAAATTCGTTG-3′.
The thermocycling conditions were as follows: 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec, and
dissociation at 60°C for 1 min and 95°C for 1 sec. UBE2T mRNA
expression was quantified using the 2−∆∆Cq method and
normalized to the internal reference gene GAPDH (11).
Western blot analysis
For western blotting, cultured cells were washed
with PBS, and total protein was extracted using
radioimmunoprecipitation lysis buffer (Shaanxi Zhonghui Hecai
Biomedical Technology Co., Ltd.) and protease inhibitors. A
bicinchoninic acid assay kit (Beyotime Institute of Biotechnology)
was used to determine the protein concentration. Next, protein
samples (30 µg/lane) were separated via SDS-PAGE on a 12% gel, and
subsequently transferred to a polyvinylidene fluoride membrane
(Immobilon; EMD Millipore). After blocking with 5% non-fat milk at
room temperature for 2 h, membranes were incubated at 4°C overnight
with primary rabbit polyclonal anti-UBE2T (cat. no. 10105-2-A;
1:1,000; ProteinTech Group, Inc.) and anti-GAPDH (cat. no. KC-5G4;
1:10,000; Kang Chen Biotech, Inc.) antibodies. Membranes were then
incubated with horseradish peroxidase-conjugated AffiniPure goat
anti-rabbit IgG (cat. no. ZB-2301; 1:2,000; OriGene Technologies,
Inc.) for 1 h at room temperature. Finally, membranes were washed
in Tris-buffered saline with Tween-20 and protein bands were
detected using the Molecular Imager ChemiDoc XRS system (Bio-Rad
Laboratories, Inc.). The intensity of the UBE2T protein bands was
determined using ImageJ software (v1.52a; National Institutes of
Health), and UBE2T expression was normalized to GAPDH.
Statistical analysis
SPSS 22.0 (IBM Corp.) software and GraphPad Prism
8.0.2 (GraphPad Software, Inc.) were used for statistical analysis.
A two-sided P<0.05 was considered to indicate statistical
significance. The χ2 test or Fisher's exact test was
used to analyze the associations between clinicopathological
characteristics and UBE2T expression. The Kaplan-Meier method and
log-rank test were used to perform survival analyses. The effects
of clinicopathological variables and UBE2T expression on survival
were analyzed by univariate and multivariate analyses using a Cox
proportional hazard regression model. Clinical variables that were
found to be statistically significant (P<0.05) in the univariate
analysis were incorporated into the multivariate analysis. One-way
analysis of variance was used to analyze differences in relative
UBE2T mRNA and protein expression levels among the cell lines (both
P<0.05), followed by Dunnett's multiple comparisons test.
Mann-Whitney U test and Wilcoxon's matched pairs tests were used to
perform non-paired and paired differential expression analyses,
respectively, for tumor and normal tissues.
Results
UBE2T expression and its associations
with clinicopathological characteristics in patients with CRC
The 50 patients included in the study ranged from 30
to 90 years in age, and had a median age at diagnosis of 67 years.
To evaluate UBE2T protein expression in CRC tissues,
paraffin-embedded samples from the 50 patients with CRC were
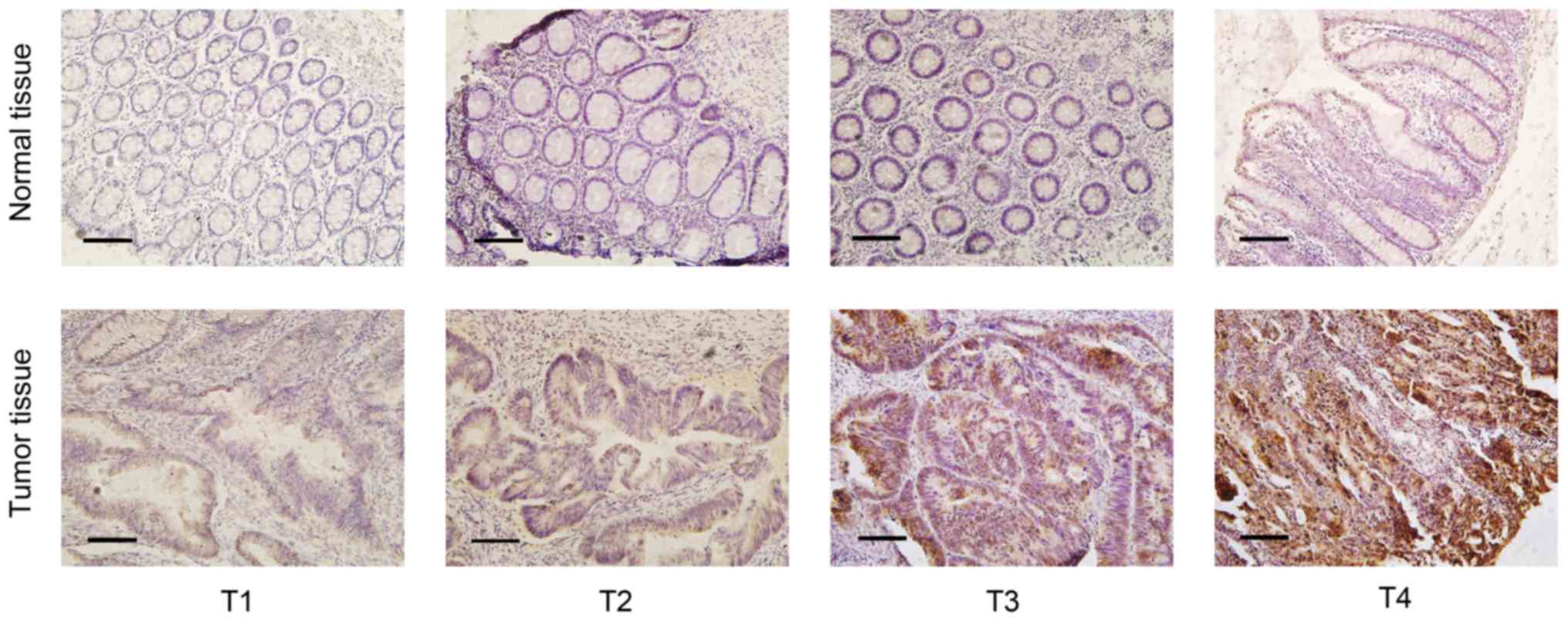
examined by IHC. The results revealed that UBE2T protein was highly
expressed in the cytoplasm of the tumor cells in 29/50 samples and
in 2/50 adjacent normal samples (Fig.
1). The χ2 analysis of the IHC scoring results
revealed that UBE2T expression in CRC tissue was significantly
higher compared with that in adjacent normal tissue (P<0.001).
Moreover, the χ2 or Fisher's exact tests were performed
to evaluate the associations between low and high UBE2T expression
in CRC tissue and clinicopathological parameters of CRC. Based on
this analysis, UBE2T expression was found to be positively
associated with the N classification (P<0.001), clinical TNM
stage (P<0.001) and histological grade (P=0.010). However, no
association was observed between UBET2 protein expression and sex,
age, tumor size, T classification or M classification (Table I).
 | Table I.Associations between UBE2T protein
expression and clinical parameters in patients with CRC. |
Table I.
Associations between UBE2T protein
expression and clinical parameters in patients with CRC.
|
|
| UBE2T
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameters | N | Low | High | P-value |
|---|
| Sex |
|
|
| 0.206 |
|
Male | 29 | 10 | 19 |
|
|
Female | 21 | 11 | 10 |
|
| Age, years |
|
|
| 0.196 |
|
>60 | 33 | 16 | 17 |
|
|
≤60 | 17 | 5 | 12 |
|
| Tumor size, cm |
|
|
| 0.179 |
|
>4 | 23 | 12 | 11 |
|
| ≤4 | 27 | 9 | 18 |
|
| Histological
grade |
|
|
| 0.010a |
|
High | 5 | 5 | 0 |
|
|
Moderate/poor | 45 | 16 | 29 |
|
| T
classification |
|
|
| 0.068 |
|
T1+T2 | 3 | 3 | 0 |
|
|
T3+T4 | 47 | 18 | 29 |
|
| N
classification |
|
|
|
<0.001a |
| N0 | 19 | 15 | 4 |
|
|
N1+N2 | 31 | 6 | 25 |
|
| M
classification |
|
|
| 0.383 |
| M0 | 45 | 20 | 25 |
|
| M1 | 5 | 1 | 4 |
|
| Clinical TNM
stage |
|
|
|
<0.001a |
|
I+II | 18 | 14 | 4 |
|
|
III+IV | 32 | 7 | 25 |
|
Survival analysis of clinical
patients
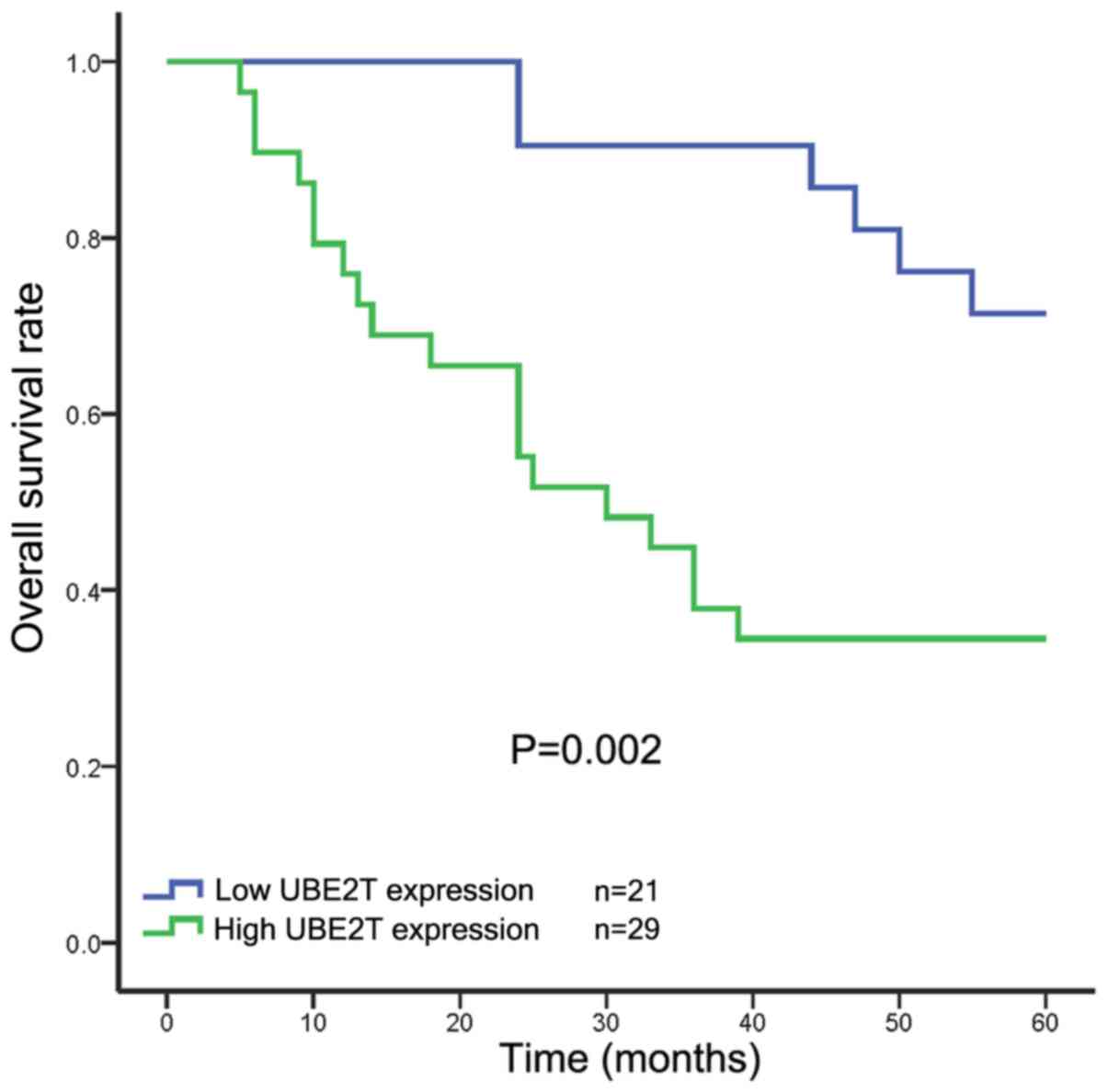
The results of Kaplan-Meier analysis and a log-rank
test indicated that patients with high levels of UBE2T protein
expression in their tumors had a significantly shorter overall
survival time compared with those with low UBE2T expression
(Fig. 2). Additionally, univariate
and multivariate Cox regression analyses were performed to explore
the independent prognostic factors for the overall survival of
patients with CRC. Univariate Cox regression analyses indicated
that UBE2T expression (P=0.006), N classification (P=0.043), M
classification (P=0.012) and clinical TNM stage (P=0.008) were
associated with the overall survival of patients with CRC.
Additionally, multivariate Cox regression analysis suggested that
UBE2T expression (P=0.018) and M classification (P=0.040) were both
independent predictors of overall survival, indicating that UBE2T
protein expression could be useful in the prediction of overall
survival for patients with CRC (Table
II).
 | Table II.Survival analysis of patients with
CRC using univariate and multivariate Cox regression analyses. |
Table II.
Survival analysis of patients with
CRC using univariate and multivariate Cox regression analyses.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinical
parameters | P-value | HR | 95% CI | P-value | HR | 95% CI |
|---|
| Age, years | 0.837 | 0.916 | 0.395 |
|
|
|
| ≤60 vs.
>60 |
|
| 2.122 |
|
|
|
| Sex | 0.786 | 1.117 | 0.502 |
|
|
|
| Male
vs. female |
|
| 2.488 |
|
|
|
| Tumor size, cm | 0.099 | 0.501 | 0.221 |
|
|
|
| ≤4 vs.
>4 |
|
| 1.138 |
|
|
|
| Histological
grade | 0.156 | 1.713 | 0.815 |
|
|
|
|
Poor/moderate vs. high |
|
| 3.603 |
|
|
|
| T
classification | 0.163 | 1.607 | 0.826 |
|
|
|
| T3+T4
vs. T1+T2 |
|
| 3.126 |
|
|
|
| Clinical TNM
stage | 0.008a | 2.310 | 1.238 | 0.604 | 0.735 | 0.229 |
| III+IV
vs. I+II |
|
| 4.309 |
|
| 2.352 |
| N
classification | 0.043a | 1.696 | 1.017 | 0.297 | 1.525 | 0.691 |
| N1+N2
vs. N0 |
|
| 2.827 |
|
| 3.366 |
| M
classification | 0.012a | 4.098 | 1.370 | 0.040a | 8.109 | 1.103 |
| M1 vs.
M0 |
|
| 12.260 |
|
| 59.628 |
| UBE2T
expression | 0.006a | 3.676 | 1.458 | 0.018a | 3.683 | 1.255 |
| High
vs. low |
|
| 9.269 |
|
| 10.813 |
UBE2T mRNA and protein expression
levels are elevated in CRC cell lines
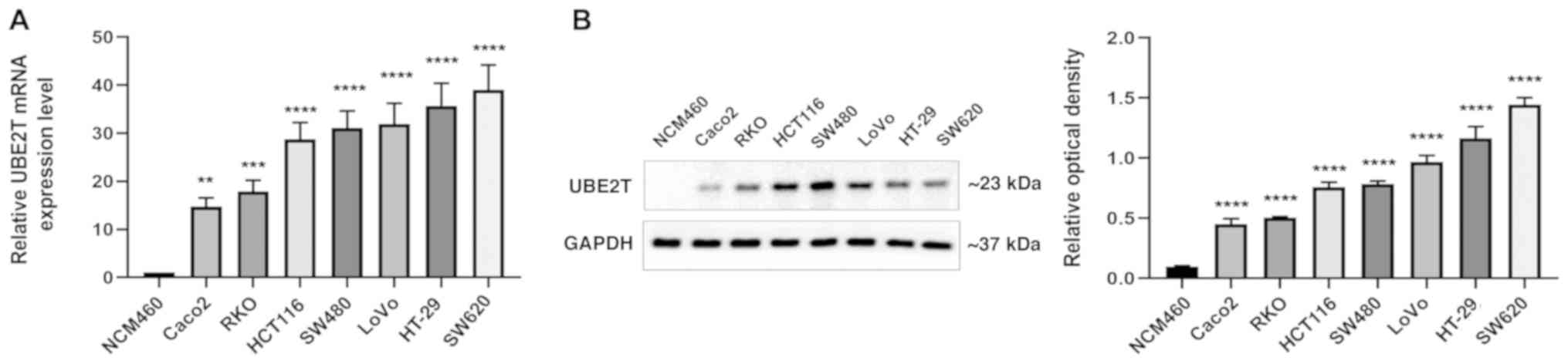
To examine the expression levels of UBE2T mRNA and
protein in normal colon and CRC cell lines, RT-qPCR and western
blotting were performed. The expression of UBE2T was examined in
the CRC cell lines SW480, SW620, Caco2, HCT116, HT-29, LoVo and
RKO, as well as in the normal colonic mucosa cell line NCM460.
UBE2T mRNA and protein expression levels varied among the different
cell lines. In the CRC cell lines, UBE2T expression was observed to
increase in the following order: Caco2, RKO, HCT116, SW480, LoVo,
HT-29 and SW620. Significantly lower UBE2T expression was detected
at both the mRNA and protein levels in the normal colonic mucosa
cell line NCM460 compared with the CRC cell lines (Fig. 3). Therefore, based on the IHC of
patient samples and the RT-qPCR and western blot analysis of cell
lines, it can be concluded that UBE2T is primarily expressed in CRC
tissues and cell lines rather than in normal colorectal tissues and
cells. Based on the UBE2T expression detected in the CRC cell lines
in the present study, SW620 cells were selected for further
experiments, as they exhibited the highest expression of UBE2T.
UBE2T expression and its clinical
significance in CRC in TCGA
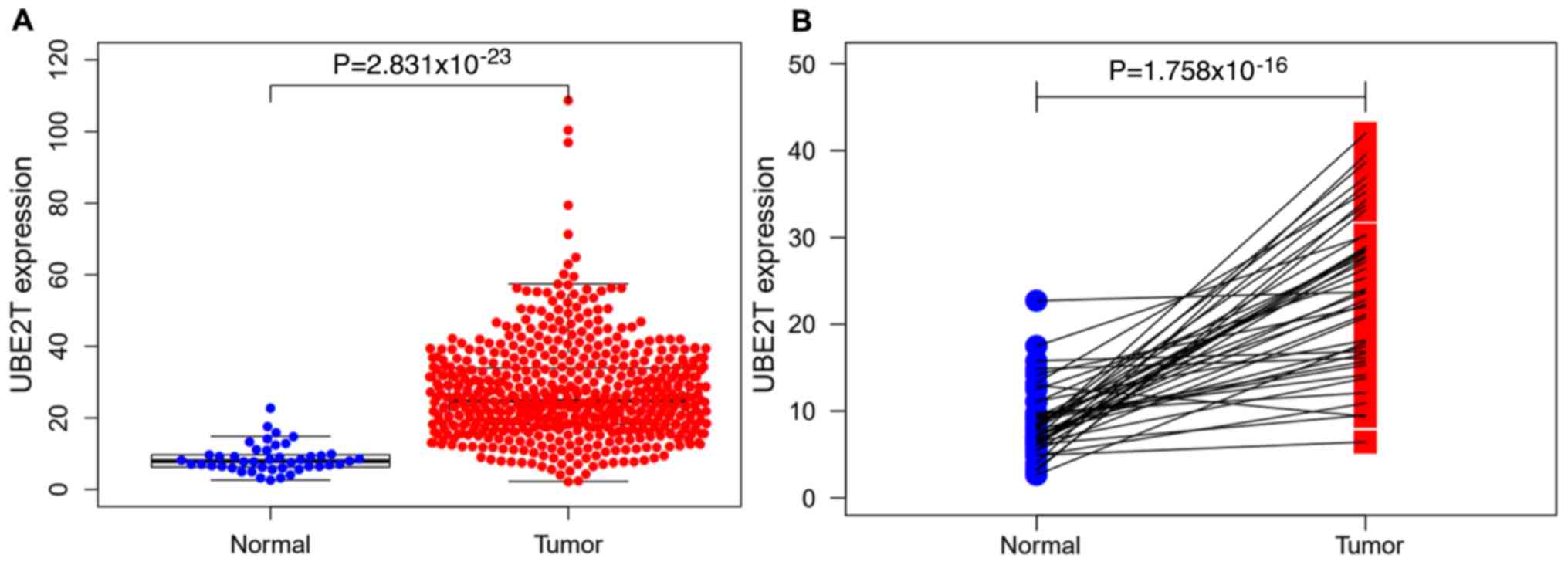
The non-paired differential expression analysis of
the 568 tumor tissues and 44 normal tissues from TCGA indicated
that there was a significant difference in the level of UBE2T
expression between tumor and normal tissues (P<0.001; Fig. 4A). As shown in Fig. 4B, a comparable result was obtained
from the paired differential expression analysis of the tumor and
normal tissues from 44 patients. After removing incomplete clinical
data, the clinical data of 480 patients with CRC and the
corresponding UBE2T expression were subjected to logistic
regression analyses to investigate the potential associations
between them. Among the 480 patients, there were 226 females and
254 males, and the median age at diagnosis was 68 years. As shown
in Table III, UBE2T expression as
a categorical dependent variable (based on a median expression of
25.01) was significantly associated with stage [II vs. I; odds
ratio (OR)=1.024, P=0.029], T classification (T2 vs. T1; OR=0.960,
P=0.030) and lymph node metastasis (N1 vs. N0; OR=0.981,
P=0.043).
 | Table III.Logistic regression analysis of the
association between UBE2T expression and clinicopathological
characteristics in patients with CRC from TCGA. |
Table III.
Logistic regression analysis of the
association between UBE2T expression and clinicopathological
characteristics in patients with CRC from TCGA.
| Clinicopathological
characteristics | OR | 95% CI | P-value |
|---|
| Stage |
| II vs.
I | 1.024 | 1.003–1.046 | 0.029a |
| III vs.
I | 1.008 | 0.984–1.031 | 0.528 |
| IV vs.
I | 1.005 | 0.980–1.032 | 0.682 |
| T
classification |
| T2 vs.
T1 | 0.960 | 0.926–0.996 | 0.030a |
| T3 vs.
T1 | 0.979 | 0.949–1.010 | 0.188 |
| T4 vs.
T1 | 0.968 | 0.932–1.005 | 0.093 |
| N
classification |
| N1 vs.
N0 | 0.981 | 0.964–0.999 | 0.043a |
| N2 vs.
N0 | 0.996 | 0.977–1.015 | 0.667 |
| M
classification |
| M1 vs.
M0 | 0.991 | 0.972–1.011 | 0.373 |
Survival analysis and Cox regression
analysis in TCGA
Kaplan-Meier survival analysis showed that there was
no significant difference in prognosis between patients in the
UBE2T-high and UBE2T-low groups (P=0.238; data not shown).
Furthermore, the same conclusion could be drawn from the results of
the univariate and multivariate Cox regression analyses (Table IV). Excluding the influence of other
confounding factors, the expression level of UBE2T was still not
associated with the prognosis patients with CRC; however, age and T
stage were associated with prognosis.
 | Table IV.Univariate and multivariate analyses
of the associations between UBE2T expression and overall survival
in patients with CRC from TCGA. |
Table IV.
Univariate and multivariate analyses
of the associations between UBE2T expression and overall survival
in patients with CRC from TCGA.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age
(continuous) | 1.036 | 1.014–1.057 | 0.001a | 1.046 | 1.024–1.069 | 0.001a |
| Sex (male vs.
female) | 1.025 | 0.668–1.573 | 0.911 | 0.875 | 0.566–1.352 | 0.546 |
| Stage (IV vs.
I) | 21.291 | 5.117–88.581 | 0.001a | 1.527 | 0.753–3.095 | 0.240 |
| T classification
(T4 vs. T1) | 8.895 | 1.191–66.464 | 0.033a | 1.811 | 1.114–2.943 | 0.017a |
| N classification
(N2 vs. N1) | 4.210 | 2.552–6.944 | 0.001a | 1.208 | 0.785–1.857 | 0.390 |
| M classification
(M1 vs. M0) | 4.698 | 3.029–7.285 | 0.001a | 1.775 | 0.676–4.831 | 0.238 |
| UBE2T
(continuous) | 0.998 | 0.981–1.014 | 0.786 | 1.001 | 0.984–1.017 | 0.952 |
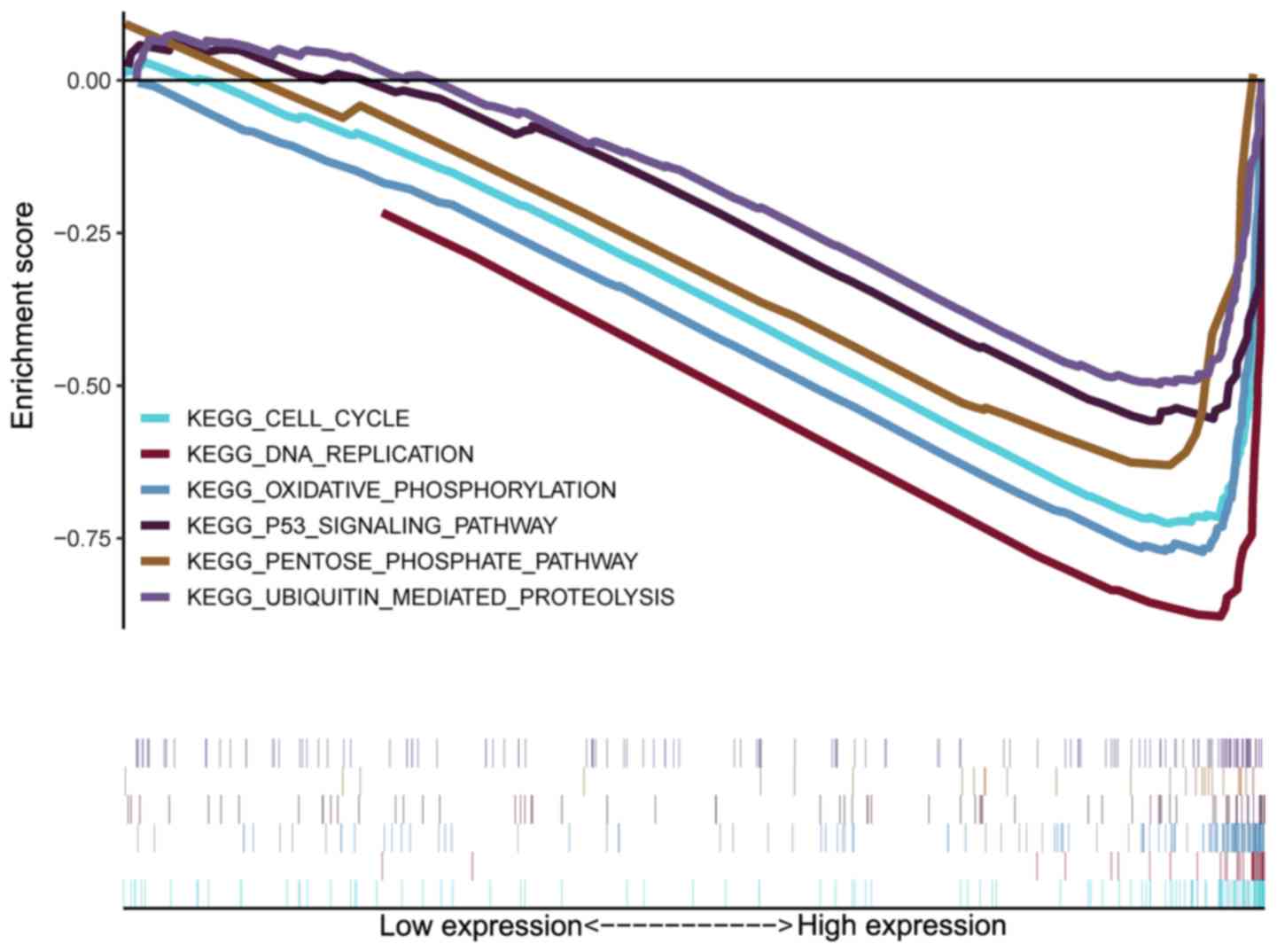
Identification of UBE2T-associated
pathways via GSEA
GSEA was conducted on the UBE2T-high and UBE2T-low
groups to explore the underlying mechanisms of UBE2T in CRC. As
shown in Fig. 5, high expression of
UBE2T was associated with the KEGG pathways ‘cell cycle’,
‘oxidative phosphorylation’, ‘DNA replication’, ‘p53 signaling
pathway’, ‘ubiquitin mediated proteolysis’ and ‘pentose phosphate
pathway’.
Discussion
CRC is one of the most common cancers, and although
great progress has been achieved in terms of its diagnosis and
treatment, the rate of overall survival for patients with advanced
CRC remains low (12). Several tumor
markers are used for the diagnosis of CRC and function as targets
for antitumor therapy, including adenomatous polyposis coli protein
(APC), carcinoembryonic antigen, carbohydrate antigen 199, vascular
endothelial growth factor and p53; however, their sensitivity and
specificity for the diagnosis and prognosis of CRC are not high
(13–15). Hence, the molecular mechanisms
associated with the development and progression of CRC must be
further explored in order to find potentially useful markers for
early CRC diagnosis and disease monitoring (16).
The ubiquitin-proteasome system, which consists of
two E1 ubiquitin-activating enzymes, ~40 E2 enzymes and >600 E3
ubiquitin ligases, plays a crucial role in numerous cellular
activities, most importantly in the degradation of intracellular
proteins (17,18). As a member of the E2 enzyme family,
UBE2T has gained considerable attention for its roles in the
promotion of tumorigenesis and mediation of several signaling
pathways (19). For example,
reported that ubiquitin E2 has been reported to stabilize
β-catenin, one of the key regulators of the Wnt/β-catenin signaling
pathway, by mediating β-catenin polyubiquitination to form
ubiquitin-β-catenin conjugates (6).
In addition, Rickman et al (7) demonstrated that UBE2T is a crucial
member of the FA pathway (20), and
Longerich et al (21)
reported that it mediates the mono-ubiquitination of Fanconi anemia
complementation group L and Fanconi anemia complementation group I.
In addition to its role in the FA pathway, UBE2T has other
important functions in cells (22).
For instance, the interaction of UBE2T with the breast cancer type
1 susceptibility protein (BRCA1)/BRCA1 associated RING domain 1
complex and high expression levels of UBE2T has been demonstrated
to be critical in the progression of breast cancer (23). According to other studies, UBE2T
expression is also upregulated in other types of tumor, including
gastric, lung, prostate, nasopharyngeal and bladder cancer
(24–27). However, few studies have examined the
association between UBE2T and CRC; one study has reported that
UBE2T may promote CRC progression (28). Based on previous reports, we
speculated that the role of UBE2T may be similar in different types
of tumor and that UBE2T could be involved in the development and
progression of CRC.
In the present study, the expression of UBE2T in CRC
tissues and matched adjacent normal tissue samples was examined via
IHC. The results suggested that UBE2T protein expression was higher
in CRC tissues than in the surrounding normal tissue. Additionally,
high expression of UBE2T protein was found to be significantly
associated with high N classification, advanced clinical TNM stage
and tumors of poor histological grade. Data from >5 years of
follow-up of the patients included in the present study showed that
patients with high UBE2T expression were more likely to have a
worse overall survival outcome. Moreover, Cox proportional hazard
regression analysis indicated that M classification and UBE2T
expression were independent prognostic factors for CRC. In addition
to UBE2T protein expression in patient tissue, UBE2T expression in
various CRC cell lines and a normal colorectal cell line was also
examined at the mRNA and protein levels. This in vitro
analysis revealed that UBE2T was upregulated in CRC cells at the
mRNA and protein levels compared with its expression in normal
colorectal cells. TCGA dataset analysis revealed that UBE2T was
highly expressed in tumors compared with normal samples, which was
consistent with the results from patient tissues. However, UBE2T
was not associated with CRC prognosis in TCGA data. GSEA analysis
revealed that high expression of UBE2T was associated with ‘cell
cycle’ and ‘DNA replication’, the abnormalities in which have been
reported to be involved in oncogenesis (29,30).
Oxidative phosphorylation may be involved in cancer progression
(31), which is consistent with the
GSEA results of the present study. In addition, the p53 signaling
pathway is involved in the development of CRC and can alter the
risk of CRC (32). Additionally,
aberrant ubiquitin-mediated proteolysis of proteins, especially
cell cycle regulatory proteins, may promote tumorigenesis (33). During tumorigenesis, the role of the
elevated pentose phosphate pathway is protection from cell death
(34), which also supports the GSEA
results obtained in the present study.
In summary, the present study demonstrated the
upregulation of UBE2T expression in CRC and its association with
patient prognosis, which is consistent with previous studies, thus
suggesting that UBE2T may have a similar role in various types of
cancer, such as breast cancer, hepatoma and cervical carcinoma
(35). Based on the clinical
results, it may be concluded that UBE2T is a valuable prognostic
marker for CRC and could provide a novel direction for the
development of targeted therapeutic strategies. However, there are
limitations of this study, which may be addressed by increasing the
clinical sample size, and examining the role of UBE2T in CRC
tumorigenesis and the underlying associations between UBE2T and
downstream molecules in further studies.
Acknowledgements
Not applicable.
Funding
This study was supported by the grants from the
Innovation Capability Support Plan of Shaanxi Science and
Technology Department-Science and Technology Innovation Team (grant
no. 2020TD-048) and 2018 Shaanxi Province Health research fund
program (grant no. 2018A003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GW conceived the experiments. XW and GL designed the
experiments. JQ prepared the samples. NC, JQ and RL performed the
experiments. JB and JH processed and analyzed the data. XW and JH
wrote the manuscript with input from all authors. HZ and TL
performed the bioinformatics analysis and article revision. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from all patients
involved in this study, which was conducted according to the
guidelines and with the approval of the Medical Ethics Committee of
Shaanxi Provincial People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Goding SA, Fedewa
SA, Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal A:
Colorectal cancer statistics, 2020. CA Cancer J Clin. 70:145–164.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Custodio A and Feliu J: Prognostic and
predictive biomarkers for epidermal growth factor receptor-targeted
therapy in colorectal cancer: Beyond KRAS mutations. Crit Rev Oncol
Hematol. 85:45–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lim KH, Song MH and Baek KH: Decision for
cell fate: Deubiquitinating enzymes in cell cycle checkpoint. Cell
Mol Life Sci. 73:1439–1455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou MJ, Chen FZ and Chen HC:
Ubiquitination involved enzymes and cancer. Med Oncol. 31:932014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rickman KA, Lach FP, Abhyankar A, Donovan
FX, Sanborn EM, Kennedy JA, Sougnez C, Gabriel SB, Elemento O,
Chandrasekharappa SC, et al: Deficiency of UBE2T, the E2 ubiquitin
ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T
subtype of fanconi anemia. Cell Rep. 12:35–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alpi A, Langevin F, Mosedale G, Machida
YJ, Dutta A and Patel KJ: UBE2T, the Fanconi anemia core complex,
and FANCD2 are recruited independently to chromatin: A basis for
the regulation of FANCD2 monoubiquitination. Mol Cell Biol.
27:8421–8430. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Joenje H and Patel KJ: The emerging
genetic and molecular basis of Fanconi anaemia. Nat Rev Genet.
2:446–457. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weiser MR: AJCC 8th edition: Colorectal
cancer. Ann Surg Oncol. 25:1454–1455. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee YT, Tan YJ and Oon CE: Molecular
targeted therapy: Treating cancer with specificity. Eur J
Pharmacol. 834:188–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nikolaou S, Qiu S, Fiorentino F, Rasheed
S, Tekkis P and Kontovounisios C: Systematic review of blood
diagnostic markers in colorectal cancer. Tech Coloproctol.
22:481–498. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bhatti I, Patel M, Dennison AR, Thomas MW
and Garcea G: Utility of postoperative CEA for surveillance of
recurrence after resection of primary colorectal cancer. Int J
Surg. 16:123–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsilimigras DI, Ntanasis-Stathopoulos I,
Bagante F, Moris D, Cloyd J, Spartalis E and Pawlik TM: Clinical
significance and prognostic relevance of KRAS, BRAF, PI3K and TP53
genetic mutation analysis for resectable and unresectable
colorectal liver metastases: A systematic review of the current
evidence. Surg Oncol. 27:280–288. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Raskov H, Pommergaard HC, Burcharth J and
Rosenberg J: Colorectal carcinogenesis-update and perspectives.
World J Gastroenterol. 20:18151–18164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bassermann F, Eichner R and Pagano M: The
ubiquitin proteasome system-implications for cell cycle control and
the targeted treatment of cancer. Biochim Biophys Acta.
1843:150–162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Teixeira LK and Reed SI: Ubiquitin ligases
and cell cycle control. Annu Rev Biochem. 82:387–414. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Alpi AF, Chaugule V and Walden H:
Mechanism and disease association of E2-conjugating enzymes:
Lessons from UBE2T and UBE2L3. Biochem J. 473:3401–3419. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hira A, Yoshida K, Sato K, Okuno Y,
Shiraishi Y, Chiba K, Tanaka H, Miyano S, Shimamoto A, Tahara H, et
al: Mutations in the gene encoding the E2 conjugating enzyme UBE2T
cause Fanconi anemia. Am J Hum Genet. 96:1001–1007. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Longerich S, San Filippo J, Liu D and Sung
P: FANCI binds branched DNA and is monoubiquitinated by
UBE2T-FANCL. J Biol Chem. 284:23182–23186. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kelsall IR, Langenick J, MacKay C, Patel
KJ and Alpi AF: The Fanconi anaemia components UBE2T and FANCM are
functionally linked to nucleotide excision repair. PLoS One.
7:e369702012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ueki T, Park JH, Nishidate T, Kijima K,
Hirata K, Nakamura Y and Katagiri T: Ubiquitination and
downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T
overexpression in human breast cancer cells. Cancer Res.
69:8752–8760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gong YQ, Peng D, Ning XH, Yang XY, Li XS,
Zhou LQ and Guo YL: UBE2T silencing suppresses proliferation and
induces cell cycle arrest and apoptosis in bladder cancer cells.
Oncol Lett. 12:4485–4492. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hao J, Xu A, Xie X, Hao J, Tian T, Gao S,
Xiao X and He D: Elevated expression of UBE2T in lung cancer tumors
and cell lines. Tumour Biol. 29:195–203. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu H, Xiang P, Pan Q, Huang Y, Xie N and
Zhu W: Ubiquitin-conjugating enzyme E2T is an independent
prognostic factor and promotes gastric cancer progression. Tumour
Biol. 37:11723–11732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu W, Xiao L, Cao C, Hua S and Wu D: UBE2T
promotes nasopharyngeal carcinoma cell proliferation, invasion, and
metastasis by activating the AKT/GSK3β/β-catenin pathway.
Oncotarget. 7:15161–15172. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu M, Li X, Huang W, Chen Y, Wang B and
Liu X: Ubiquitin-conjugating enzyme E2T(UBE2T) promotes colorectal
cancer progression by facilitating ubiquitination and degradation
of p53. Clin Res Hepatol Gastroenterol. Jul 29–2020.(Online ahead
of print). View Article : Google Scholar
|
|
29
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kastan MB and Bartek J: Cell-cycle
checkpoints and cancer. Nature. 432:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sica V, Bravo-San Pedro JM, Stoll G and
Kroemer G: Oxidative phosphorylation as a potential therapeutic
target for cancer therapy. Int J Cancer. 146:10–17. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Slattery ML, Mullany LE, Wolff RK, Sakoda
LC, Samowitz WS and Herrick JS: The p53-signaling pathway and
colorectal cancer: Interactions between downstream p53 target genes
and miRNAs. Genomics. 111:762–771. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bashir T and Pagano M: Aberrant
ubiquitin-mediated proteolysis of cell cycle regulatory proteins
and oncogenesis. Adv Cancer Res. 88:101–144. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Patra KC and Hay N: The pentose phosphate
pathway and cancer. Trends Biochem Sci. 39:347–354. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mamrak NE, Shimamura A and Howlett NG:
Recent discoveries in the molecular pathogenesis of the inherited
bone marrow failure syndrome Fanconi anemia. Blood Rev. 31:93–99.
2017. View Article : Google Scholar : PubMed/NCBI
|