Introduction
Thyroid cancer is the most common thyroid
malignancy, accounting for 2.6% of all malignant tumors (1). The majority of thyroid cancer types
originate from follicular epithelial cells, and are mainly divided
into papillary carcinoma and follicular adenocarcinoma, according
to their pathological subtypes (2).
Available treatment methods exist, such as surgical treatment
(3), thyroid hormone suppression
therapy (4) and radioiodine ablation
(5) that may effectively inhibit
tumors. However, secondary recurrence is common (6). Thus, it is important to identify
effective biomarkers and targets to improve the survival rate of
patients with thyroid cancer.
Circular RNAs (circRNAs) are non-coding RNAs that
play important roles in the development of several diseases, such
as cardiovascular disease, nervous system disease and cancer, as
well as regulate specific cellular functions, including
development, proliferation and invasion (7,8). Lee
et al (7) demonstrated the
roles of circRNAs in the development of human diseases.
Furthermore, Ma et al (8)
revealed the potential regulatory role of circRNA-000284 in
cervical cancer cell proliferation and invasion by targeting
microRNA (miRNA/miR)-506. Previous studies have reported that
circRNAs contain several miRNA binding sites, which are involved in
the regulation of gene expressions by sponging miRNAs (9,10).
circRNA_0000285 has been identified as a novel circRNA, which
promotes the metastasis and invasion of several types of cancer by
targeting specific miRNAs (11).
However, the roles and underlying molecular mechanism of
circRNA_0000285 in thyroid cancer remain unclear.
miRNAs are a class of endogenous small RNAs with
vital regulatory roles in cells (12). It has been reported that miRNAs are
promising markers of several biological processes, including cell
proliferation, migration and invasion (13). For example, miR-211 triggers an
autophagy-dependent apoptotic response in cervical cancer cells by
regulating B-cell lymphoma 2 (Bcl-2) (14). Furthermore, miR-654-3p is a key
regulator of cell proliferation and migration that promotes the
induction of gene reprogramming (15). miR-654-3p is a tumor inhibitory
factor that is downregulated in hepatocellular carcinoma (16). Pu et al (17) highlighted that miR-654-3p suppresses
non-small cell lung cancer tumorigenesis by inhibiting polo like
kinase 4. However, whether miR-654-3p is associated with the
development of thyroid cancer remains unknown. Notably, a previous
study revealed the interaction sites between circRNA_0000285 and
miR-654-3p, and circRNA_0000285 functions as a sponge of miR-654-3p
in diabetic nephropathy (18).
However, the association between circRNA_0000285 and miR-654-3p in
thyroid cancer has not been studied till now.
The present study aimed to investigate the roles and
underlying molecular mechanism of circRNA_0000285, and determine
the association between circRNA_0000285 and miR-654-3p in thyroid
cancer to identify novel strategies for thyroid cancer
treatment.
Materials and methods
Cell culture
The cell lines, TPC-1, FTC133 and Nthy-ori 3-1, were
purchased from the American Type Culture Collection and maintained
in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (HyClone; Cytiva) and 1%
penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.), at
37°C with 5% CO2.
Dual-luciferase reporter assay
StarBase version 2.0 (http://starbase.sysu.edu.cn/index.php) was used to
predict the binding sites between circRNA_0000285 and miR-654-3p.
The dual-luciferase reporter assay was performed to confirm the
binding sites between circRNA_0000285 and miR-654-3p. The wild-type
3′-untranslated region (UTR) of circRNA_0000285 (WT circ_0000285:
5′-GGUGAUGCUUUUCAGCAGACAUU-3′), which contains the miR-654-3p
binding site or the mutated target site (MUT circ_0000285:
5′-GGUGAUGCUUUUCAGGUCUGUAU-3′) was generated by PCR (using a
Transcriptor First Strand cDNA Synthesis kit (cat. no. 04896866001;
Roche Molecular Diagnostics) (incubation for 5 min at 25°C followed
by 60 min at 42°C) from total RNA preps extracted from TPC-1 cells
(electrophoresis analysis, 1% agarose gel; visualisation method,
ethidium bromide) and cloned into the pMIR vectors (Ambion; Thermo
Fisher Scientific, Inc.) to construct the reporter vector
circRNA_0000285 wild-type or the circRNA_0000285 mutant-type. 293T
cells (American Type Culture Collection) were transfected with
circRNA_0000285 wild-type or the mutant sequence, and with
miR-654-3p mimic (forward, 5′-UGGUGGGCCGCAGAACAUGUGC-3′ and
reverse, 5′-ACAUGUUCUGCGGCCCACGAAU-3′) or mimic control (forward,
5′-UUCUCCGAACGUGUCACGUUU-3′ and reverse,
5′-ACGUGACACGUUCGGAGAAUU-3′; both purchased from Shanghai
GenePharma, Co., Ltd.) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48 h,
according to the manufacturer's instruction. Following cell
transfection, 48 h later the Dual-Luciferase Reporter Assay system
(Promega Corporation) was applied to measure luciferase activity
and Renilla luciferase was used as a normalization
control.
RNA pull-down assay
The pull-down assay was performed as previously
described (19). Briefly, the
biotinylated circRNA_0000285 probe and oligo probe (Shanghai
GenePharma Co., Ltd.) were incubated with M-280 Streptavidin
magnetic beads (cat. no. 60210; Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instruction for 2
h at room temperature to generate probe-coated beads. Subsequently,
the biotinylated RNA coated beads were separated with a magnet for
3 min and washed with 1X B&W Buffer [2X B&W Buffer: 10 mM
Tris-HCl (pH 7.5) 1 mM EDTA 2 M NaCl]. TPC-1 and FTC133 cells
(1×107 cells) were harvested, lysed in 500 µl lysis
buffer (Beyotime Institute of Biotechnology), sonicated and
incubated with probe-coated beads overnight at 4°C. After washing
with 1X B&W Buffer, the RNA complexes bound to the beads were
eluted and extracted using the RNA isolation kit (Thermo Fisher
Scientific, Inc.), and analyzed via reverse
transcription-quantitative (RT-q)PCR analysis.
The pull-down efficiency was verified in TPC-1 and
FTC133 cells transfected with circRNA_0000285 (human
circRNA_0000285 cDNA was synthesized and cloned into the
plenti-ciR-GFP-T2A vector) or vector (plenti-ciR-GFP-T2A vector;
IGE Biotech Co., Ltd.) using Lipofectamine RNAiMax®
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instruction.
RNA immunoprecipitation (RIP)
assay
The RIP assay is a powerful tool used to assess the
binding of RNA molecules to proteins in cells and to assess the
dynamic process of post transcriptional regulatory networks
(20). Thyroid cancer cells were
transfected with miR-654-3p mimic and mimic control at 37°C for 48
h, and were lysed using RIP buffer (Beyotime Institute of
Biotechnology). The magnetic beads were coated with anti-Ago2
(1:50; cat. no. ab186733; Abcam) or IgG (1:300; cat. no. ab109489;
Abcam) antibodies and the cell lysate was incubated with the
magnetic beads at 4°C for 1 h, according to the manufacturer's
instructions.
RT-qPCR
RNA was extracted from thyroid cancer cells and
normal thyroid cells using the RNA isolation kit (Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
cDNA was synthesized using the PrimeScript RT kit (Takara Bio,
Inc.) and amplified via qPCR on an ABI PRISM 7900 sequence
detection system (Applied Biosystems; Thermo Fisher Scientific,
Inc.), with SYBR Premix Ex-Taq (Takara Bio, Inc.). The
thermocycling conditions were as follows: Initial denaturation at
95°C for 5 min, followed by 38 cycles of denaturation at 95°C for
15 sec and annealing/elongation at 60°C for 30 sec. The following
primer sequences (Sangon Biotech Co., Ltd.) were used for qPCR: U6
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′; GAPDH forward,
5′-TCAACGACCACTTTGTCAAGCTCA-3′ and reverse,
5′-GCTGGTGGTCCAGGGGTCTTACT-3′; miR-654-3p forward,
5′-GGGATGTCTGCTGACCA-3′ and reverse, 5′-CAGTGCGTGTCGTGGA-3′; and
circRNA_0000285 forward, 5′-TACCTCTGCAGGCAGGAACT-3′ and reverse,
5′-TCACATGAATTTAGGTGGGACTT-3′. Relative expression levels were
calculated using the 2−ΔΔCq method (21).
Cell transfection
Control-small interfering (si)RNA (1 µg,
5′-AAGACAUUGUGUGUCCGCCTT-3′), circRNA_0000285-siRNA (1 µg,
5′-CCCCAGCUAUUCAAGUGUAAA−3′), inhibitor control (50 nM,
5′-AAGUCAGGUGAUGGACAGCAUA-3′) and miR-654-3p inhibitor (50 nM,
5′-AAGGUGAUGGUCAGCAGACAUA-3′) (all purchased from Shanghai
GenePharma Co., Ltd.) were transfected into TPC-1 and FTC133 cells
using Lipofectamine 2000® (Thermo Fisher Scientific,
Inc.) at 37°C for 48 h, according to the manufacturer's
instructions. After 48 h, RT-qPCR analysis was performed to assess
the efficiency of transfection.
MTT assay
Following transfection, TPC-1 and FTC133 cells were
cultured for 48 h and seeded into 96-well plates. The plates were
incubated for 24, 48 and 72 h at 37°C. Subsequently, cells were
treated with 10 µl MTT (5 mg/ml) solution and incubated for an
additional 4 h. Following treatment, the solution was removed and
100 µl DMSO was added to each well to dissolve the formazan
product. Samples were vibrated using a Microplate Shaker (Thermo
Fisher Scientific, Inc.) and optical density was measured at a
wavelength of 570 nm, using a multifunctional plate reader (BioTek
Instruments, Inc.), according to the manufacturer's
instructions.
Flow cytometry
Following incubation of transfected TPC-1 and FTC133
cells for 48 h, the induction of apoptosis was measured using the
Annexin V-fluorescein isothiocyanate/PI apoptosis detection kit (BD
Biosciences), according to the manufacturer's instructions. Flow
cytometry was performed to detect cell apoptosis (early apoptotic
cells + late apoptotic cells; upper right quadrant + lower right
quadrant) and the results were analyzed using Kaluza Analysis
(version 2.1.1.20653; Beckman Coulter, Inc.).
Western blotting
Following transfection, TPC-1 and FTC133 cells were
incubated for 48 h and total proteins were extracted using RIPA
buffer (Invitrogen; Thermo Fisher Scientific, Inc.). BCA assay
(Thermo Fisher Scientific, Inc.) was used to measure the protein
concentrations. Equal amounts of proteins (40 µg/lane) were
separated via 12% SDS-PAGE, transferred onto PVDF membranes and
blocked with 5% skimmed milk in PBS-0.1% Tween-20 at room
temperature for 1.5 h. The membranes were incubated with primary
antibodies against Bcl-2 (cat. no. 4223), Bax (cat. no. 5023) or
GAPDH (cat. no. 5174) (all 1:1,000 and purchased from Cell
Signaling Technology, Inc.) at 4°C overnight. Following the primary
incubation, membranes were washed with PBST 3 times and incubated
with anti-rabbit IgG horseradish peroxidase-linked antibody (cat.
no. 7074; 1:2,000; Cell Signaling Technology, Inc.) at room
temperature for 1.5 h. Protein bands were visualized using ECL
detection system reagents (MilliporeSigma), according to the
manufacturer's instructions. ImageJ v.2.0 software (National
Institutes of Health) was used to quantify the band intensity.
Statistical analysis
Statistical analysis was performed using SPSS 20.0
software (IBM Corp.). All experiments were performed in triplicate
and data are presented as the mean ± SD. Unpaired Student's t-test
was used to compare differences between two groups, while one-way
ANOVA followed by Tukey's post hoc test were used to compare
differences between multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-654-3p directly interacts with
circRNA_0000285
Previous studies have reported that circRNAs are
involved in cancer development by regulating the expression of
specific target genes (8,11). Initially, bioinformatics analysis was
performed to predict the target gene of circRNA_0000285. miR-654-3p
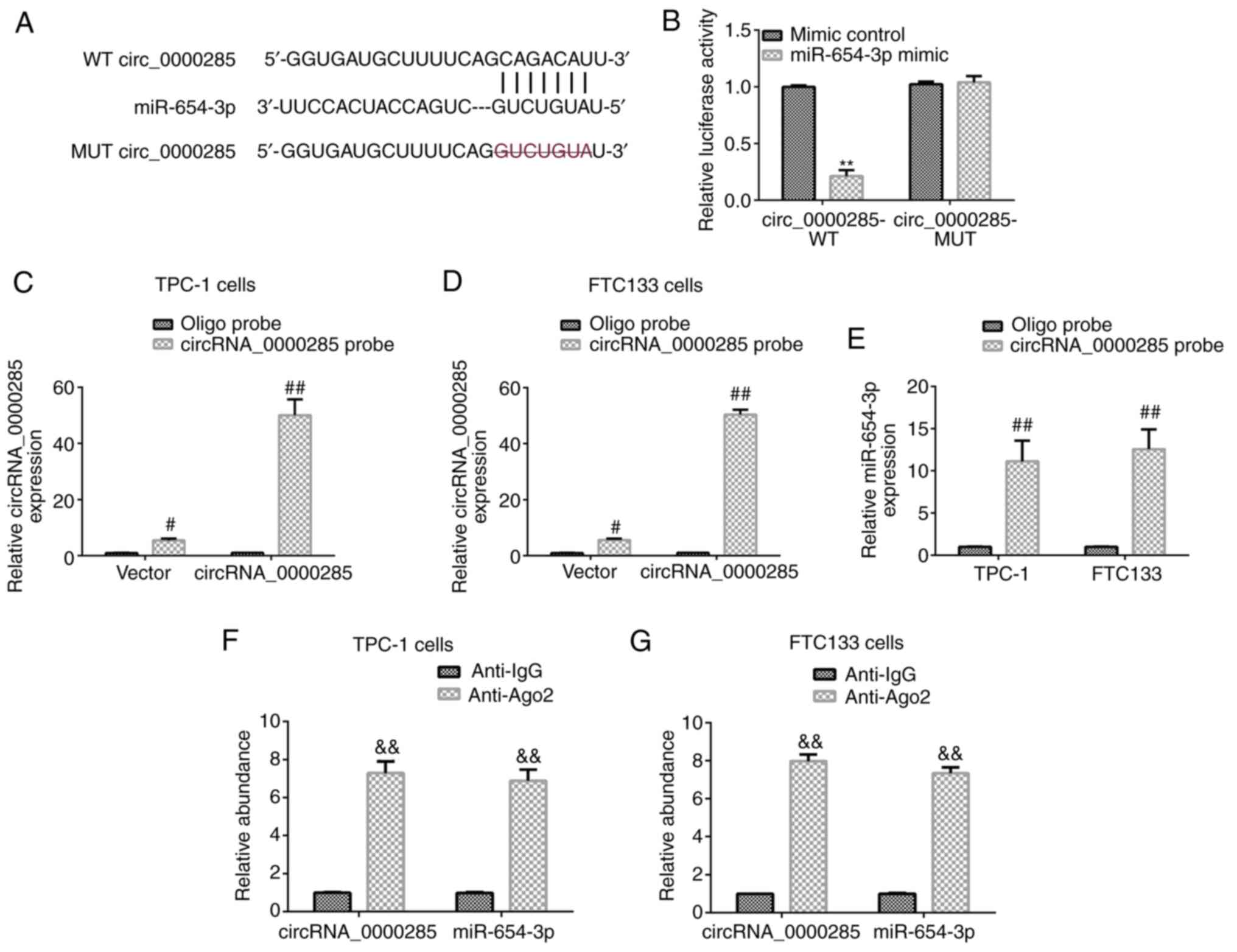
was revealed to be a latent target of circRNA_0000285 (Fig. 1A). Subsequently, the association
between circRNA_0000285 and miR-654-3p was confirmed via the
dual-luciferase reporter assay (Fig.
1B). Compared with the cells co-transfected with
circRNA_0000285 wild-type and mimic control, the luciferase
activity of cells co-transfected with circRNA_0000285 wild-type and
miR-654-3p mimic were significantly reduced (P<0.01; Fig. 1B). While no significant changes were
observed of the luciferase activity in cells co-transfected with
circRNA_0000285 wild-type and mimic control and cells
co-transfected with circRNA_0000285 wild-type and miR-654-3p mimic
(Fig. 1B). The RNA pull-down assay
was performed to confirm the binding between circRNA_0000285 and
miR-654-3p, and the pull-down efficiency was verified in TPC-1 and
FTC133 cells transfected with circRNA_0000285 or vector (Fig. 1C and D). Compared with Oligo probe,
circRNA_0000285 probe significantly enhanced circRNA_0000285 level
in TPC-1 and FTC133 cells (all P<0.01; Fig. 1C and D). miR-654-3p pulled down by
biotinylated probes were purified and analyzed via RT-qPCR
analysis. The results demonstrated that miR-654-3p was notably
pulled down by circRNA_0000285 probe in both TPC-1 and FTC133 cells
(Fig. 1E). The results of the RIP
assay (Fig. 1F and G) verified that
circRNA_0000285 can directly target miR-654-3p, evidenced by
significant enhancement of circRNA_0000285 and miR-654-3p in the
Anti-Ago2 group in TPC-1 and FTC133 cells compared with in the (all
P<0.01; Fig. 1F and G). Taken
together, these results suggest that circRNA_0000285 is a direct
target of miR-654-3p.
circRNA_0000285 expression is
upregulated and miR-654-3p expression is downregulated in thyroid
carcinoma cells
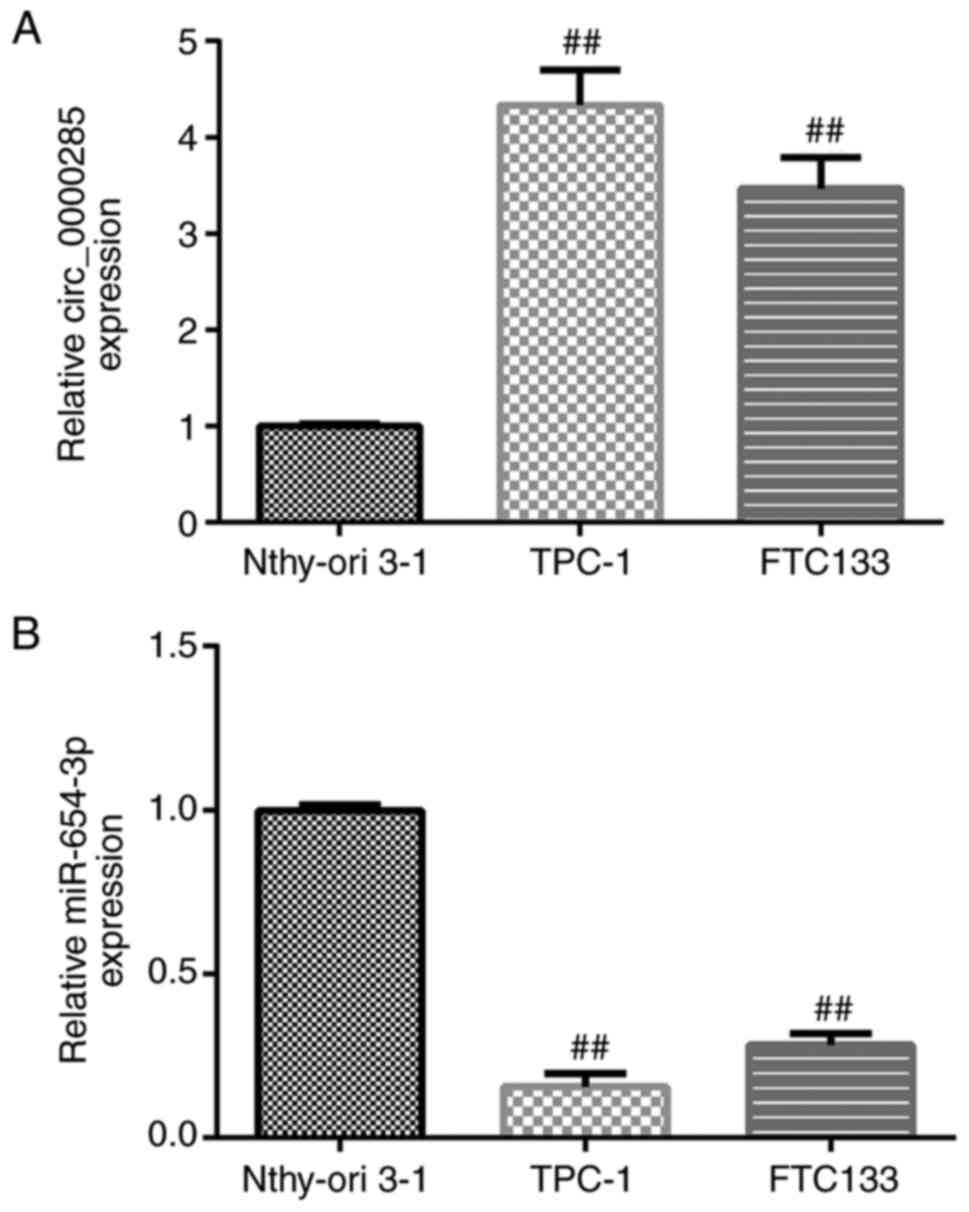
The expression levels of circRNA_0000285 and
miR-654-3p in thyroid cancer cell lines were detected via RT-qPCR
analysis. circRNA_0000285 expression was significantly upregulated
(P<0.01; Fig. 2A) and miR-654-3p
expression was significantly downregulated (P<0.01; Fig. 2B) in the thyroid cancer cell lines
(TPC-1 and FTC133) compared with Nthy-ori 3-1 cells.
circRNA_0000285 negatively regulates
miR-654-3p expression in thyroid cancer cells
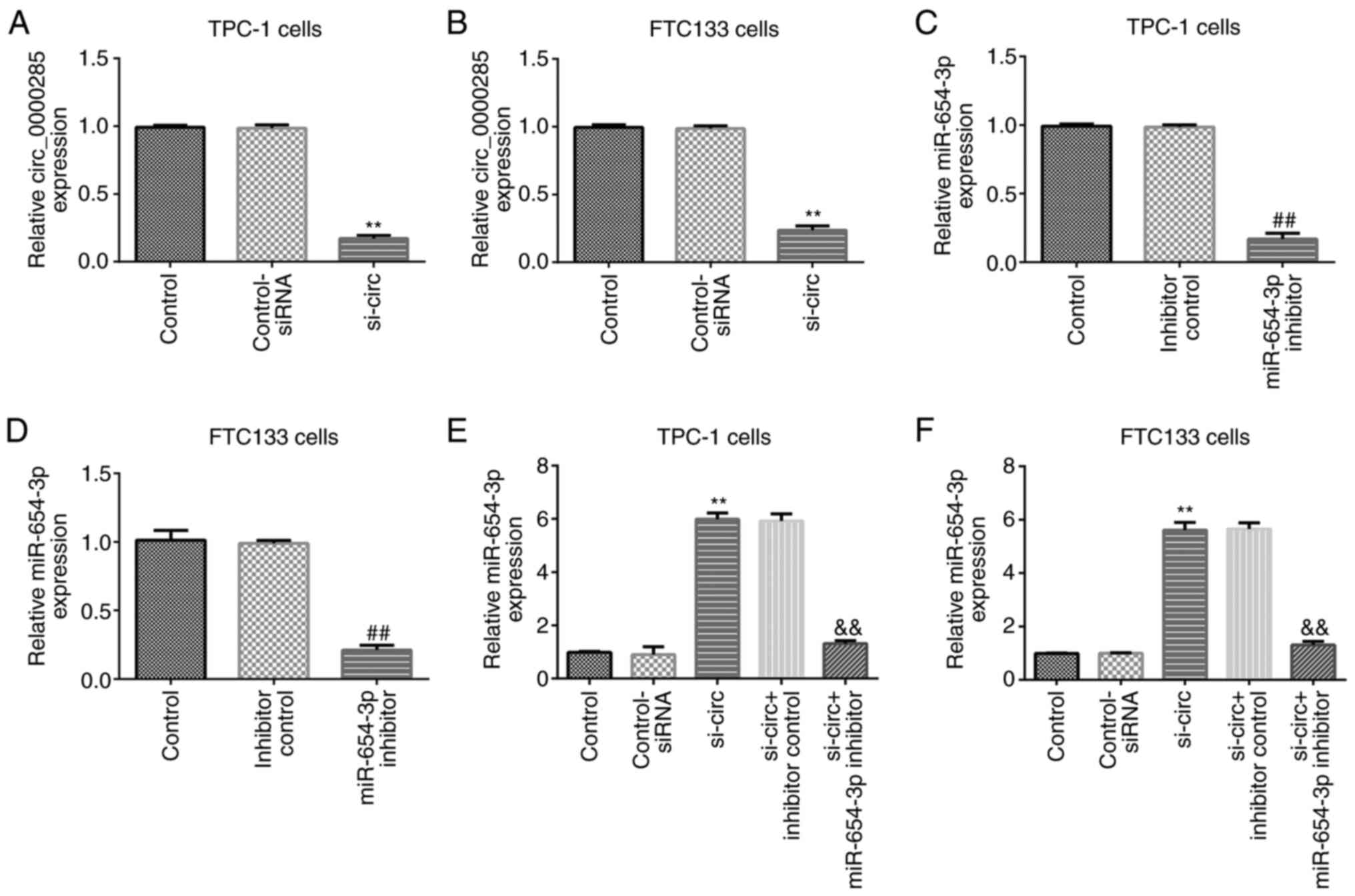
To verify the role of circRNA_0000285 in thyroid
cancer, control-siRNA, circRNA_0000285-siRNA, inhibitor control and
miR-654-3p inhibitor sequences were transfected into TPC-1 and
FTC133 cells for 48 h. RT-qPCR analysis demonstrated that
circRNA_0000285 expression significantly decreased in TPC-1 and
FTC133 cells following transfection with circRNA_0000285-siRNA
compared with the control-siRNA group (P<0.01; Fig. 3A and B). In addition, miR-654-3p
expression significantly decreased in TPC-1 and FTC133 cells
transfected with miR-654-3p inhibitor compared with the inhibitor
control group (P<0.01; Fig. 3C and
D). Notably, miR-654-3p expression significantly increased
following transfection with circRNA_0000285-siRNA, the effect of
which was reversed following addition of miR-654-3p inhibitor
(P<0.01; Fig. 3E and F).
Collectively, these results suggest that miR-654-3p interferes with
circRNA_0000285 expression in thyroid cancer cells.
circRNA_0000285-siRNA inhibits cell
proliferation and induces apoptosis by increasing miR-654-3p
expression in TPC-1 cells
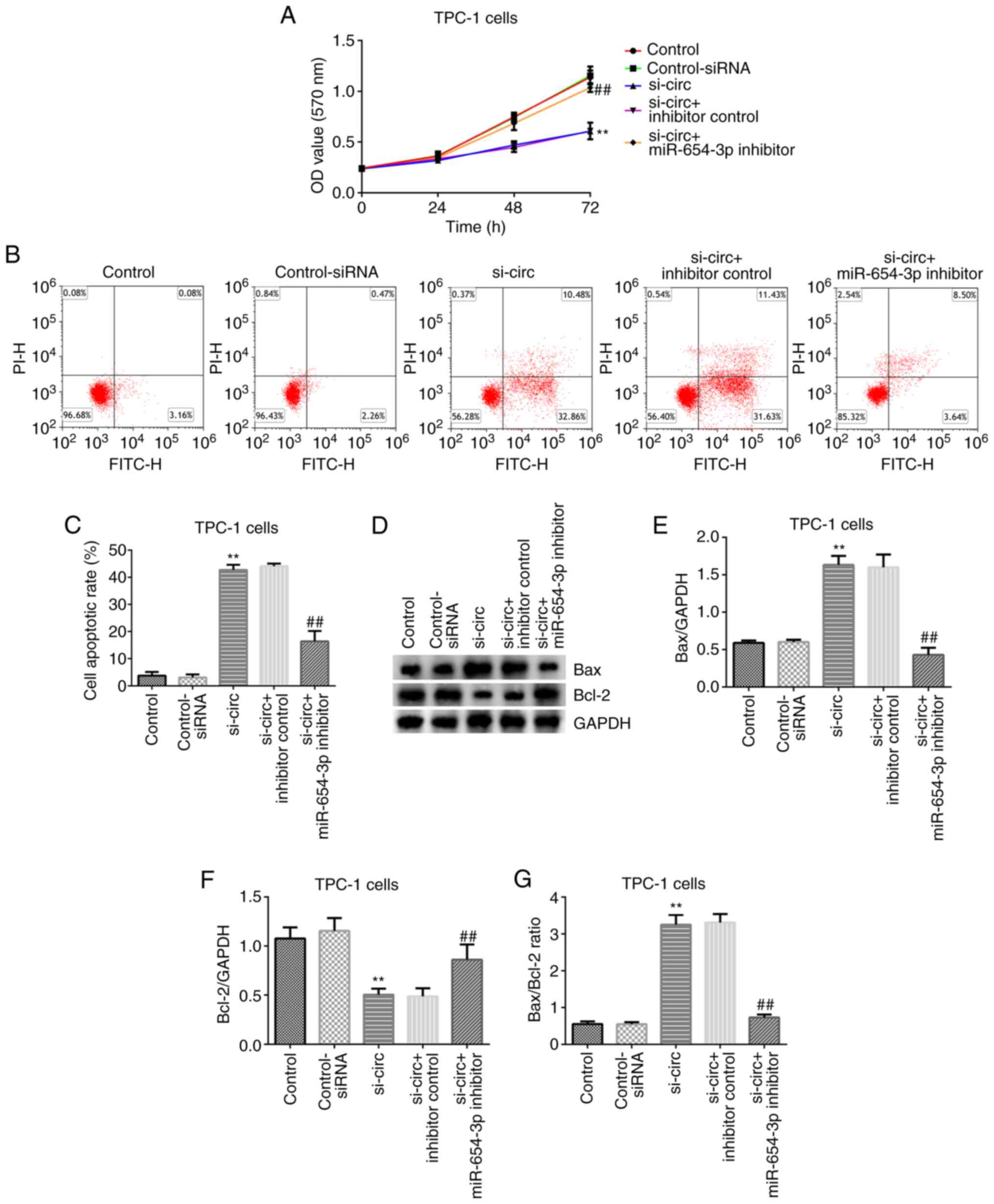
To further assess the underlying molecular mechanism
between miR-654-3p and circRNA_0000285, control-siRNA,
circRNA_0000285-siRNA, inhibitor control and miR-654-3p inhibitor
sequences were transfected into TPC-1 cells for 48 h to determine
the effects of circRNA 0000285-siRNA on cell proliferation and
apoptosis. The results from the MTT assay and flow cytometric
analysis demonstrated that circRNA_0000285-siRNA significantly
inhibited TPC-1 cell proliferation and promoted apoptosis (all
P<0.01; Fig. 4A-C). The
expression levels of the apoptosis-associated proteins, Bax and
Bcl-2, were detected via western blot analysis. The results
demonstrated that transfection with circRNA_0000285-siRNA
significantly increased Bax expression and decreased Bcl-2
expression (P<0.01; Fig. 4D-F),
and the ratio of Bax/Bcl-2 significantly increased compared with
the control-siRNA group (P<0.01; Fig.
4G). Notably, these effects were reversed following
transfection with circRNA_0000285-siRNA + miR-654-3p inhibitor
(Fig. 4).
circRNA_0000285-siRNA inhibits cell
proliferation and induces apoptosis by increasing miR-654-3p
expression in FTC133 cells
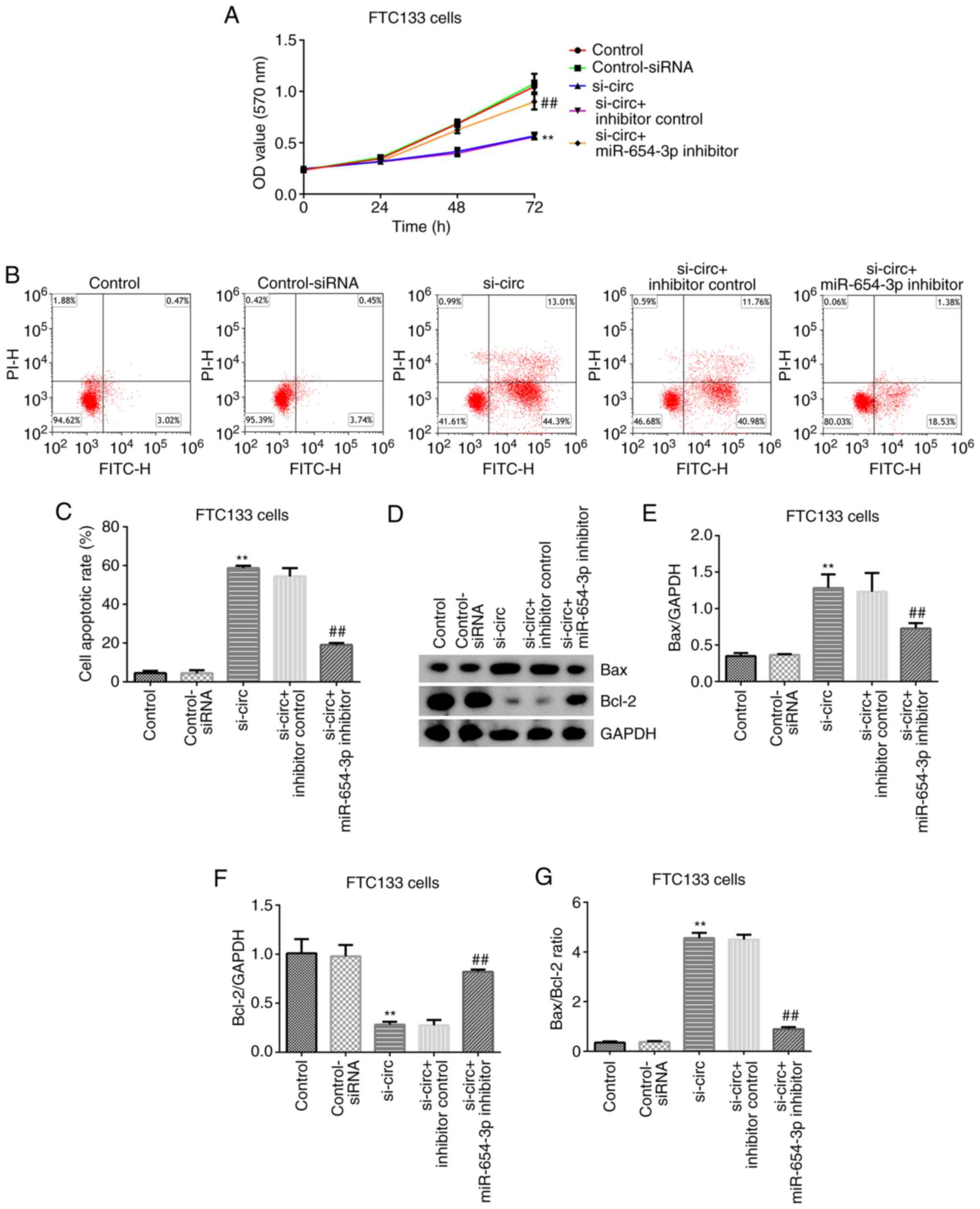
The effects of circRNA_0000285-siRNA were assessed
on FTC133 cell proliferation and apoptosis. Control-siRNA,
circRNA_0000285-siRNA, inhibitor control and miR-654-3p inhibitor
sequences were transfected into FTC133 cells for 48 h. Transfection
with circRNA_0000285-siRNA significantly decreased TPC-1 cell
proliferation and promoted induction of TPC-1 cell apoptosis
(P<0.01; Fig. 5A-C). Western blot
analysis demonstrated that circRNA_0000285-siRNA significantly
enhanced Bax expression (P<0.01; Fig.
5D and E) and decreased Bcl-2 expression (all P<0.01;
Fig. 5D and F), and significantly
increased the ratio of Bax/Bcl-2 compared with the control-siRNA
group (P<0.01; Fig. 5G). Notably,
these effects were reversed following transfection with
circRNA_0000285-siRNA + miR-654-3p inhibitor. Taken together, these
results suggest that circRNA 0000285-siRNA suppresses cell
proliferation and induces apoptosis in thyroid cancer cells by
targeting miR-654-3p.
Discussion
Thyroid cancer is an endocrine tumor with high
morbidity and incidence rates (22)
and 255,490 incident cases and 41,240 deaths were reported in 2017
worldwide (23). Over the past
decade, increasing numbers of thyroid cancer cases have been
reported (24). According to
previous report, it is estimated that >64,300 newly diagnosed
cases of thyroid cancer will emerge in the United States in 2016
(25). Thus, effective therapeutic
methods are required to reduce the burden of patients with thyroid
cancer.
circRNAs are a type of non-coding RNAs that play
important roles in oncogenesis, including thyroid cancer
development (26,27). Increasing evidence suggest that
circRNAs are involved in cellular processes, such as cell
proliferation, apoptosis, differentiation and invasion, and that
they may be used as novel biomarkers or therapeutic targets for
disease treatment. For example, Zhang et al (28) confirmed that circ_0067934 depletion
accelerates cell apoptosis and represses cell proliferation,
migration and invasion in thyroid cancer by sponging miR-1304 and
regulating C-X-CR motif chemokine receptor 1 expression.
hsa_circ_0000285 is a novel circRNA that is dysregulated in various
cancers and disease types, including osteosarcoma (29), diabetic nephropathy (30) and laryngocarcinoma (31). However, the specific role and
molecular mechanism of circ_0000285 in thyroid cancer have not yet
been fully investigated. Thus, the present study investigated the
underlying molecular mechanism of circRNA_0000285 in thyroid cancer
to identify novel treatment methods for this disease.
A previous study revealed the interaction sites
between circRNA_0000285 and miR-654-3p (18). However, the association between
circRNA_0000285 and miR-654-3p in thyroid cancer remain unknow. To
the best of our knowledge, the present study was the first to
investigate the association between circRNA_0000285 and miR-654-3p
in thyroid cancer. The present study confirmed the binding between
circRNA_0000285 and miR-654-3p via the dual-luciferase reporter and
RIP assays. The results revealed that miR-654-3p directly
interacted with circRNA_0000285. The present study also assessed
the role of circRNA_0000285 in thyroid cancer cells. Its expression
was detected, along with miR-654-3p expression, in the thyroid
cancer cell lines, TPC-1 and FTC133, and Nthy-oir 3-1 cells. The
results demonstrated that circRNA_0000285 expression was
upregulated, whereas miR-654-3p expression was downregulated in
thyroid cancer cells compared with Nthy-ori 3-1 cells. Taken
together, these results suggest that circ_0000285 functions as an
oncogene in thyroid tumors by regulating miR-654-3p expression.
Previous studies have reported that abnormal
regulation of miRNAs is associated with the development and
aggressiveness of several tumors (32–34).
Altered expression levels of circRNA_0000285 or miR-654-3p may
affect the functions of thyroid cancer cells. Control-siRNA,
circRNA_0000285-siRNA, inhibitor control and miR-654-3p inhibitor
sequences were transfected into TPC-1 and FTC133 cells for 48 h.
Transfection efficiency was assessed via RT-qPCR analysis. The
results demonstrated that circRNA_0000285-siRNA suppressed
circRNA_0000285 expression in TPC-1 and FTC133 cells. In addition,
transfection with miR-654-3p inhibitor significantly inhibited
miR-654-3p expression, whereas circRNA_0000285-siRNA promoted
miR-654-3p expression in TPC-1 and FTC133 cells compared with the
control-siRNA group. Notably, these results were reversed following
transfection with miR-654-3p inhibitor. Collectively, these results
suggest that circRNA_0000285 negatively regulates miR-654-3p
expression in TPC-1 and FTC133 cells.
Defective apoptosis is a main characteristic of
tumor cells, whereby induction of apoptosis may prevent cancer
development (35). It has been
reported that long non-coding RNA urothelial cancer associated 1
promotes gastric cancer proliferation, migration and inhibits
apoptosis by sponging antitumor miRNAs (36). In the present study, transfection of
thyroid cancer cells with circRNA_0000285-siRNA decreased cell
viability, which was accompanied by increased cell apoptosis in the
circRNA_0000285-siRNA group. These effects were reversed following
transfection of cells with miR-654-3p inhibitor.
Bax is a pro-apoptotic protein that is a
transcriptional target for p53 (37). Bcl-2 can inhibit the activity of Bax
and induce cell apoptosis (38). To
further investigate the molecular mechanism of circRNA_0000285 and
miR-654-3p in thyroid cancer cells, the expression levels of Bax
and Bcl-2 were detected in the different groups. The results
demonstrated that circRNA_0000285-siRNA enhanced Bax expression,
decreased Bcl-2 expression and increased the ratio of Bax/Bcl-2
compared with the control-siRNA group. Notably, these effects were
reversed following transfection with circRNA_0000285-siRNA +
miR-654-3p inhibitor. Taken together, these results suggest that
circRNA_0000285-siRNA inhibits thyroid cancer cell proliferation
and induces apoptosis by increasing miR-654-3p expression.
The present study was an in vitro preliminary
study of the role of circRNA_0000285 in the proliferation and
apoptosis of thyroid cancer cells. To confirm the role of
circRNA_0000285 in thyroid cancer, in-depth experiments are
required. For example, determining the role of miR-654-3p alone in
thyroid cancer cells is required to verify the results presented
here. In addition, the specific molecular mechanism of
circRNA_0000285/miR-654-3p affecting the viability of thyroid
cancer cells requires investigation. Prospective studies are also
required to investigate other target genes downstream of
circRNA_0000285 or miR-654-3p in thyroid cancer. Furthermore, the
role of circRNA_0000285 in thyroid cancer should be determined
in vivo.
In conclusion, the results of the present study
demonstrated that circRNA_0000285-siRNA repressed thyroid cancer
cell proliferation and induced apoptosis by regulating miR-654-3p
expression. These findings may provide evidence for a novel
biomarker and a potential therapeutic target for thyroid
cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
RG contributed to the study design, data collection,
statistical analysis, data interpretation and manuscript
preparation. HY contributed to data collection, statistical
analysis and manuscript preparation. QG, NW, YZ and HD contributed
to data collection and statistical analysis. RG and HY performed
the experiments. RG and HY confirmed the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Peng C, Li Z, Gao H, Zou X, Wang X, Zhou C
and Niu J: Synchronous primary sigmoid colon cancer and primary
thyroid cancer followed by a malignant tumor of the kidney: Case
report of multiple primary cancer and review of the literature.
Oncol Lett. 17:2479–2484. 2019.PubMed/NCBI
|
|
2
|
Kaptan E, Sancar-Bas S, Sancakli A, Bektas
S and Bolkent S: The effect of plant lectins on the survival and
malignant behaviors of thyroid cancer cells. J Cell Biochem.
119:6274–6287. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chow TL: Quality of life after surgical
treatment for thyroid cancer. JAMA Otolaryngol Head Neck Surg.
145:8732019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iniguez-Ariza NM, Stan MN and Bible KC:
Effect of thyroid hormone suppression on control of advanced
well-differentiated thyroid cancer. Endocrine. 59:228–229. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Barakat S, Odem J, Batanian JR, Raza S and
Khan UZ: Papillary thyroid cancer in struma testis with malignant
transformation in the lung associated with trisomy 17 successfully
treated with total thyroidectomy and radioiodine ablation. Case Rep
Oncol. 7:751–757. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luo X and Wu ACYZYJBZJ: Analysis of risk
factors for postoperative recurrence of thyroid cancer. J BUON.
24:813–818. 2019.PubMed/NCBI
|
|
7
|
Lee E, Elhassan S, Lim GPL, Kok WH, Tan
SW, Leong EN, Tan SH, Chan EWL, Bhattamisra SK, Rajendran R and
Candasamy M: The roles of circular RNAs in human development and
diseases. Biomed Pharmacother. 111:198–208. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma HB, Yao YN, Yu JJ, Chen XX and Li HF:
Extensive profiling of circular RNAs and the potential regulatory
role of circRNA-000284 in cell proliferation and invasion of
cervical cancer via sponging miR-506. Am J Transl Res. 10:592–604.
2018.PubMed/NCBI
|
|
9
|
Panda AC: Circular RNAs act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dori M and Bicciato S: Integration of
bioinformatic predictions and experimental data to identify
circRNA-miRNA associations. Genes (Basel). 10:6422019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang W and Zhang S: Down-regulation of
circRNA_0000285 suppresses cervical cancer development by
regulating miR197-3p-ELK1 Axis. Cancer Manag Res. 12:8663–8674.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shirjang S, Mansoori B, Asghari S, Duijf
P, Mohammadi A, Gjerstorff M and Baradaran B: MicroRNAs in cancer
cell death pathways: Apoptosis and necroptosis. Free Radic Biol
Med. 139:1–15. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carroll AP, Goodall GJ and Liu B:
Understanding principles of miRNA target recognition and function
through integrated biological and bioinformatics approaches. Wiley
Interdiscip Rev RNA. 5:361–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu S, Wang H, Mu J, Wang H, Peng Y, Li Q,
Mao D and Guo L: miRNA-211 triggers an autophagy-dependent
apoptosis in cervical cancer cells: Regulation of Bcl-2. Naunyn
Schmiedebergs Arch Pharmacol. 393:359–370. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Geraldo MV, Nakaya HI and Kimura ET:
Down-regulation of 14q32-encoded miRNAs and tumor suppressor role
for miR-654-3p in papillary thyroid cancer. Oncotarget.
8:9597–9607. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang J, Zhang Z, Chen S, Dou W, Xie R and
Gao J: miR-654-3p predicts the prognosis of hepatocellular
carcinoma and inhibits the proliferation, migration, and invasion
of cancer cells. Cancer Biomark. 28:73–79. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pu JT, Hu Z, Zhang DG, Zhang T, He KM and
Dai TY: miR-654-3p suppresses non-small cell lung cancer
tumourigenesis by inhibiting PLK4. Onco Targets Ther. 13:7997–8008.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yao T, Zha D, Hu C and Wu X: circ_0000285
promotes podocyte injury through sponging miR-654-3p and activating
MAPK6 in diabetic nephropathy. Gene. 747:1446612020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q,
Wang Q, Xie R, Su Y, Yang M, et al: Circular RNA ACVR2A suppresses
bladder cancer cells proliferation and metastasis through
miR-626/EYA4 axis. Mol Cancer. 18:952019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bierhoff H: Analysis of lncRNA-protein
interactions by RNA-Protein Pull-Down assays and RNA
immunoprecipitation (RIP). Methods Mol Biol. 1686:241–250. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pajamaki N, Metso S, Hakala T, Ebeling T,
Huhtala H, Ryödi E, Sand J, Jukkola-Vuorinen A, Kellokumpu-Lehtinen
PL and Jaatinen P: Long-term cardiovascular morbidity and mortality
in patients treated for differentiated thyroid cancer. Clin
Endocrinol (Oxf). 88:303–310. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deng Y, Li H, Wang M, Li N, Tian T, Wu Y,
Xu P, Yang S, Zhai Z, Zhou L, et al: Global burden of thyroid
cancer from 1990 to 2017. JAMA Netw Open. 3:e2087592020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burton BN, Okwuegbuna O, Jafari A,
Califano J, Brumund KT and Gabriel RA: Association of preoperative
anemia with 30-day morbidity and mortality among patients with
thyroid cancer who undergo thyroidectomy. JAMA Otolaryngol Head
Neck Surg. 145:124–1231. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mao Y and Xing M: Recent incidences and
differential trends of thyroid cancer in the USA. Endocr Relat
Cancer. 23:313–322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu G, Zhou W, Pan X, Sun Z, Sun Y, Xu H,
Shi P, Li J, Gao L and Tian X: Circular RNA profiling reveals
exosomal circ_0006156 as a novel biomarker in papillary thyroid
cancer. Mol Ther Nucleic Acids. 19:1134–1144. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cui W and Xue J: Circular RNA DOCK1
down-regulates microRNA-124 to induce the growth of human thyroid
cancer cell lines. Biofactors. 46:591–599. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang H, Ma XP, Li X and Deng FS: Circular
RNA circ_0067934 exhaustion expedites cell apoptosis and represses
cell proliferation, migration and invasion in thyroid cancer via
sponging miR-1304 and regulating CXCR1 expression. Eur Rev Med
Pharmacol Sci. 23:10851–10866. 2019.PubMed/NCBI
|
|
29
|
Long Z, Gong F, Li Y, Fan Z and Li J:
circ_0000285 regulates proliferation, migration, invasion and
apoptosis of osteosarcoma by miR-409-3p/IGFBP3 axis. Cancer Cell
Int. 20:4812020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yao T, Zha D, Hu C and Wu X: circ_0000285
promotes podocyte injury through sponging miR-654-3p and activating
MAPK6 in diabetic nephropathy. Gene. 747:1446612020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qin JB, Chang W, Yuan GH, Huang L and Qiu
ZF: Circular RNA hsa_circ_0000285 acts as an oncogene in
laryngocarcinoma by inducing Wnt/beta-catenin signaling pathway.
Eur Rev Med Pharmacol Sci. 24:97732020.PubMed/NCBI
|
|
32
|
Bao B, Ali S, Ahmad A, Li Y, Banerjee S,
Kong D, Aboukameel A, Mohammad R, Van Buren E, Azmi AS and Sarkar
FH: Differentially expressed miRNAs in cancer-stem-like cells:
Markers for tumor cell aggressiveness of pancreatic cancer. Stem
Cells Dev. 23:1947–1958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Buruiană A, Florian ȘI, Florian AI, Timiș
TL, Mihu CM, Miclăuș M, Oșan S, Hrapșa I, Cataniciu RC, Farcaș M
and Șușman S: The roles of miRNA in glioblastoma tumor cell
communication: Diplomatic and aggressive negotiations. Int J Mol
Sci. 21:19502020. View Article : Google Scholar
|
|
34
|
Donnarumma E, Fiore D, Nappa M, Roscigno
G, Adamo A, Iaboni M, Russo V, Affinito A, Puoti I, Quintavalle C,
et al: Cancer-associated fibroblasts release exosomal microRNAs
that dictate an aggressive phenotype in breast cancer. Oncotarget.
8:19592–19608. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao Y, Ponnusamy M, Dong Y, Zhang L, Wang
K and Li P: Effects of miRNAs on myocardial apoptosis by modulating
mitochondria related proteins. Clin Exp Pharmacol Physiol.
44:431–440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY,
Liu Q, Zhao G and Zhang ZZ: The lncRNA UCA1 promotes proliferation,
migration, immune escape and inhibits apoptosis in gastric cancer
by sponging anti-tumor miRNAs. Mol Cancer. 18:1152019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee SB, Lee S, Park JY, Lee SY and Kim HS:
Induction of p53-dependent apoptosis by prostaglandin
A2. Biomolecules. 10:4922020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Azimian H, Dayyani M, Toossi M and
Mahmoudi M: Bax/Bcl-2 expression ratio in prediction of response to
breast cancer radiotherapy. Iran J Basic Med Sci. 21:325–332.
2018.PubMed/NCBI
|