Introduction
MiT/TFE transcriptional factors encodes four
distinct genes: MITF, TFEB, transcription factor binding to IGHM
enhancer 3 (TFE3) and TFEC (1–5).
Structurally, MiT/TFE genes encode a protein with a double-helix
leucine zipper motif, a transactivating zone and a domain
responsible for DNA contact and binding (1–5). The
TFE3 gene is 14,749-bp long and is located on chromosome Xp11.22
(6). The functional domain of the
TFE3 gene can fuse with the promoter region of other genes, usually
housekeeping genes, resulting in the constitutive overexpression of
the TFE3 protein and a condition referred to as
Xp11.2-translocation renal cell carcinoma (tRCC) (7,8). The
TFE3 protein plays an important role in the development of Xp11.2
tRCC, as well as other types of renal cell carcinoma (9,10).
Indeed, high levels of TFE3 protein tend to indicate poor prognosis
and fast progression (8). Previous
studies have identified that TFE3 binds to coordinated lysosomal
expression and regulation sequence elements to induce lysosomal
biogenesis and autophagy (11,12).
TFE3 also plays a role in cellular energy metabolism. For instance,
TFE3 governs insulin signaling and glucose metabolism by
upregulating insulin receptor 1 substrate (IRS)-2 and hexokinase
enzymes, thus inhibiting lipogenesis and increasing glycogen
synthesis (13,14). In addition, TFE3 may promote
oncogenesis by regulating the cell cycle in Xp11.2 tRCC (15,16).
Nuclear respiratory factor-1 (NRF-1) is a
transcription factor that functions primarily as a positive
regulator of genes involved in mitochondrial biogenesis and
oxidative phosphorylation (17,18).
NRF-1 belongs to the NRF family and, together with NRF-2,
participates in the regulation of the expression of components
related to respiratory chain subunits, mitochondrial replication
and transcription (16). NRF-1 plays
a vital role in maintaining mitochondrial oxidative respiration and
energy production, as well as regulating cell proliferation and
growth (19). In addition, a
previous study demonstrated that NRF-1, the main regulator of
various genes in the nervous system, is associated with
neurodegenerative diseases (20). A
previous study identified 2,470 NRF-1 target genes in SK-N-SH human
neuroblastoma cells using chromatin immunoprecipitation sequencing
(ChIP-Seq) (18). The molecular
pathways of these genes involve regulation of RNA metabolism,
splicing, cell cycle, DNA damage repair, protein translation
initiation and ubiquitin-mediated protein degradation, along with
mitochondrial respiratory function. TFE3 is also a target gene of
NRF-1 (21).
It is well known that tumorigenesis results in a
parallel change in cellular metabolism, which is closely related to
the biosynthesis of mitochondria and linked to NRF-1 (22,23).
Moreover, TFE3 itself is involved in cellular energy metabolism.
Previous reports indicated that TFE3 may be one of the downstream
target genes of NRF-1 (1,8,19,22).
Therefore, the aim of the present study was to determine whether
NRF-1 had a direct regulatory effect on TFE3 and whether it could
mediate the occurrence and development of tumors through TFE3.
Materials and methods
Cell culture
The cell lines used in this study included the 786-O
human kidney adenocarcinoma cell line (CRL-1932™) and the 293T
human embryonic kidney cell line (CRL11268™), which were obtained
from the American Type Culture Collection. DMEM supplemented with
10% FBS (both from Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.) was used to culture cells in a 5% CO2 humidified
incubator at 37°C.
Gene silencing and plasmid
constructs
In the present study, lentiviral vectors were used,
and the virus assembly and sequence design were completed by OBiO
Technology (Shanghai) Corp. The selected interference vector was
pLKD-CMV-Puro-U6-shRNA vector [OBiO Technology (Shanghai) Corp.].
The ccdB toxic gene downstream of the U6 promoter was cut with
AgeI and EcoR and the shRNA sequence to be
constructed was inserted. Lentiviruses were produced by the
transfection of 293T cells [OBiO Technology (Shanghai) Corp.] with
Gag/Pal along with VSV-G [OBiO Technology (Shanghai) Corp.]. Cells
at 50–60% confluence were transduced with lentivirus carrying short
hairpin RNA (shRNA) targeting the human TFE3 or NRF1 genes
[multiplicity of infection, 3; virus titer, 1×108
transduction units/ml (TU)]. The transfection reagent was HitransG
(cat. no. GCD0252780; Shanghai Genechem Co., Ltd.). The sequences
used were as follows: i) NRF1-shRNA, 5′-GTAAGTACAAGAGCATGAT-3′; ii)
TFE3-shRNA, 5′-GCTCCGAATTCAGGAACTA-3′; and iii) negative control
(NC) shRNA, 5′-TTCTCCGAACGTGTCACGT-3′. The samples were incubated
at 37°C for 12–16 h, the medium was changed to complete medium and
then incubation was continued. After 48-h incubation in a 5%
CO2 humidified incubator at 37°C. The infected cells
were selected using 2 µg/ml of puromycin (cat. no. P8833;
Sigma-Aldrich; Merck KGaA) for stable clones.
For overexpression, the cDNA encoding TFE3 was
subcloned into pcDNA3.1 (V790-20; Invitrogen; Thermo Fisher
Scientific Inc.) with ClonExpress II One Step Cloning kit (cat. no.
C112; Vazyme Biotech Co., Ltd.). The primers used for constructing
the TFE3 overexpression plasmid were as follows: forward,
5′GCCACCATGTCTCATGCGGCCGAA-3′; reverse,
5′-CAGGACTCCTCTTCCATGCTG-3′. Cells in the logarithmic growth phase
(density, 40–50%) were plated in a 6-well plate with 3.5 µg plasmid
per well. The plasmid was transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions. The
plasmid suspension and Lipofectamine ®2000 suspension
were mixed gently and allowed to stand at room temperature for 20
min followed by incubation in a 5% CO2 humidified
incubator at 37°C for 48 h. The expression plasmids for NRF-1 were
constructed according to a previous report (24).
ChIP
ChIP was performed according to the protocol from
the Pierce™ Agarose ChIP kit (Thermo Fisher Scientific, Inc.; cat.
no. 26156). Briefly, 293T cells were fixed with in 1% formaldehyde
in culture medium at room temperature for 5 min. The remaining
unreacted formaldehyde was quenched with glycine. Cells were lysed
in Buffer 1, centrifuged at 9,000 × g for 3 min and the supernatant
was removed. Pellets were resuspended in Buffer 1, homogenized on
ice, then pelleted again. Next, the pellet was resuspended in
Buffer 2. The chromatin fraction was sheared by sonication in 1.5
ml siliconized microcentrifuge tubes. The sheared chromatin was
immunoprecipitated with either anti-NRF-1 antibody (10 µg/sample;
cat. no. ab175932; Abcam) or normal rabbit IgG (1–2 µl; cat. no.
26156; Thermo Fisher Scientific Inc.) overnight at 4°C with
constant rotation. The isolated complexes were washed with IP Wash
Buffer 2 and IP Wash Buffer 3 prior to elution. The DNA fragments
were then separated from complexes and recovered through column
purification (Thermo Fisher Scientific, Inc.; cat. no. 26156). The
co-precipitated DNA fragments were identified by quantitative PCR
(qPCR) as detailed below using primers. Specific for the TFE3
promoter region: forward primer, 5′-GGTCGTCCGGGGTTAGGTT-3′; reverse
primer, 5′-TCCGCTAAGCCATGGAGCTA-3′.
Luciferase reporter assay
The promoter region online analysis software JASPER
(http://jaspar.genereg.net/cgi-bin/jaspar_db.pl) was
used to analyze NRF-1 binding sites in the promoter region of TFE3.
The TFE3 promoter was obtained from the NCBI and cloned into the
pGL4.10 vector (Shanghai Obio Technology Co., Ltd.). The amplified
plasmid was verified by restriction enzyme digestion and the
monoclonal was in accordance with the theoretical design (Fig. 1C). The sequences of the plasmids were
verified using sequencing. The plasmid was transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's recommendations.
Plasmid constructs were co-transfected with pGMLR-TK Renilla
luciferase-containing plasmid (cat. no. 11558ES03; Shanghai Yeason
Biotechnology Co., Ltd.) as an internal control. Reporter activity
was detected using Dual Luciferase Reporter Gene Assay Kit
(Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. The TFE3 luciferase reporter gene
plasmid was transfected into 293T cells, interfered with NRF1-shRNA
and luciferase activity was detected 48 h later. The cells were
then lysed, and luciferase activity was detected using the Dual-Glo
luciferase assay system. Renilla luciferase was used the
internal control, the RLU value was obtained by the firefly
luciferase measurement divided by the RLU value obtained by the
Renilla luciferase measurement. According to the obtained
ratio, the activation degree of the target reporter gene was
assessed among different samples. The sequences of shRNA used were
as follows: NRF1-shRNA, 5′-GTAAGTACAAGAGCATGAT-3′; and negative
control (NC) shRNA, 5′-TTCTCCGAACGTGTCACGT-3′. The virus assembly
and sequence design were completed by Dharmacon.
Western blot analysis
Total cellular proteins were treated with extraction
buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1% NP-40 Lysis Buffer;
0.1% SDS; 1X protease inhibitor cocktail). Whole cell lysates were
centrifuged at 12,000 × g at 4 °C. Total protein was quantified via
the BCA assay. Total cellular protein (40 µg/lane) was subjected to
10% SDS-PAGE, then transferred to a PVDF membrane (Roche
Diagnostics). Blots were blocked with 5% non-fat milk in TBS +
0.05% Tween-20 (Sigma-Aldrich; MerckKGaA) (TBST). The membranes
were incubated with primary antibodies overnight at 4°C. After
washing with TBST, HRP-conjugated secondary antibodies were applied
at room temperature for 2 h. After antibody incubation, the blots
were washed with TBST 5 times for 5 min each time. The protein
bands were visualized using an enhanced chemiluminescence detection
kit (cat. no. P0018A; Beyotime Institute of Biotechnology) and
recorded on a radiographic film (Alpha Innotech Corporation). The
following primary antibodies were: Anti-TFE3 (1:2,000; cat. no.
HPA023881; Sigma-Aldrich; Merck KGaA); anti-NRF-1 (1:2,000; cat.
no. ab175932; Abcam); anti-mTOR (1:5,000; cat. no. ab134903;
Abcam); anti-S6 (1:1,000; cat. no. CST 54D2; Cell Signaling
Technology Inc.); anti-p-S6 (1:2,000; cat. no. CST D57.2.2E; Cell
Signaling Technology Inc.); anti-AKT (1:1,000; cat. no. CST 9272;
Cell Signaling Technology Inc.); anti-p-AKT (1:2,000; cat. no. CST
4060; Cell Signaling Technology Inc.); and anti-GAPDH (1:2,500;
cat. no. BM3876; Wuhan Boster Biological Technology, Ltd.). The
following secondary antibodies were used: anti-Rabbit IgG (1:5,000;
BA1054; Wuhan Boster Biological Technology, Ltd.) and anti-Mouse
IgG (1:5,000; cat. no. BA1051; Wuhan Boster Biological Technology,
Ltd.). Semi-quantitative analysis was performed using ImageJ
v.1.8.0 software (National Institutes of Health).
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the 786-O and 293T
cells at room temperature using Trizol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and
reverse-transcribed using HiScript Q RT Supermix for qPCR (Vazyme
Biotech Co., Ltd., cat. no. R122-01) according to the
manufacturer's instructions. In RT process, reaction system
including RNA was incubated at 42°C for 60 min to synthesize cDNA,
then incubated at 80°C for 10 min to inactivate the reverse
transcriptase and terminate the RT reaction. For qPCR analysis,
SYBR Green ER (Roche Diagnostics) was used according to the
manufacturer's protocol. Total RNA samples were prepared from
control and various treated cells and analyzed by real-time PCR
analysis using a 7300 system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The thermocycling conditions were as follows:
amplification was done for 25 cycles, each with denaturation at
95°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C
for 30 sec. The Cq values were analyzed using the 2−ΔΔCq
method (25). Amplification of the
reference endogenous gene GAPDH was used to normalize the data. The
primer sequences used for RT-qPCR are listed in Table I.
 | Table I.Primer sequences used for
RT-qPCR. |
Table I.
Primer sequences used for
RT-qPCR.
| Gene name | Primers
(5′-3′) |
|---|
| GAPDH | F:
GGAGCGAGATCCCTCCAAAAT |
|
| R:
GGCTGTTGTCATACTTCTCATGG |
| TFE3 | F:
TGTGTACAGTAGTCAAGGCGT |
|
| R:
AGTGCCCAGTTCCTTGATCC |
| NRF-1 | F:
GTACAAGAGCATGATCCTGGA |
|
| R:
GCTCTTCTGTGCGGACATC |
| mTOR | F:
TCCGAGAGATGAGTCAAGAGG |
|
| R:
CACCTTCCACTCCTATGAGGC |
| CCND1 | F:
GCTGCGAAGTGGAAACCATC |
|
| R:
CCTCCTTCTGCACACATTTGAA |
| CCND2 | F:
CTGTCTCTGATCCGCAAGCAT |
|
| R:
GGTGGGTACATGGCAAACTTAAA |
| AKT | F:
GTCATCGAACGCACCTTCCAT |
|
| R:
AGCTTCAGGTACTCAAACTCGT |
| S6 | F:
AGGGTTATGTGGTCCGAATCA |
|
| R:
TTGGTCTGTAACAGGAATGCC |
Cell cycle analysis
786-O cells with different treatments were fixed
with 70% ethanol for 1 h at 4°C and stained with 50 µg/ml propidium
iodide for 30 min at 4°C in the dark. Cell cycle data was acquired
using an LSR II flow cytometer (BD Biosciences). Quantification of
cells in each phase of the cell cycle was carried out using FlowJo
(version 10; FlowJo LLC).
Apoptosis analysis
To quantify apoptosis, annexin V and propidium
iodide staining was carried out, followed by flow cytometry. The
786-O cells with different treatments were collected and washed
twice with PBS. Subsequently, Binding Buffer was added to 400 µl of
cell suspension, stained with 5 µl of Annexin V-FITC and propidium
iodide (PI) staining solution at room temperature for 15 min in the
dark (all part of a kit; cat. no. C1062S; Beyotime Institute of
Biotechnology). Data was acquired using an LSR II flow cytometer
(BD Biosciences). Quantification of apoptotic cells was carried out
using FlowJo (version 10; FlowJo LLC).
Measurement of mitochondria using
MitoTimer
MitoTimer is a novel tool for monitoring real-time
mitochondrial aging, turnover and biogenesis, which can be used to
evaluate individual mitochondria or mitochondrial populations
within a cell (26). MitoTimer
encodes a ‘timer fluorescent protein’ that is targeted to the
mitochondrial matrix (26).
MitoTimer green fluorescence enters the mitochondrial matrix and
matures into a red fluorescent protein (26,27). In
this study, MitoTimer plasmid (cat. no. 50547; Biovector NTCC Inc.)
was transfected into 293T cells. The plasmid was transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's recommendations.
Cells in the logarithmic growth phase (density, 40–50%) were plated
in a 6-well plate with 3.5 µg plasmid/well and incubated in a 5%
CO2 humidified incubator at 37°C. Subsequently, 24 h
later production was induced with doxycycline (Dox) (2 µg/ml) at
37°C for 24 h. Mitophagy was then observed using a Olympus confocal
microscope.
Cell Counting Kit-8 (CCK-8)
analysis
The target cells were digested and seeded into a
96-well plate at a density of 3×103 cells. After 24 h of
incubation in an incubator, 10 µl CCK-8 reagent was added to each
well. After incubation in the incubator for another 2 h, the
absorbance was measured at 450 nm wavelength.
Statistical analysis
Data analysis was performed using Excel (version
16.11.1; Microsoft Corporation) or SPSS for Windows (version 13.0;
SPSS, Inc.). Differences between groups were analyzed using
Student's unpaired t-tests or one-way ANOVA followed by Tukey's
post hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
NRF-1 has functional binding sites in
the promoter region of TFE3
The promoter region online analysis software JASPER
was used to analyze whether there are NRF-1 binding sites in the
promoter region of TFE3. A total of 12 potential binding sites for
NRF-1 were identified in the promoter region of TFE3. Primers for
the two sites with the highest scores were designed for subsequent
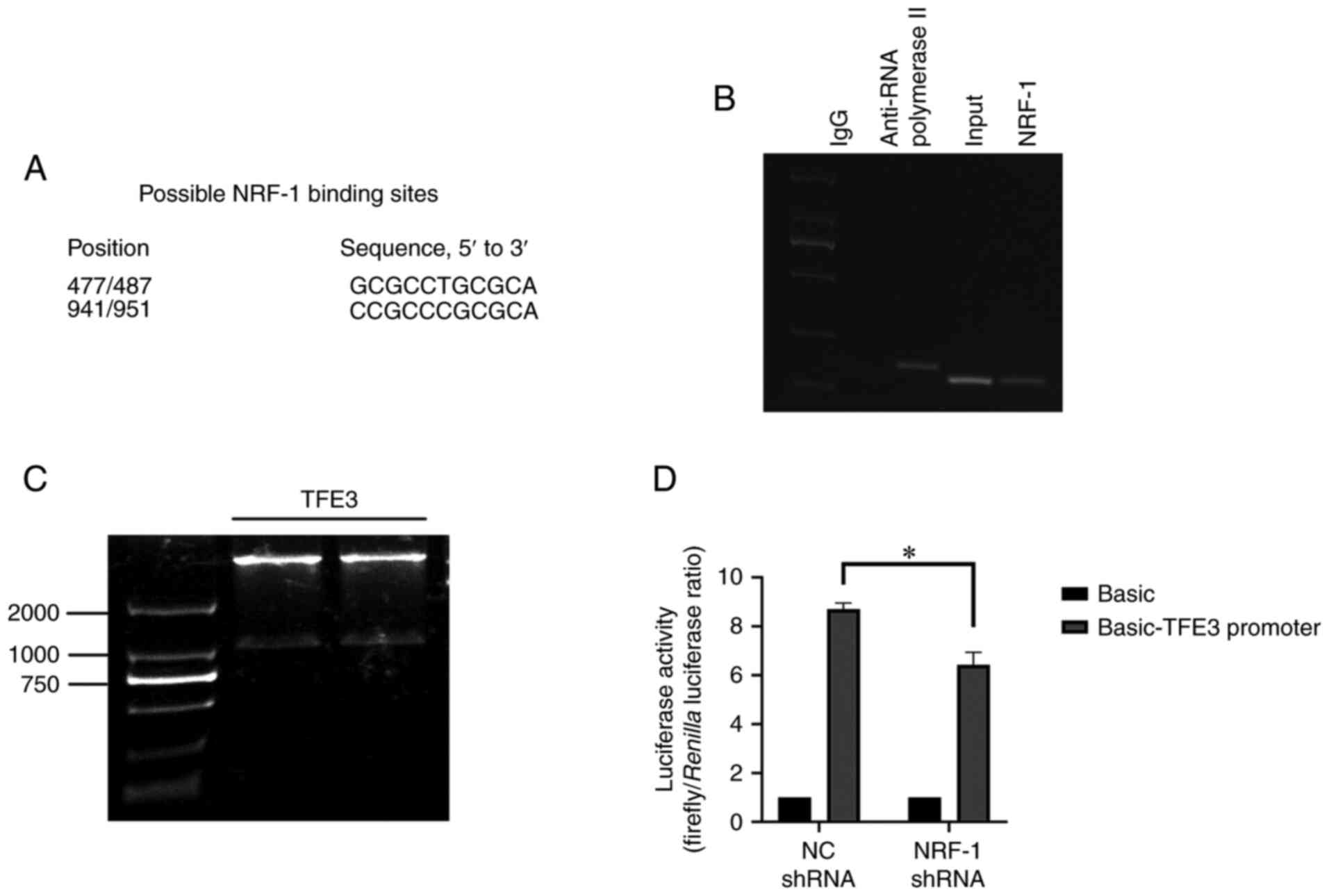
experiments (Fig. 1A).
To determine whether the TFE3 promoter interacted
with NRF-1, ChIP was performed using genomic DNA and NRF-1-specific
antibodies, and qPCR was used to analyzed the target DNA fragments.
The ChIP results indicated that NRF-1 could directly bind to the
TFE3 promoter region at position 477–487 (Fig. 1B).
The results of the luciferase assay demonstrated
luciferase activity of the TFE3 gene promoter in the NRF-1 shRNA
group decreased by ~20% compared with the control group (Fig. 1D). This confirmed that NRF-1 could
bind the promoter region of the TFE3 gene, which may have a
positive regulatory effect on the transcription of TFE3 gene. Thus,
the potential regulatory effect of NRF-1 on TFE3 was further
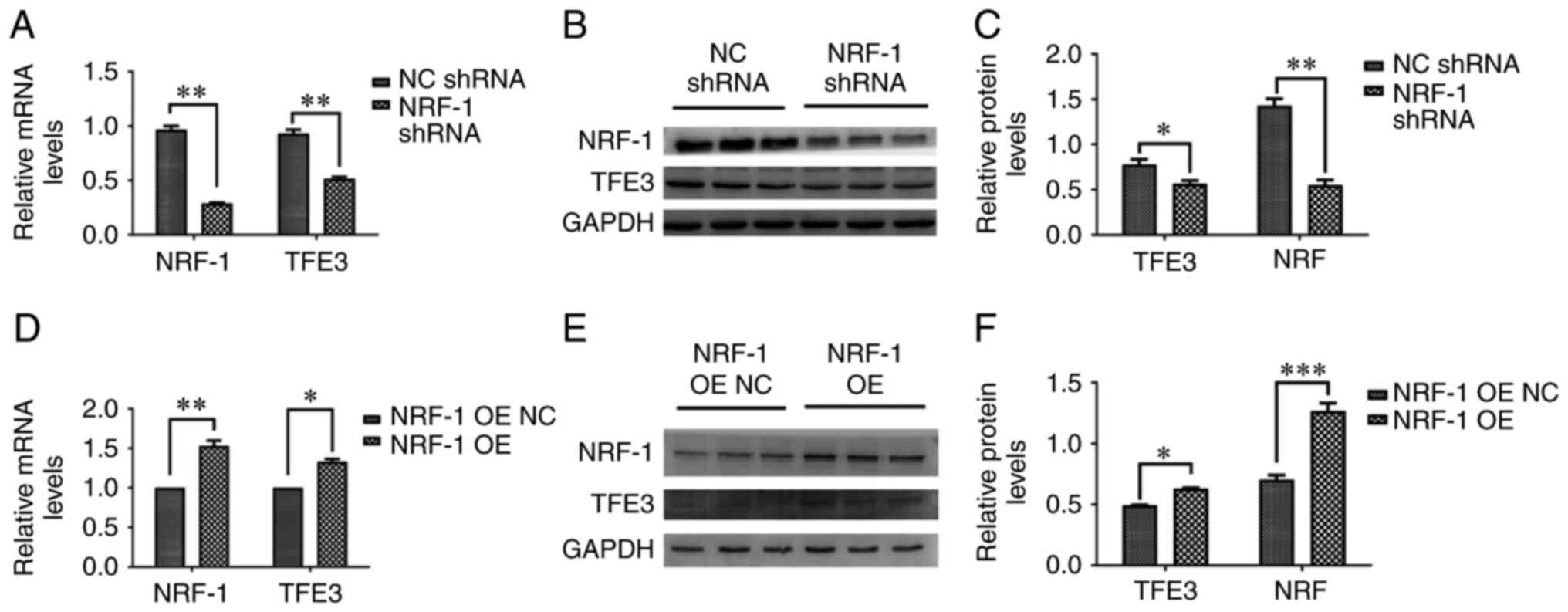
examined. NRF-1 expression was silenced using shRNA, and the mRNA
and protein expression levels of TFE3 were measured. The results
indicated that NRF-1 silencing reduced the expression of TFE3
compared with NRF-1 NC (Fig. 2A-C).
Moreover, the mRNA and protein expression levels of TFE3 were
measured following NRF-1 overexpression. The mRNA and protein
levels of TFE3 were upregulated following NRF-1 overexpression,
which was consistent with the aforementioned results (Fig. 2D-F).
Function of NRF-1 in kidney cancer
cells
TFE3 have also been implicated in mTOR signaling, a
major regulator of protein synthesis contributing to the growth of
several tumor types, including RCC (28). NRF-1 expression was silenced in 786-O
cells using shRNA and the expression of mTOR-related indicators
(mTOR, AKT/p-AKT and S6/p-S6) was measured. The protein levels of
p-AKT and p-S6 were decreased compared with the control group and
the results showed that the expression levels of components of the
mTOR pathway were downregulated following NRF-1 shRNA transfection
(Fig. 3A-C). In order to further
verify that NRF-1 can regulate the mTOR pathway through TFE3, the
TFE3 protein was overexpressed in NRF-1 shRNA 786-O cells. The
overexpression plasmid of TFE3 was verified using western blot
analysis (Fig. S1). The results
showed that the overexpression of the TFE3 protein together with
NRF-1 silencing restored the expression mTOR pathway-associated
proteins compared with NRF-1 or TFE3 silencing alone (Fig. 3D and E).
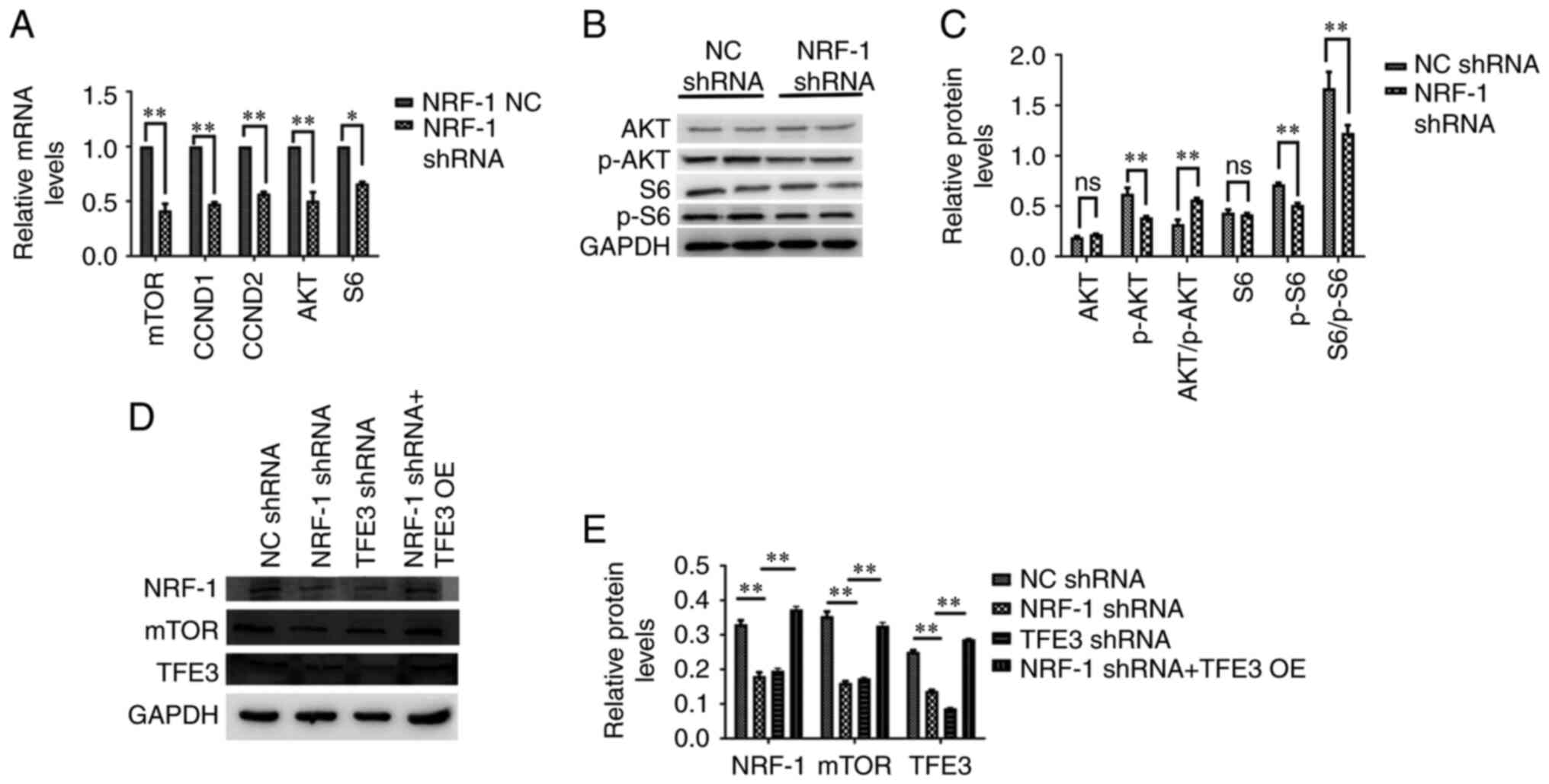 | Figure 3.NRF-1 regulates the mTOR pathway. (A)
mRNA expression levels of mTOR, CCND1, CCND2, AKT and S6 following
shRNA-mediated NRF-1 knockdown. (B and C) AKT and S6
phosphorylation levels following shRNA-mediated NRF-1 knockdown. (D
and E) NRF-1, mTOR and TFE3 protein expression levels following
NRF-1 knockdown with or without TFE3 OE. Data are presented as the
mean ± SEM. n=3. *P<0.05, **P<0.01. CCND, cyclin D; S6, S6
ribosomal protein; NRF-1, nuclear respiratory factor-1; TFE3,
transcription factor binding to IGHM enhancer 3; NC, negative
control; shRNA, short hairpin RNA; OE, overexpression; ns, not
significant; p-, phosphorylated. |
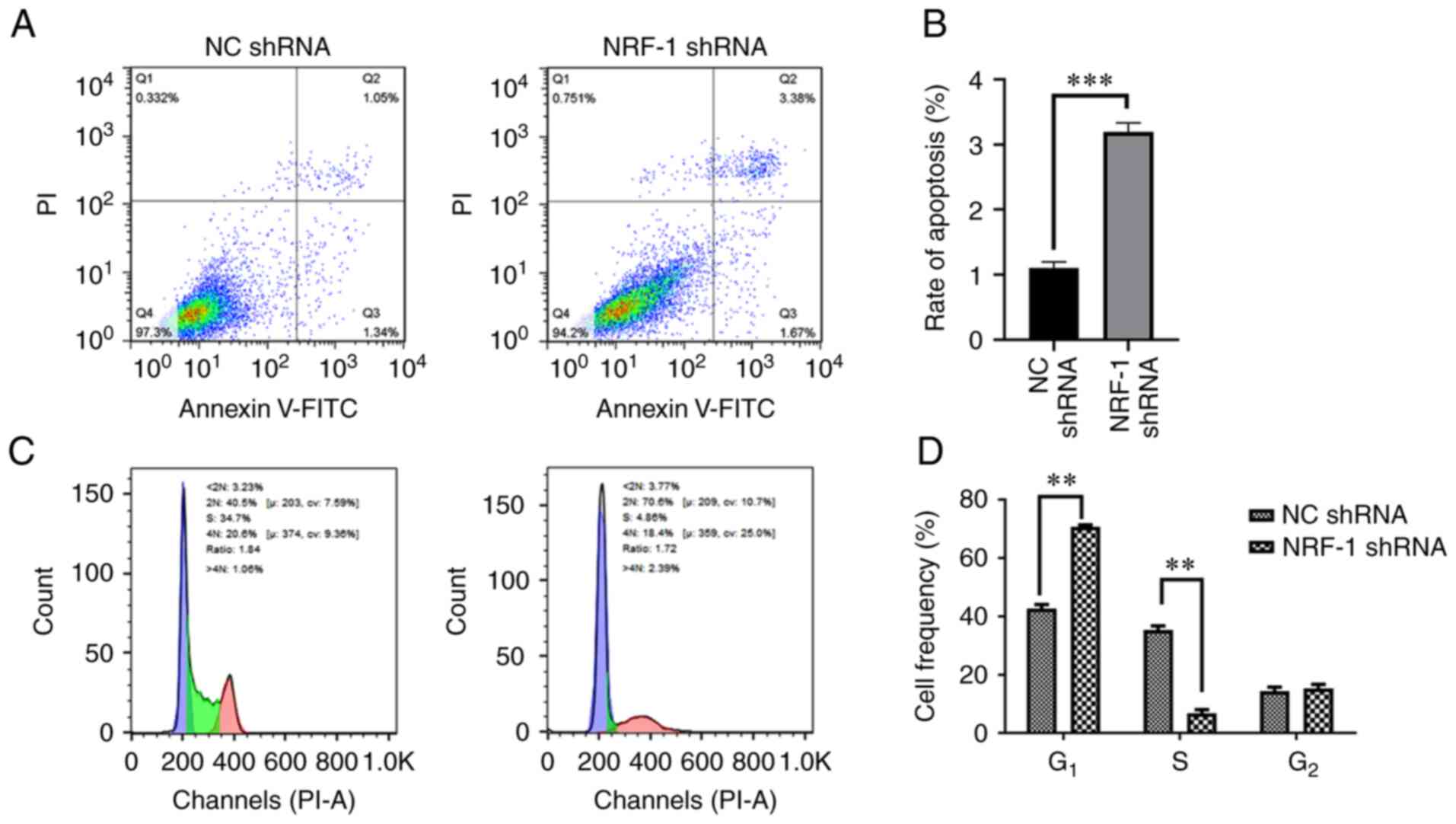
In order to detect the effect of NRF-1 on tumor cell
apoptosis and proliferation, flow cytometry was used following
NRF-1 silencing using shRNA. The results demonstrated that NRF-1
silencing promoted the apoptosis of 786-O cells (Fig. 4A and B). Compared with the control
group, NRF-1 shRNA 786-O cells showed an increase in the fraction
of cells in the G1 phase and a decrease in the
G2 phase of the cell cycle. These results indicated that
NRF-1 silencing resulted in G1 cycle arrest (Fig. 4C and D).
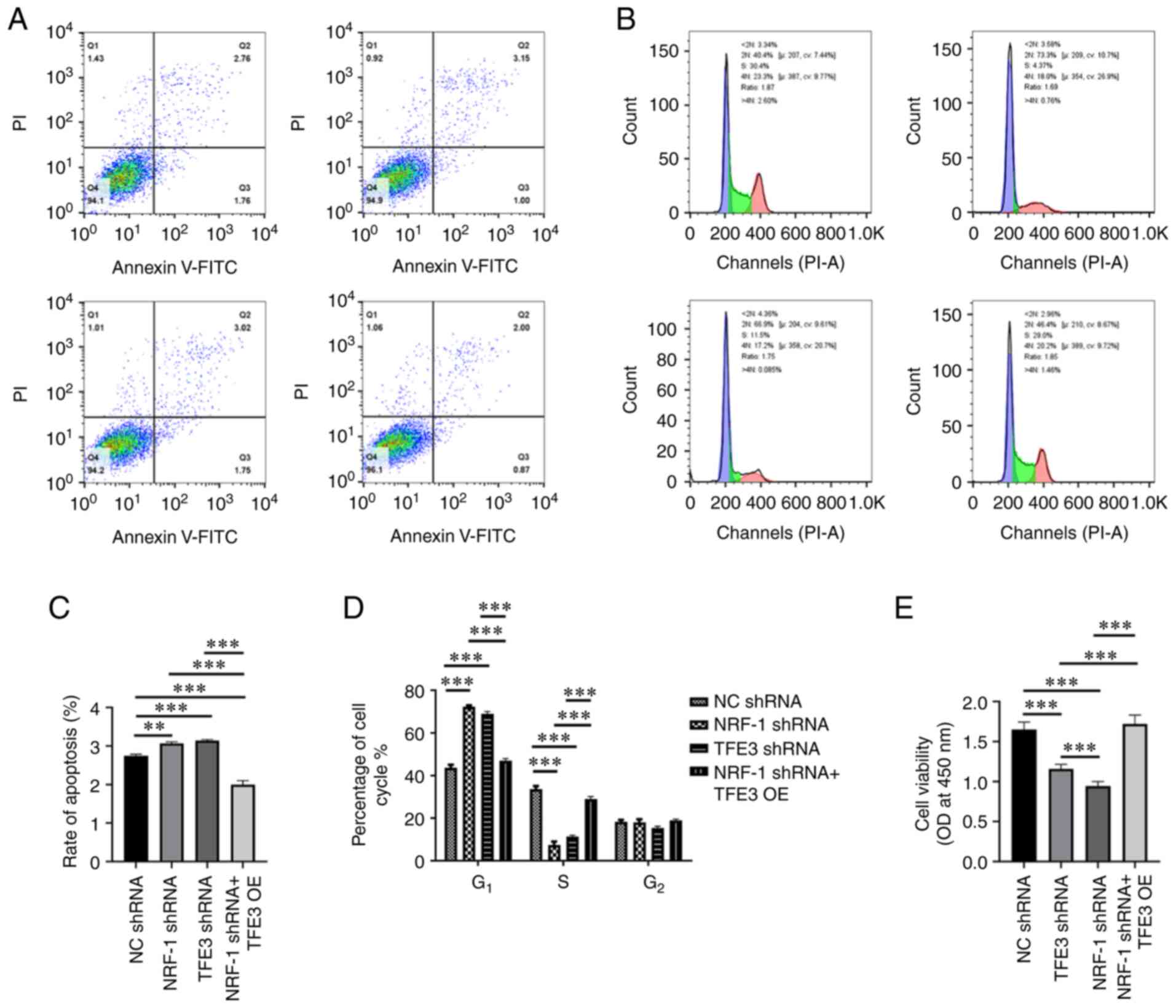
Further experiments showed that TFE3 and NRF-1
silencing promote cell apoptosis to varying degrees. Indeed,
following simultaneous NRF-1 silencing and TFE3 overexpression, the
apoptosis of 786-O cells was significantly reduced compared with
all other groups (Fig. 5A and C).
These results indicated that TFE3 played a key role in tumor cell
apoptosis. NRF-1 can participate in the regulation of cell
apoptosis by directly regulating the expression of TFE3
protein.
NRF-1 and TFE3 silencing alone resulted in
G1-phase cell cycle arrest in 786-O cells. By contrast,
following NRF-1 silencing and TFE3 overexpression, cell cycle
arrest in G1 phase was suppressed, restoring the
fraction of cells to levels similar to those of the control group
(Fig. 5B and D).
In order to detect changes in cell proliferation,
CCK-8 was used. Compared with the control group, NRF-1 and TFE3
silencing alone inhibited cell proliferation. However, following
TFE3 overexpression in NRF-1 shRNA cells, proliferation increased
compared with NRF-1 and TFE3 silencing alone (Fig. 5E).
Effect of NRF-1 expression on
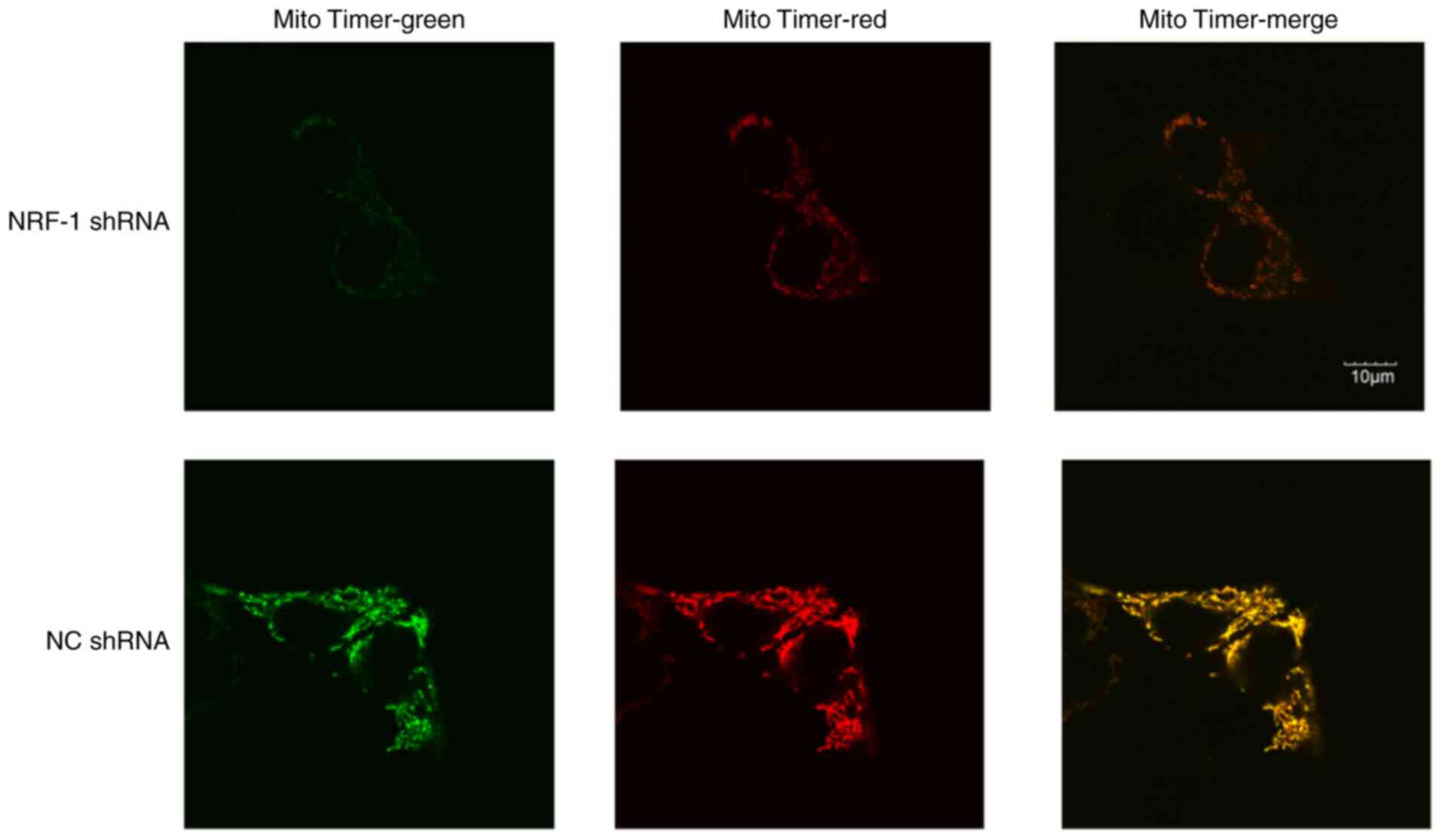
mitochondria production of tumor cells
To determine how changes in NRF expression might
affect mitochondria, the MitoTimer plasmid to detect changes in
mitochondrial formation in cells. Under the same shooting
conditions, significantly reduced fluorescence signals were
observed in the NRF-1 shRNA group, and down-regulation of
mitochondrial generation was observed. The results showed that the
fluorescence of cells in the NRF-1 shRNA group showed a significant
decrease compared with the NC (Fig.
6). Thus, the mitochondrial generation rate was slowed down
following knockdown of NRF-1.
Discussion
In the present study, the expression of the TFE3
gene was directly regulated by NRF-1, a crucial transcription
factor involved in oxidative phosphorylation and mitochondrial
biogenesis (19). The role of TFE3
and its fusion gene in renal cancer has not been fully elucidated
since its first discovery (9,10). The
occurrence and progression of tumors are closely related to energy
metabolism and NRF-1 coordinates synaptic activity and energy
metabolism (22,29). Previous reports have indicated that
TFE3 may be one of the downstream target genes of NRF-1 (30). Therefore, the aim of the present
study was to confirm the regulatory relationship between NRF-1 and
TFE3, in order to explore the role of NRF-1/TFE3 in the process of
tumorigenesis.
A previous study on NRF-1 in the literature have
focused on mitochondria and energy metabolism (31). Autophagy, as well as tumor-related
autophagy and even cellular immunity (32–35).
However, there is less research on the interaction between NRF-1
and TFE3. In the present study, NRF-1 directly regulated the
expression of the TFE3 protein. The present findings confirmed that
NRF-1 could functionally bind to the TFE3 promoter region, thereby
regulating the expression of the TFE3 protein. The mTOR signaling
pathway is activated in a variety of cancer types (36). It regulates the metabolism of amino
acids, glucose, nucleotides, fatty acids and lipids by changing the
expression and/or activity of several key metabolic enzymes and
participates in the control of cell growth and metabolism (36). Moreover, studies have shown that
metabolic changes, such as increased glucose or amino acid intake,
can affect mTOR signaling (37,38). The
present results indicated that following NRF-1 knockdown, the
expression levels of TFE3 and components of the mTOR signaling
pathway were also downregulated. The rate of apoptosis also
increased, while cell proliferation was suppressed. These results
indicated that NRF-1 regulated the mTOR pathway through TFE3.
A variety of signaling pathways are mediated by
TFE3, the dysregulation of which might contribute to renal
carcinogenesis (39). A previous
study have suggested that TFE3 can be a target of the mTOR
signaling pathway and is regulated by mTOR signaling (40). However, a previous study involving
CHIP-Seq detection of the SFPQ-TFE3 fusion gene have identified
other target genes that were related to PI3K/AKT/mTOR, including
PI3KCA, TSC1, AKT3, PTEN, 14-3-3, ITGB1, IGFR1 and IRS-1 genes
(41). This may also explain why, in
addition to being regulated by mTOR, TFE3 can also positively
regulate the expression of mTOR. In the present study, simultaneous
overexpression of TFE3 and NRF-1 silencing restored the
downregulated expression of molecules involved in the mTOR
signaling pathway, inhibited apoptosis, and increased
proliferation. These results indicated that the TFE3 protein itself
could also play a positive regulatory role on the mTOR signaling
pathway. The TFE3 protein is directly related to the apoptosis and
growth of cells. This is also consistent with a previous study
(42). There may be a circular
relationship between TFE3 and mTOR. Moreover, the present study
also confirmed the direct regulation of NRF-1 on the TFE3 gene.
Further experiments are needed to determine whether there is a
circular relationship between TFE3 and NRF-1.
NRF-1 itself is involved in the production of
mitochondria and the regulation of cell energy metabolism under
various conditions (43), and there
are few studies on the role of NRF-1 in renal cell carcinoma
(23). For tumor cells, metabolic
remodeling is an important feature (44). This allows tumor cells to adapt to
various environments and survive under the body's immune system
(22). TFE3 plays a key role in
certain renal cell carcinoma types, and NRF-1, as its upstream
regulatory gene, is highly related to its expression level
(23). These findings may provide
insight into TFE3-positive renal cancer treatment and suggest that
NRF-1 may become a new target for the treatment of this
condition.
Given the important role of TFE3 in tumors,
particularly tRCC, TFE3 may be an important target for researching
tumor treatment measures. As a direct upstream regulatory gene of
TFE3, NRF-1 may also play a key role in this process. This study
showed that NRF-1 directly regulates TFE3 and may influence
downstream tumorigenesis and progression through TFE3. Therefore,
understanding the specific regulatory effect of NRF-1 on TFE3 may
help discover new therapeutic targets for renal tumors.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by The National Natural
Science Foundation of China (grant no. 81572512) and The Beijing
Ronghe Medical Development Foundation. The funders had no role in
study design, data collection and analysis, decision to publish, or
preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WDG, CNZ and HQG conceived and designed the
experiments. WYZ, XD, BW and NL performed the experiments and
analyzed data. WYZ and XD wrote the manuscript. WDG and WYZ
confirmed the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Betschinger J, Nichols J, Dietmann S,
Corrin PD, Paddison PJ and Smith A: Exit from pluripotency is gated
by intracellular redistribution of the bHLH transcription factor
Tfe3. Cell. 153:335–347. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fisher DE, Carr CS, Parent LA and Sharp
PA: TFEB has DNA-binding and oligomerization properties of a unique
helix-loop-helix/leucine-zipper family. Genes Dev. 5:2342–2352.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hemesath TJ, Steingrímsson E, Mcgill G,
Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG,
Jenkins NA and Fisher DE: microphthalmia, a critical factor in
melanocyte development, defines a discrete transcription factor
family. Genes Dev. 8:2770–2780. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Muhle-Goll C, Gibson T, Schuck P, Schubert
D, Nalis D, Nilges M and Pastore A: The dimerization stability of
the HLH-LZ transcription protein family is modulated by the leucine
zippers: A CD and NMR study of TFEB and c-Myc. Biochemistry.
33:11296–11306. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vivian P, Ogmundsdóttir MH,
Bergsteinsdóttir K, Schepsky A, Phung B, Deineko V, Milewski M,
Steingrímsson E and Wilmanns M: Restricted leucine zipper
dimerization and specificity of DNA recognition of the melanocyte
master regulator MITF. Genes Dev. 26:2647–2658. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Raben N and Puertollano R: TFEB and TFE3:
Linking lysosomes to cellular adaptation to stress. Annu Rev Cell
Dev Biol. 32:255–278. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiong L, Chen X, Liu N, Wang Z, Miao B,
Gan W, Li D and Guo H: PRCC-TFE3 dual-fusion FISH assay: A new
method for identifying PRCC-TFE3 renal cell carcinoma in
paraffin-embedded tissue. PLoS One. 12:e01853372017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Magers MJ, Udager AM and Mehra R: MiT
family translocation-associated renal cell carcinoma: A
contemporary update with emphasis on morphologic, immunophenotypic,
and molecular mimics. Arch Pathol Lab Med. 139:1224–1233. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Martina JA, Diab HI, Brady OA and
Puertollano R: TFEB and TFE3 are novel components of the integrated
stress response. EMBO J. 35:479–495. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pastore N, Vainshtein A, Klisch TJ, Armani
A, Huynh T, Herz NJ, Polishchuk EV, Sandri M and Ballabio A: TFE3
regulates whole-body energy metabolism in cooperation with TFEB.
EMBO Mol Med. 9:605–621. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Martina JA, Diab HI, Lishu L, Jeong-A L,
Patange S, Raben N and Puertollano R: The nutrient-responsive
transcription factor TFE3 promotes autophagy, lysosomal biogenesis,
and clearance of cellular debris. Sci Signal. 7:ra92014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ploper D, Taelman VF, Robert L, Perez BS,
Titz B, Chen HW, Graeber TG, von Euw E, Ribas A and De Robertis EM:
MITF drives endolysosomal biogenesis and potentiates Wnt signaling
in melanoma cells. Proc Natl Acad Sci USA. 112:E420–E429. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iwasaki H, Naka A, Iida KT, Nakagawa Y,
Shimano H, Matsuzaka T, Ishii KA, Kobayashi K, Takahashi A, Yatoh
S, et al: TFE3 regulates muscle metabolic gene expression,
increases glycogen stores, and enhances insulin sensitivity in
mice. Am J Physiol Endocrinol Metab. 302:E896–E902. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nakagawa Y, Shimano H, Yoshikawa T, Ide T,
Tamura M, Furusawa M, Yamamoto T, Inoue N, Matsuzaka T, Takahashi
A, et al: TFE3 transcriptionally activates hepatic IRS-2,
participates in insulin signaling and ameliorates diabetes. Nat
Med. 12:107–113. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nijman SMB, Hijmans EM, Messaoudi SE, van
Dongen MMW, Sardet C and Bernards R: A functional genetic screen
identifies TFE3 as a gene that confers resistance to the
anti-proliferative effects of the retinoblastoma protein and
transforming growth factor-beta. J Biol Chem. 281:21582–21587.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Muller-Hocker J, Babaryka G, Schmid I and
Jung A: Overexpression of cyclin D1, D3, and p21 in an infantile
renal carcinoma with Xp11.2 TFE3-gene fusion. Pathol Res Pract.
204:589–597. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scarpulla RC: Nuclear control of
respiratory gene expression in mammalian cells. J Cell Biochem.
97:673–683. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Scarpulla RC: Nuclear control of
respiratory chain expression by nuclear respiratory factors and
PGC-1-related coactivator. Ann N Y Acad Sci. 1147:321–334. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Virbasius CA, Virbasius JV and Scarpulla
RC: NRF-1, an activator involved in nuclear-mitochondrial
interactions, utilizes a new DNA-binding domain conserved in a
family of developmental regulators. Genes Dev. 7:24311993.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang M, Yang Y, Yu J, Qiu J, Chen P, Wu Y,
Wang Q, Xu Z, Ge J, Yu K and Zhuang J: Tetramethylpyrazine in a
murine alkali-burn model blocks NFκB/NRF-1/CXCR4-signaling-induced
corneal neovascularization. Invest Ophthalmol Vis Sci.
59:2133–2141. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Satoh J, Kawana N and Yamamoto Y: Pathway
analysis of chip-seq-based NRF1 target genes suggests a logical
hypothesis of their involvement in the pathogenesis of
neurodegenerative diseases. Gene Regul Syst Bio. 7:139–152.
2013.PubMed/NCBI
|
|
22
|
Kimmelman AC and White E: Autophagy and
tumor metabolism. Cell Metab. 25:1037–1043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alam C, Hoque MT, Sangha V and Bendayan R:
Nuclear respiratory factor 1 (NRF-1) upregulates the expression and
function of reduced folate carrier (RFC) at the blood-brain
barrier. FASEB J. 34:10516–10530. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Solecki D, Bernhardt G, Lipp M and Wimmer
E: Identification of a nuclear respiratory Factor-1 binding site
within the core promoter of the human polio virus receptor/CD155
Gene. J Biol Chem. 275:12453–12462. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hernandez G, Thornton C, Stotland A, Lui
D, Sin J, Ramil J, Magee N, Andres A, Quarato G, Carreira RS, et
al: MitoTimer: A novel tool for monitoring mitochondrial turnover.
Autophagy. 9:1852–1861. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Williams JA, Zhao K, Jin S and Ding WX:
New methods for monitoring mitochondrial biogenesis and mitophagy
in vitro and in vivo. Exp Biol Med (Maywood). 242:781–787. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Motzer RJ, Escudier B, Oudard S, Hutson
TE, Porta C, Bracarda S, Grünwald V, Thompson JA, Figlin RA,
Hollaender N, et al: Efficacy of everolimus in advanced renal cell
carcinoma: A double-blind, randomised, placebo-controlled phase III
trial. Lancet. 372:449–456. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Scarpulla RC: Metabolic control of
mitochondrial biogenesis through the PGC-1 family regulatory
network. Biochim Biophys Acta. 1813:1269–1278. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jun-Ichi S, Natsuki K and Yoji Y: Pathway
analysis of ChIP-Seq-Based NRF1 target genes suggests a logical
hypothesis of their involvement in the pathogenesis of
neurodegenerative diseases. Gene Regul Syst Bio. 7:139–152.
2013.PubMed/NCBI
|
|
31
|
Evans MJ and Scarpulla RC: NRF-1: A
trans-activator of nuclear-encoded respiratory genes in animal
cells. Genes Dev. 4:1023–1034. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Taniguchi M, Nadanaka S, Tanakura S,
Sawaguchi S, Midori S, Kawai Y, Yamaguchi S, Shimada Y, Nakamura Y,
Matsumura Y, et al: TFE3 is a bHLH-ZIP-type transcription factor
that regulates the mammalian Golgi stress response. Cell Struct
Funct. 40:13–30. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zanocco-Marani T, Vignudelli T, Parenti S,
Gemelli C, Condorelli F, Martello A, Selmi T, Grande A and Ferrari
S: TFE3 transcription factor regulates the expression of MAFB
during macrophage differentiation. Exp Cell Res. 315:1798–1808.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zanocco-Marani T, Vignudelli T, Gemelli C,
Pirondi S, Testa A, Montanari M, Parenti S, Tenedini E, Grande A
and Ferrari S: Tfe3 expression is closely associated to macrophage
terminal differentiation of human hematopoietic myeloid precursors.
Exp Cell Res. 312:4079–4089. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shi CS, Shenderov K, Huang NN, Kabat J,
Abu-Asab M, Fitzgerald KA, Sher A and Kehrl JH: Activation of
autophagy by inflammatory signals limits IL-1β production by
targeting ubiquitinated inflammasomes for destruction. Nat Immunol.
13:255–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Murugan AK: mTOR: Role in cancer,
metastasis and drug resistance. Semin Cancer Biol. 59:92–111. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Saxton RA and Sabatini DM: mTOR signaling
in growth, metabolism, and disease. Cell. 168:960–976. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y, Kwok-Shing Ng P, Kucherlapati M,
Chen F, Liu Y, Tsang YH, de Velasco G, Jeong KJ, Akbani R,
Hadjipanayis A, et al: A Pan-cancer proteogenomic Atlas of
PI3K/AKT/mTOR pathway alterations. Cancer Cell. 31:820–832.e3.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Argani P, Hicks J, De Marzo AM, Albadine
R, Illei PB, Ladanyi M, Reuter VE and Netto GJ: Xp11 translocation
renal cell carcinoma (RCC): Extended immunohistochemical profile
emphasizing novel RCC markers. Am J Surg Pathol. 34:1295–1303.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li J, Wada S, Weaver LK, Biswas C, Behrens
EM and Arany Z: Myeloid Folliculin balances mTOR activation to
maintain innate immunity homeostasis. JCI Insight.
5:e1269392019.PubMed/NCBI
|
|
41
|
Damayanti NP, Budka JA, Khella HWZ, Ferris
MW, Ku SY, Kauffman E, Wood AC, Ahmed K, Chintala VN,
Adelaiye-Ogala R, et al: Therapeutic targeting of
TFE3/IRS-1/PI3K/mTOR axis in translocation renal cell carcinoma.
Clin Cancer Res. 24:5977–5989. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kauffman EC, Ricketts CJ, Rais-Bahrami S,
Yang Y, Merino MJ, Bottaro DP, Srinivasan R and Linehan WM:
Molecular genetics and cellular features of TFE3 and TFEB fusion
kidney cancers. Nat Rev Urol. 11:465–475. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang D, Zhang J, Lu Y, Luo Q and Zhu L:
Nuclear respiratory factor-1 (NRF-1) regulated hypoxia-inducible
factor-1α (HIF-1α) under hypoxia in HEK293T. IUBMB Life.
68:748–755. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li X, Wenes M, Romero P, Huang CC, Fendt
SM and Ho PC: Navigating metabolic pathways to enhance antitumour
immunity and immunotherapy. Nat Rev Clin Oncol. 16:425–441. 2019.
View Article : Google Scholar : PubMed/NCBI
|