Introduction
Breast cancer (BC) is the most common type of
malignant tumor (1); two million
new BC cases were diagnosed in 2018 (23% of all cancers), and the
disease ranks second in mortality rate overall worldwide (10.9% of
all cancers) (2). The majority of
patients initially respond well to treatment; however, BC cells
gradually develop resistance to conventional radiotherapy and
chemotherapy, which can result in treatment failure (3). Drug resistance during BC treatment
was found to be regulated by several different mechanisms,
including somatic mutations, epigenetic modifications and aberrant
changes in signaling pathways, as well as influences from tumor
microenvironment-induced modifications to drug transport systems
(4–6).
MicroRNAs (miRNAs/miRs) are a type of non-coding
endogenous RNA of 20–25 nucleotides in length, that bind to the
3′-untranslated region of target mRNAs to induce degradation or
translational inhibition, thereby negatively regulating gene
expression (7–9). Previous studies have reported that miRNAs play
an important role in reversing tumor drug resistance (10). In fact, evidence has suggested that
the abnormal expression of miRNAs, including miRNA-449, miR-140 and
miR-200a, promotes drug resistance in BC (11–13). Furthermore, the
abnormal expression of multiple miRNAs in BC was found to be
pathologically associated with radiotherapy and multidrug
resistance, including resistance to chemotherapy, endocrine therapy
and targeted therapy (14,15). Among the different miRNAs
identified to date, miR-320b was discovered to be involved in the
development of colorectal cancer, nasopharyngeal carcinoma and
osteosarcoma by regulating several processes, such as tumor cell
proliferation, metastasis and invasion, via targeting various
signaling pathways (16–18). Therefore, further investigations into
the specific roles of miRNAs in the development of drug resistance
in BC are likely to be of great clinical significance.
RNA binding proteins (RBPs) have a high affinity
towards miRNAs, and are involved in RNA splicing, polyadenylation,
sequence editing, RNA transport, maintenance of RNA stability and
degradation, intracellular localization and translation regulation
(19). RNA binding motif protein
38 (RBM38) is located at chromosome 20q13 and belongs to the RNA
recognition motif RBP family (20). Previous studies have reported that
RBM38 suppresses cellular migration and invasiveness in BC by
directly binding to estrogen receptor-α transcripts or the tumor
suppressor mutant, p53 (21,22).
Adriamycin (ADM) is the current drug used for combination adjuvant
BC treatment (23). However, ADM
resistance frequently occurs following BC treatment (24). Although studies have demonstrated
that RBM38 is closely associated with the inhibition of BC
tumorigenesis (21,25), the present study aimed to further
determine whether RBM38 facilitated ADM resistance in BC. By
utilizing PCR, western blotting, Transwell assays and flow
cytometry, regulation of the miR-320b/RBM38 signaling axis in
drug-resistant BC was further investigated.
Materials and methods
Patient studies
A total of 5 ADM-sensitive and 5 ADM-resistant BC
samples were obtained from patients with BC (age range, 33–55
years), cryopreserved in liquid nitrogen and then stored at −80°C.
These patients only treated with ADM or ADM combined therapy and
underwent surgery at the Affiliated Hospital of Nantong University
(Nantong, China) between January 2017 and December 2018 were
included in this study. The experimental protocol was approved by
the ethics committee of The Affiliated Hospital of Nantong
University (approval no. 2019032174; Nantong, China), and was
performed in accordance with the principles of the 1964 Declaration
of Helsinki. All patients provided written informed consent prior
to the commencement of the study.
Cell lines and culture
The human ADM-sensitive BC cell line, MCF-7/S, and
human ADM-resistant BC cell line, MCF-7/A, were both purchased from
the American Type Culture Collection. The cells were cultured in
DMEM supplemented with 10% FBS and 100 U/ml penicillin (all Gibco;
Thermo Fisher Scientific, Inc.), and maintained at 37°C with 5%
CO2.
Reverse transcription-quantitative
(RT-q) PCR
For RT-qPCR analysis, MCF-7/S and MCF-7/A cells were
harvested in the logarithmic growth phase, and the clinical BC
samples were ground using liquid nitrogen. Total RNA was extracted
from the cells and tissue samples using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) for 5 min at 25°C.
Then, 200 µl chloroform was added to the solution for an additional
15 min at 25°C. The mixture was centrifuged at 12,000 × g at 4°C
for 15 min to obtain the RNA sediment, and 0.5 ml isopropyl alcohol
was then added. The RNA extract was dried and stored at −80°C until
required.
Total RNA was reverse transcribed into cDNA using
the PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol. qPCR analysis of RBM38
and miR-320b expression was subsequently performed using a SYBR
Prime Script RT-PCR kit (Takara Biotechnology Co., Ltd.), according
to the manufacturer's instructions. The following thermocycling
conditions were used for amplification: 94°C for 10 min, followed
by 40 cycles of 94°C for 30 sec, 55–58°C for 30 sec, and 72°C for
45 sec, and a final cycle at 72°C for 10 min. Relative expression
levels of RBM38 and miR-320b were calculated using the
2−ΔΔCq method (26),
and RBM38 expression was normalized to that of β-actin expression,
while miR-320b was normalized to U6 expression. The primers used
were: RBM38 forward 5′-TGAACTTTGACGGGAGGAGC-3′ and reverse,
5′-TGATGGGGTTCGGGTCTTTG-3′; β-actin forward, 5′-CTTCGCGGGCGACGAT-3′
and reverse, 5′-ACATAGGAATCCTTCTGACCCA-3′; miR-320b forward,
5′-GATGCTGAAAAGCTGGGTTG-3′ and reverse,
5′-TATGGTTGTTCTGCTCTCTGTCTC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Transfection
Small interfering RNA (siRNA/si) targeting RBM38
(siR-RBM38, 20 nM), siR-negative control (NC, 20 nM),
pcDNA3.1-RBM38 overexpression vector (4 µg/ml), pcDNA3.1 empty
vector (acting as the overexpression NC, 4 µg/ml), miR-320b
inhibitor (50 nM), inhibitor NC (50 nM), miR-320b mimics (50 nM)
and mimics NC (50 nM) were purchased from Shanghai GenePharma Co.,
Ltd. The constructs were transfected into MCF-7/A cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 48 h according to the manufacturer's
protocol. The sequences used were: siR-NC, forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′; siR-RBM38, forward,
5′-CCAGACGGGCUUUGCCAUUTT-3′ and reverse,
5′-AAUGGCAAAGCCCGUCUGGTT-3′; miR-320b inhibitor,
5′-UUGCCCUCUCAACCCAGCUUUU-3′; inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′; miR-320b mimics, forward,
5′-AAAAGCUGGGUUGAGAGGGCAA-3′ and reverse,
5′-GCCCUCUCAACCCAGCUUUUUU-3′; and mimics NC, forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′. The following studies were conducted
48 h after transfection.
MTT assay
Briefly, 5×103 MCF-7/A cells/well were
seeded into 96-well plates and incubated with 100 µl DMEM at 37°C
(5% CO2). After a 24-h incubation period, the cells were
treated with increasing concentrations of ADM (1, 2.5, 5, 10, 20,
40, 80,160 and 320 µM) in DMEM containing 10% FBS for 48 h at 37°C.
Subsequently, 20 µl MTT reagent was added per well, and the cells
cultured for a further 4 h at 37°C. Following incubation, 150 µl
DMSO was added to each well to dissolve the purple formazan
crystals, and the absorbance was measured within 10 min at a
wavelength of 570 nm (OD570) using a microplate reader.
Transwell assay
The invasive ability of MCF-7/A cells was analyzed
using a Transwell assay. Briefly, 200 µl cell suspension
(1×105 cells/well) was plated into the
Matrigel-precoated upper chamber (cat. no. 356234; BD Biosciences)
of a 24-well Transwell plate (Corning, Inc.; pore size, 8-µm). The
upper chamber was filled with 50 µl serum-free DMEM medium
supplemented with 10 g/l BSA (Thermo Fisher Scientific, Inc.),
while the lower chamber was filled with DMEM containing 10% FBS.
Following a 72 h-incubation period at 37°C, the invasive cells were
fixed with 4% phosphate-buffered neutral formalin at room
temperature for 20 min and then washed with PBS for three times,
then stained by crystal violet for 5 min at room temperature.
Invasive cells were visualized in five randomly selected fields of
view using an inverted light microscope (Olympus Corporation).
Western blotting
Total protein was extracted from MCF-7/A cells using
RIPA lysis buffer (Beyotime Institute of Biotechnology)
supplemented with 1% PMSF. Total protein was quantified using a BCA
protein assay kit, 50 µg protein was resolved by 5 or 10% SDS-PAGE,
and then transferred to PVDF membranes (EMD Millipore). The
membranes were blocked with 5% non-fat milk in 0.1% Tris-buffered
saline-Tween (TBST) at room temperature for 2 h, and then incubated
with primary antibodies (all from Abcam) against RBM38 (cat. no.
ab200403), phosphorylated (p)-PI3K (cat. no. ab182651), PI3K (cat.
no. ab32089), p-AKT (cat. no. ab38449), AKT (cat. no. ab8805), Bax
(cat. no. ab32503), Bcl-2 (cat. no. ab32124), breast cancer
resistance protein (BCRP; cat. no. ab207732), multiple drug
resistant protein 1 (MDR1; cat. no. ab170904) and β-actin (cat. no.
ab8226) diluted at 1:1,000 at 4°C overnight with gentle rocking.
β-actin served as the internal loading control. After washing with
TBST, the membranes were incubated with the appropriate horseradish
peroxidase-conjugated secondary antibodies (SA00001-1 or SA00001-2;
ProteinTech Group, Inc.) diluted at 1:2,000 for 2 h at 37°C. After
extensive washing with TBST, the proteins were visualized using an
ECL detection kit in accordance with the manufacturer's
recommendations (EMD Millipore). The integrated density of the
bands was quantified using Image Lab software version 6.1 (Bio-Rad
Laboratories, Inc.).
Flow cytometric analysis of apoptosis
and the cell cycle
Cell cycle distribution and apoptosis levels were
analyzed via flow cytometry after transfection. Both the Annexin
V-FITC Apoptosis Detection and Cell Cycle Detection kits were
purchased from Nanjing KeyGen Biotech Co., Ltd., and used according
to the manufacturers' protocols. The staining buffer contained
RNase and the cells were permeabilized for cell cycle assay. The
apoptosis ratio was calculated as the percentage of early plus late
apoptotic cells.
Dual luciferase reporter assay
The binding microRNAs of RBM38 were predicted by
miRanda algorithm (http://www.microrna.org/microrna/home.do) and the
interaction was demonstrated by dual luciferase reporter assay. The
wild-type (WT) or mutated (Mut) RBM38 sequences were inserted into
the pGL3 vector (Promega Corporation) to construct luciferase
reporter plasmids, which were subsequently co-transfected with a
miR-320b mimics or mimic NC into cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Following 24 h of incubation, the relative Firefly luciferase
activity was determined using a Dual Luciferase Reporter assay
system (Promega Corporation), and normalized to that of
Renilla luciferase.
Statistical analysis
Statistical analysis was performed using SPSS 13.0
(SPSS, Inc.) or GraphPad Prism 7.0 (GraphPad Software, Inc.)
software. Data are presented as the mean ± SD of triplicate
measurements. Two-tailed unpaired Student's t-test was used to
determine statistical differences between two groups, while
multiple comparisons between groups were performed using one-way
ANOVA followed by a Tukey's post hoc test. P<0.05 was considered
to indicate a statistically significant difference.
Results
RBM38 expression levels are
upregulated in ADM-resistant BC tissues and cell lines
Although a previous study reported that RBM38
expression levels were upregulated in BC tissues (25), to the best of our knowledge, the
present study was the first to identify differences in RBM38
expression between ADM-sensitive and ADM-resistant BC tissues and
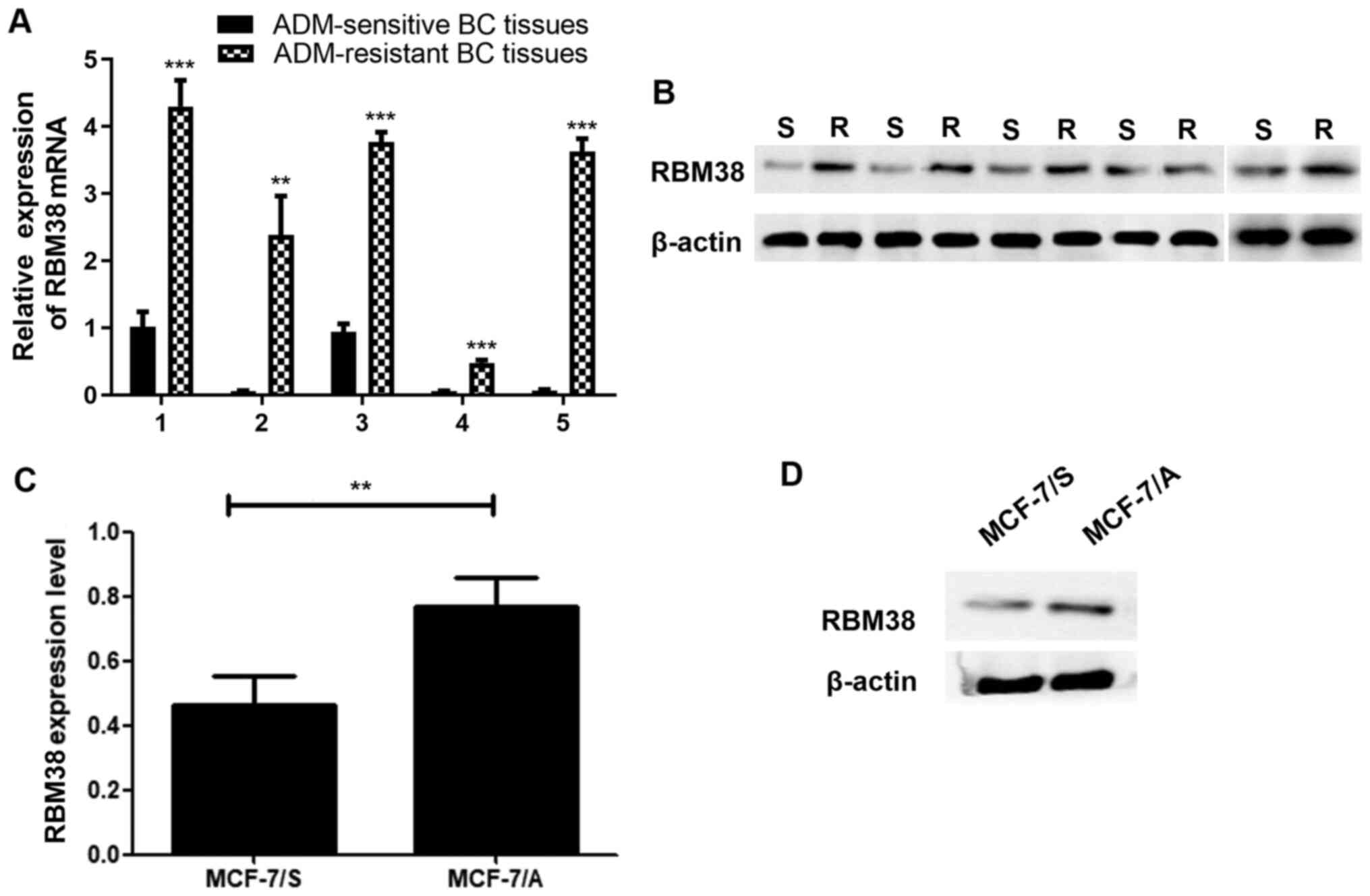
cells. The expression levels of RBM38 in ADM-resistant BC tissues
were first analyzed using RT-qPCR. The results revealed that RBM38
expression was significantly upregulated in ADM-resistant BC
tissues compared with that in ADM-sensitive BC tissues (Fig. 1A and B). MCF-7 cells are a common
cell line used to study BC, and numerous studies have used MCF-7/S
and MCF-7/A cells to investigate the underlying mechanism of ADM
resistance therein (27–29). Therefore, these cell lines were also
used in the present study. The differences in proliferation between
MCF-7/S and MCF-7/A cells were initially determined. Notably, the
proliferative ability of MCF-7/A cells was found to be
significantly increased compared with that of MCF-7/S cells
(Fig. S1). The expression levels
of RBM38 in both MCF-7/S and MCF-7/A cells were also determined.
The results demonstrated that RBM38 mRNA and protein expression
levels were upregulated in MCF-7/A cells compared with those in
MCF-7/S cells (Fig. 1C and D).
These results prompted further investigation into the association
between RBM38 and ADM resistance in BC.
Overexpression of RBM38 increases the
resistance of MCF-7/A cells to ADM
To further investigate the role that RBM38 plays in
ADM-resistant BC, the MCF-7/A cell line was used in subsequent
experiments. siR-NC and siR-RBM38, or pcDNA3.1 and pcDNA3.1-RBM38,
were separately transfected into MCF-7/A cells for RBM38-knockdown
or overexpression experiments, respectively; transfection
efficiency was determined using RT-qPCR and western blotting. The
results revealed that transfection with siR-RBM38 significantly
downregulated the expression levels of RBM38, while transfection
with pcDNA3.1-RBM38 upregulated expression levels, compared with
the respective controls (Fig. 2A and
B). Next, the IC50 values of ADM across the
different groups were determined using an MTT assay.
RBM38-knockdown cells exhibited lower IC50 values
compared with the corresponding RBM38-overexpressing cells
(Fig. 2C), which suggested that
RBM38 may affect the sensitivity of BC cells to ADM. A Transwell
assay was also used to investigate the effect of RBM38 on the
invasiveness of ADM-resistant BC cells. Invasion ability was
significantly decreased in siR- RBM38 group compared with siR-NC
group, while the invasive ability was markedly increased in the
pcDNA3.1-RBM38 group compared with the pcDNA3.1 group (Fig. 2D), which suggested that RBM38 may
decrease ADM resistance in BC cells by increasing their invasive
capacity.
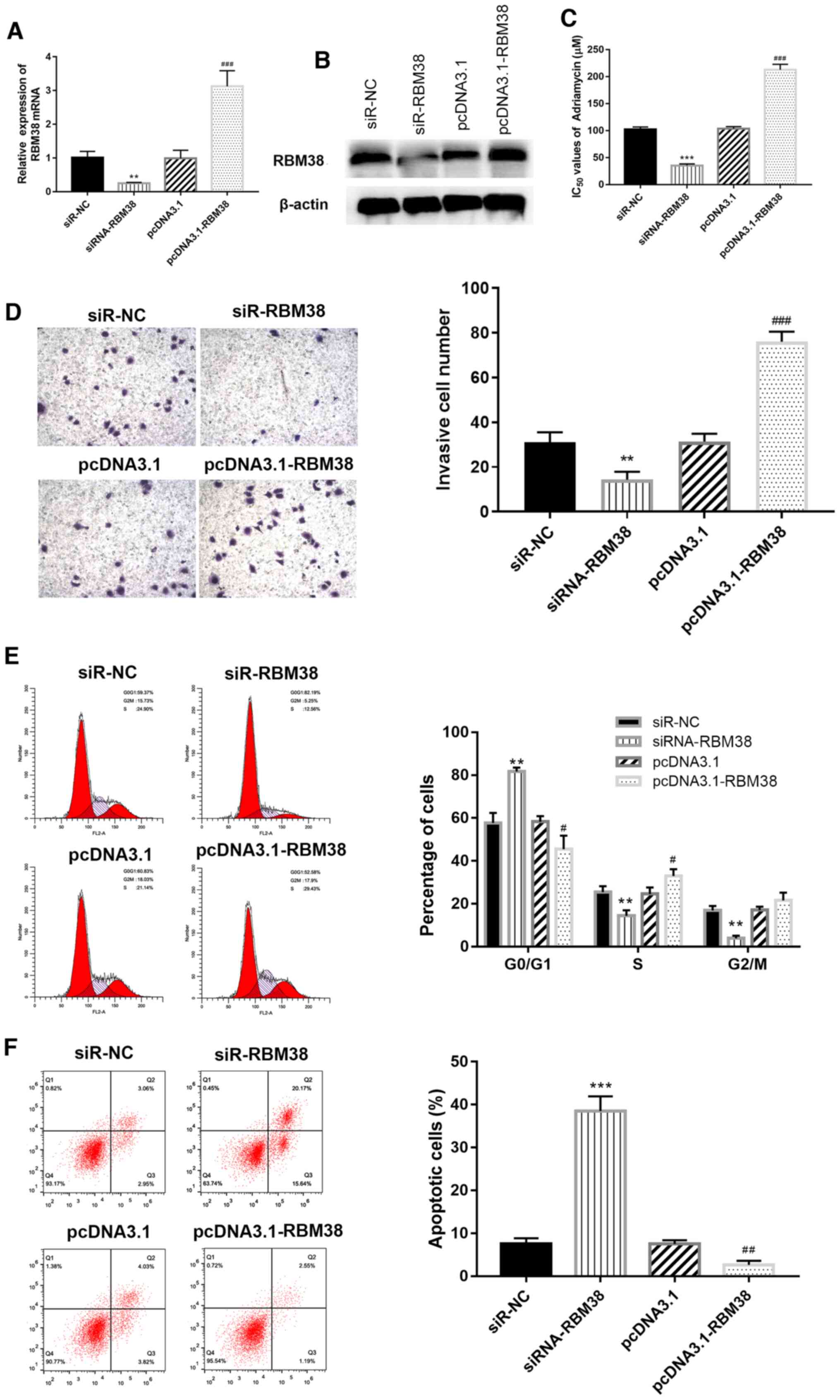 | Figure 2.RBM38 impairs the sensitivity of BC
cells to ADM, and inhibits the invasive ability of ADM-resistant BC
cells. (A) mRNA and (B) protein expression levels of RBM38 in
MCF-7/A cells. (C) MCF-7/A cells overexpressing RBM38 were less
susceptible to ADM treatment. Furthermore, knockdown of RBM38
decreased the IC50 value of ADM. (D) Invasive ability of
si-RBM38-transfected MCF-7/A cells, magnification, ×100. (E) Cell
cycle distribution and (F) apoptosis analyses were performed using
flow cytometry. **P<0.01 and ***P<0.001 vs. si-NC;
#P<0.05, ##P<0.01 and
###P<0.001 vs. pcDNA3.1. RBM38, RNA binding motif
protein 38; BC, breast cancer; ADM, Adriamycin; A, ADM-resistant
cells; si, small interfering RNA; NC, negative control. |
Furthermore, the effects of RBM38 overexpression or
silencing on cell cycle distribution were determined using flow
cytometry. As shown in Fig. 2E,
the overexpression of RBM38 decreased the percentage of cells in
the G0/G1 phase compared with the pcDNA3.1
group, while the knockdown of RBM38 increased the
G0/G1 phase ratio compared with the siR-NC
group (P<0.01). Similarly, flow cytometric detection of
apoptosis demonstrated that the overexpression of RBM38
significantly reduced apoptosis levels compared with the pcDNA3.1
group, while RBM38 silencing exerted the opposite effect (Fig. 2F). These findings suggested that
RBM38 may promote BC cell proliferation by affecting apoptosis and
the cell cycle.
miR-320b negatively regulates
RBM38
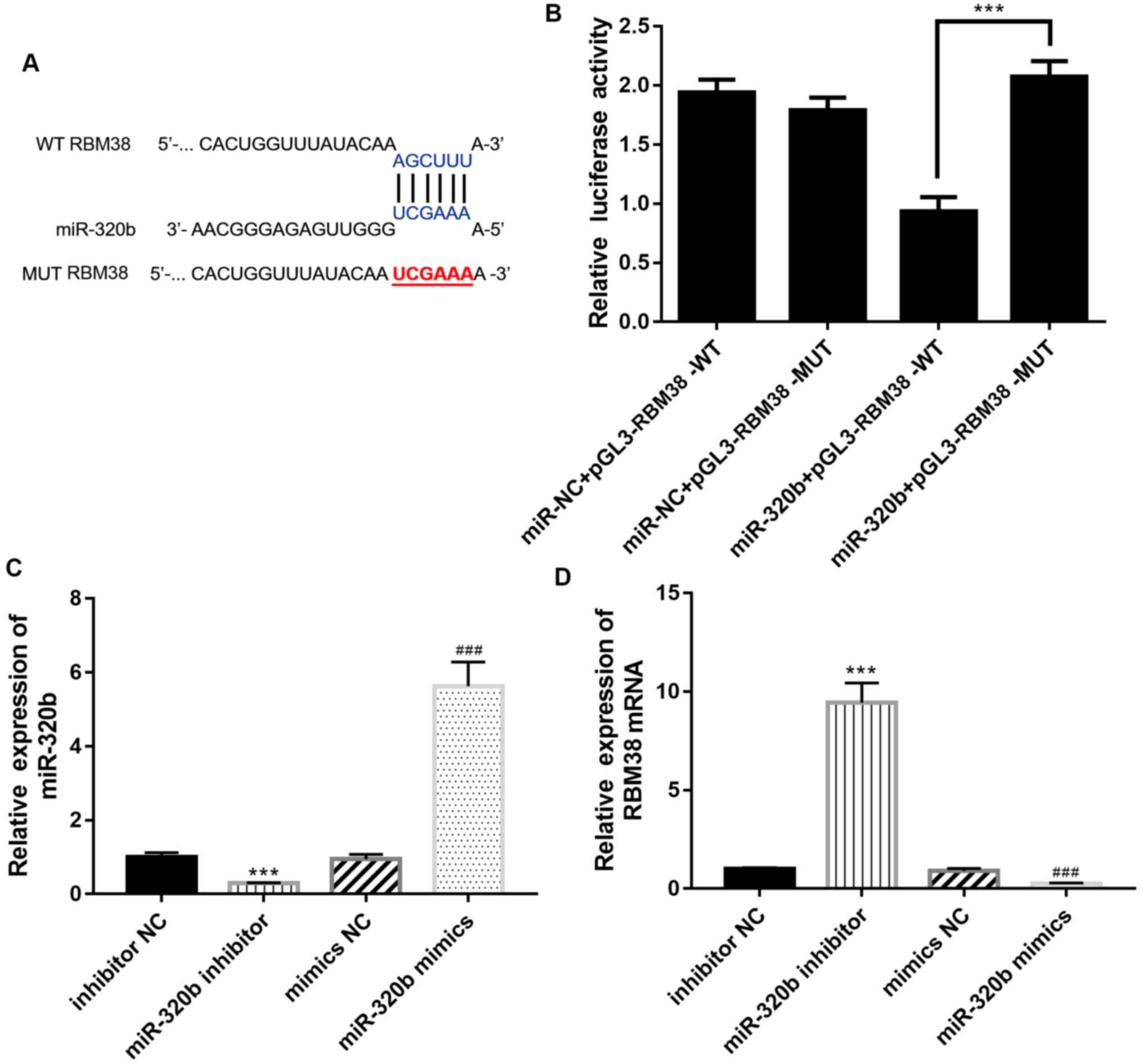
The miRanda algorithm (http://www.microrna.org/microrna/home.do) was used to
predict the presence of a WT RBM38 binding site in the miR-320b
sequence (Fig. 3A), which
suggested the existence of an interaction between RBM38 and
miR-320b during the progression of ADM-resistant BC. To verify this
prediction, a dual luciferase reporter assay was used to confirm
the binding relationship between miR-320b and RBM38. The results
indicated that WT RBM38 was able to directly bind to miR-320b to
significantly reduce relative luciferase activity (Fig. 3B).
Transfection efficiencies of the miR-320b mimics and
inhibitor in BC cells were determined using RT-qPCR; the results
revealed that compared with the inhibitor NC group, the expression
levels of miR-320b were downregulated in the miR-320b inhibitor
group; conversely, compared with the mimics NC group, the
expression levels of miR-320b were upregulated in the miR-320b
mimics group (Fig. 3C). The
association between RBM38 and miR-320b was determined using RT-qPCR
with MCF-7/A cells transfected with the miR-320b inhibitor or
miR-320b mimics. Following the transfection of cells with the
miR-320b inhibitor, the expression levels of RBM38 were upregulated
compared with the inhibitor NC group (Fig. 3D), while the expression levels of
RBM38 were downregulated following transfection with the miR-320b
mimics compared with the mimics NC group (P<0.001), suggesting
that RBM38 may be negatively regulated by miR-320b.
miR-320b restores sensitivity to ADM
and inhibits the resistance capacity of MCF-7/A cells
The expression levels of miR-320b in ADM-resistant
BC tissues and cells (MCF-7/A) were also analyzed using RT-qPCR.
The results demonstrated that miR-320b expression was downregulated
in ADM-resistant BC tissues and cells compared with ADM-sensitive
BC tissues and cells, respectively (Fig. 4A and B), which indicated that
miR-320b may also affect drug resistance in BC. Subsequently,
western blotting was performed to identify changes in RBM38
expression levels following transfection with either the miR-320b
inhibitor or mimics. The results revealed that the miR-320b
inhibitor upregulated the expression levels of RBM38 in MCF-7/A
cells compared with the inhibitor NC group, while the
overexpression of miR-320b downregulated RBM38 expression (Fig. 4C). To investigate whether miR-320b
may be a favorable anti-resistance factor for ADM-resistant BC, an
MTT assay was conducted to determine changes in the cell inhibition
rate of ADM in MCF-7/A cells. Notably, the silencing of miR-320b
increased the IC50 value of ADM compared with the
inhibitor NC group, while miR-320b mimics significantly decreased
the IC50 value compared with the mimic NC group
(Fig. 4D). These results indicated
that the overexpression of miR-320b may effectively potentiate cell
sensitivity towards ADM.
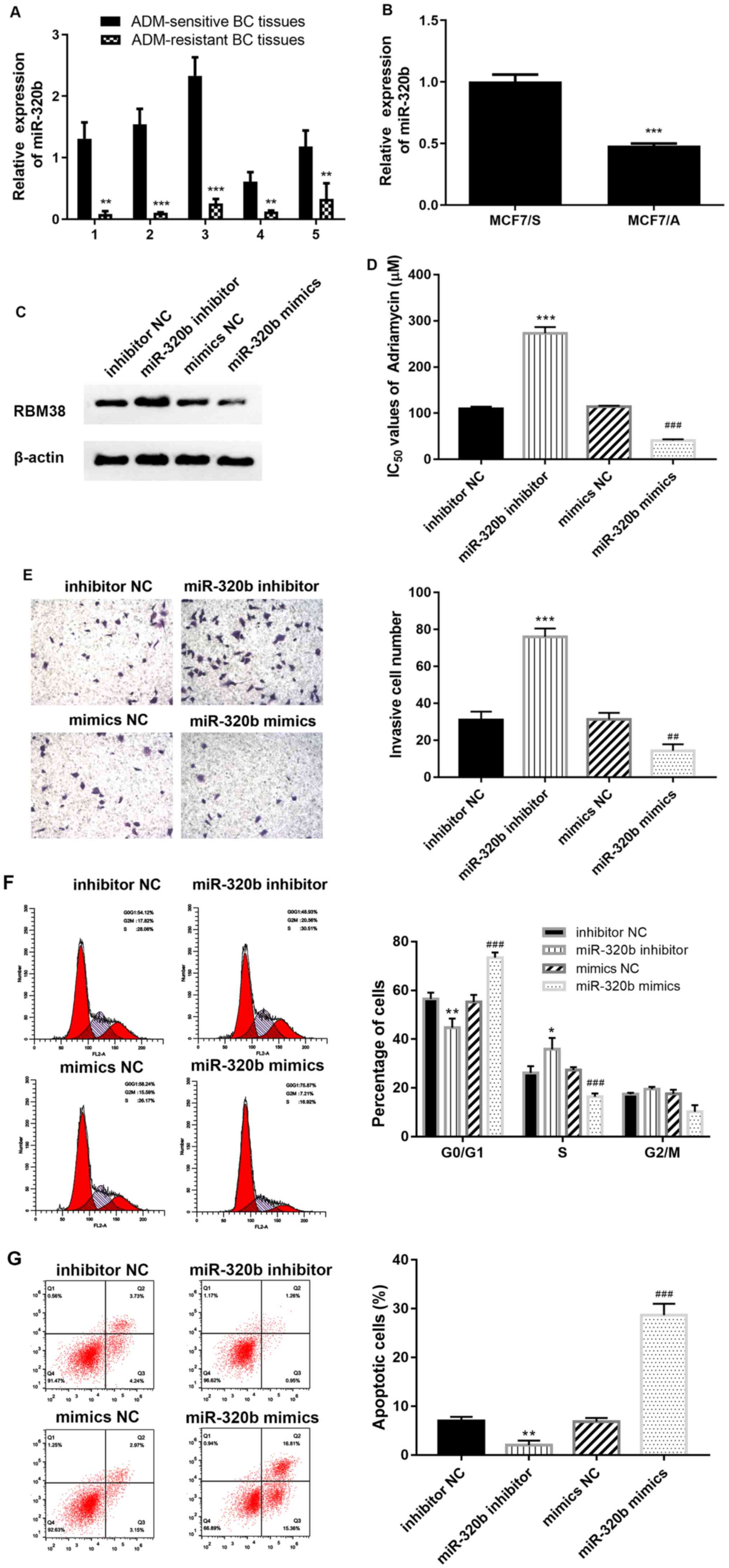 | Figure 4.miR-320b promotes the resistance of
BC cells to ADM. miR-320b expression levels were downregulated in
(A) ADM-resistant BC tissues and (B) MCF-7/A cells compared with
ADM-sensitive BC tissues and MCF-7/S cells, respectively. (C)
Western blotting demonstrated that RBM38 expression was
downregulated in the miR-320b mimics group. (D) miR-320b-inhibited
MCF-7/A cells were less susceptible to ADM, and overexpression of
miR-320b decreased the IC50 value of ADM. (E) Transwell
assays indicated that miR-320b inhibited the invasive ability of
ADM-resistant BC cells, magnification, ×100. (F) Cell cycle
distribution and (G) apoptosis were analyzed via flow cytometry.
*P<0.05, **P<0.01 and ***P<0.001 vs. inhibitor NC, or as
indicated; ##P<0.01 and ###P<0.001 vs.
mimic NC. miR, microRNA; BC, breast cancer; ADM, Adriamycin; S,
ADM-sensitive cells; A, ADM-resistant cells; RBM38, RNA binding
motif protein 38; NC, negative control. |
A Transwell assay was subsequently conducted to
determine the effect of miR-320b on cellular invasion capacity.
Compared with the respective NC groups, the number of invasive
cells was increased in the miR-320b inhibitor group, but decreased
in the miR-320b mimics group (Fig.
4E). These results suggested that miR-320b may be a favorable
factor for inhibiting cellular invasiveness and restoring the
sensitivity of resistant BC cells to ADM. Next, flow cytometry was
used to investigate the cell cycle distribution and apoptosis
levels. The results revealed that the ratio of cells in the
G0/G1 phase was decreased (Fig. 4F) and apoptosis was reduced
(Fig. 4G) following the inhibition
of miR-320b compared with the inhibitor NC group. However, the
overexpression of miR-320b significantly increased the number of
cells in the G0/G1 phase (Fig. 4F), as well as the apoptotic rate
(Fig. 4G) compared with the mimic
NC group. These findings suggested that miR-320b may be a crucial
factor involved in promoting the apoptosis, and inhibiting the
proliferation, of ADM-resistant BC cells.
RBM38 rescues miR-230b-induced
resistance by mediating the expression levels of proteins
associated with apoptosis, drug resistance and the cell cycle
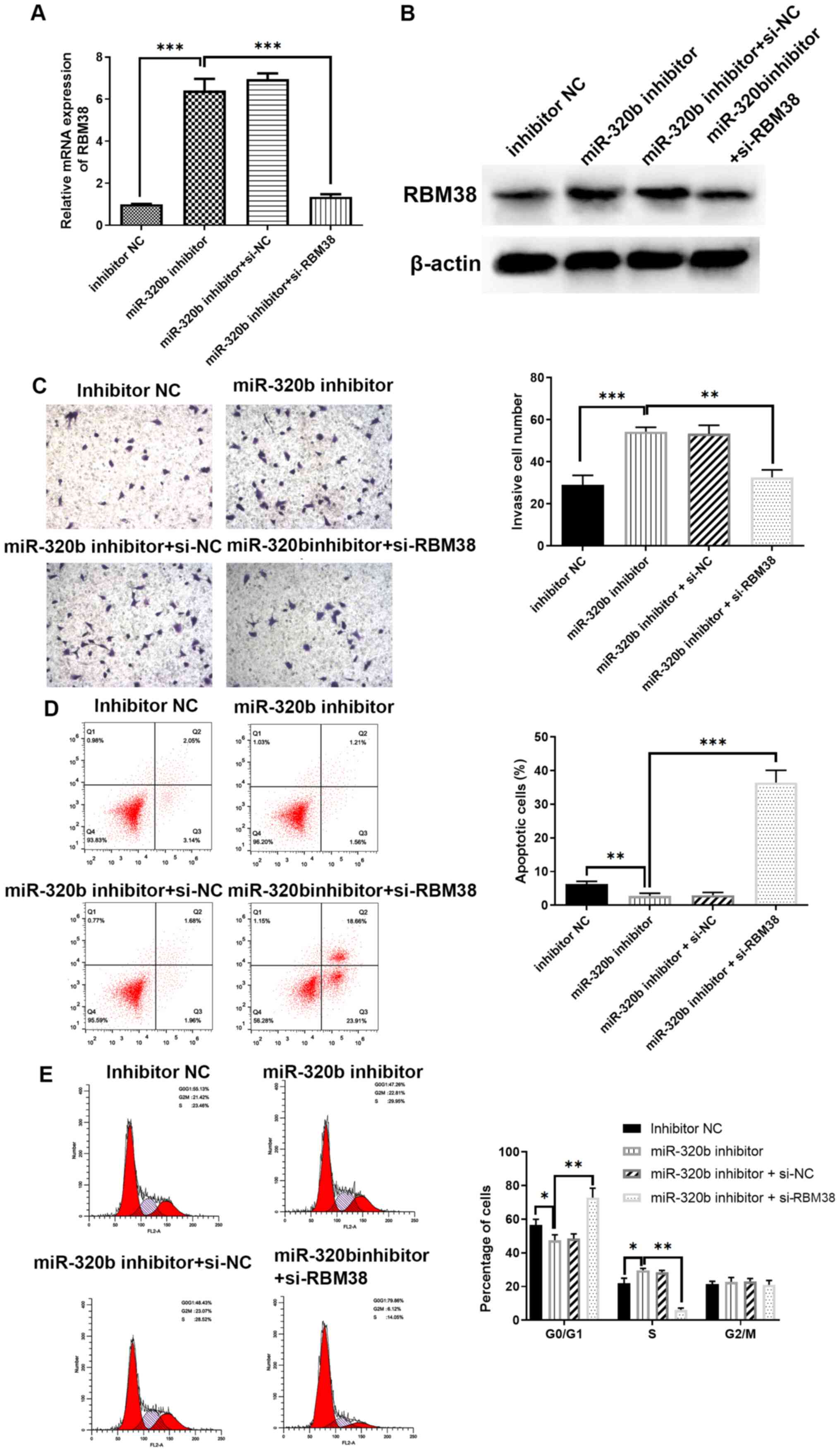
The association between miR-320b and RBM38 was
further investigated by transfecting MCF-7/A cells with an miR-320b
inhibitor, or co-transfection with an miR-320b inhibitor and
si-RBM38. The transfection efficiency was determined using RT-qPCR.
miR-320b inhibition significantly upregulated the level of RBM38
mRNA compared with the inhibitor NC group (Fig. 5A), and siR-RBM38 markedly reduced
this trend compared with the miR-320b inhibitor group (Fig. 5A); the resultant trend was also
demonstrated by western blotting (Fig.
5B). Subsequently, Transwell assays were conducted to detect
any alterations in the invasive capacity of the different groups.
The results revealed that miR-320b inhibition significantly
increased the invasive cell number compared with the inhibitor NC
group (Fig. 5C). However, the
combined knockdown of RBM38 reversed the effect of miR-320b
silencing compared with the miR-320b inhibitor alone group
(Fig. 5C). Furthermore, flow
cytometric analysis demonstrated that apoptotic rate was notably
increased in the miR-320b inhibitor and si-RBM38 co-transfection
group compared with the miR-320b inhibitor alone group (Fig. 5D). Cell cycle analysis also
demonstrated that the inhibition of miR-320b inhibited
G0/G1 phase arrest, while the silencing of
RBM38 further rescued the effect induced by miR-320b inhibition
Fig. 5E).
The exact underlying mechanism of action of the
RBM38/miR-320b axis in ADM resistance remains to be determined.
Hence, the present study further investigated various common tumor
modulatory signaling pathways and the expression levels of proteins
related to the cell cycle, apoptosis and drug resistance, to
determine their degree of contribution to the regulatory effects of
the miR-320b/RBM38 signaling axis. The protein expression levels of
the selected factors were indicated in Fig. 6A, and the quantification results
revealed that the expression levels of MDR1 (Fig. 6B) and BCRP (Fig. 6C) were upregulated in miR-320b
inhibitor-transfected cells compared with the inhibitor NC group,
and downregulated in RBM38-silenced cells compared with the
miR-320b inhibitor group. The levels of Bax were downregulated
(Fig. 6D), while those of Bcl-2
were upregulated (Fig. 6E),
following miR-320b inhibition. . In addition, western blot analysis
demonstrated that transfection with the miR-320b inhibitor
upregulated the p-PI3K/PI3K (Fig.
6F) and p-AKT/AKT (Fig. 6G)
ratios compared with the inhibitor NC group. Conversely,
co-transfection with si-RBM38 and the miR-320b inhibitor
significantly downregulated the p-PI3K/PI3K and p-AKT/AKT ratios
compared with the miR-320b inhibitor alone group. These findings
indicated that the miR-320b/RBM38 signaling axis may regulate the
activation of the PI3K/AKT signaling pathway in ADM-resistant BC
cells. Collectively, the knockdown of miR-320b was found to promote
BC cell resistance to ADM by influencing the PI3K/AKT signaling
pathway and apoptosis, as well as the expression levels of drug
resistance-related proteins, while silencing RBM38 reversed the
effects induced by the miR-320b inhibitor.
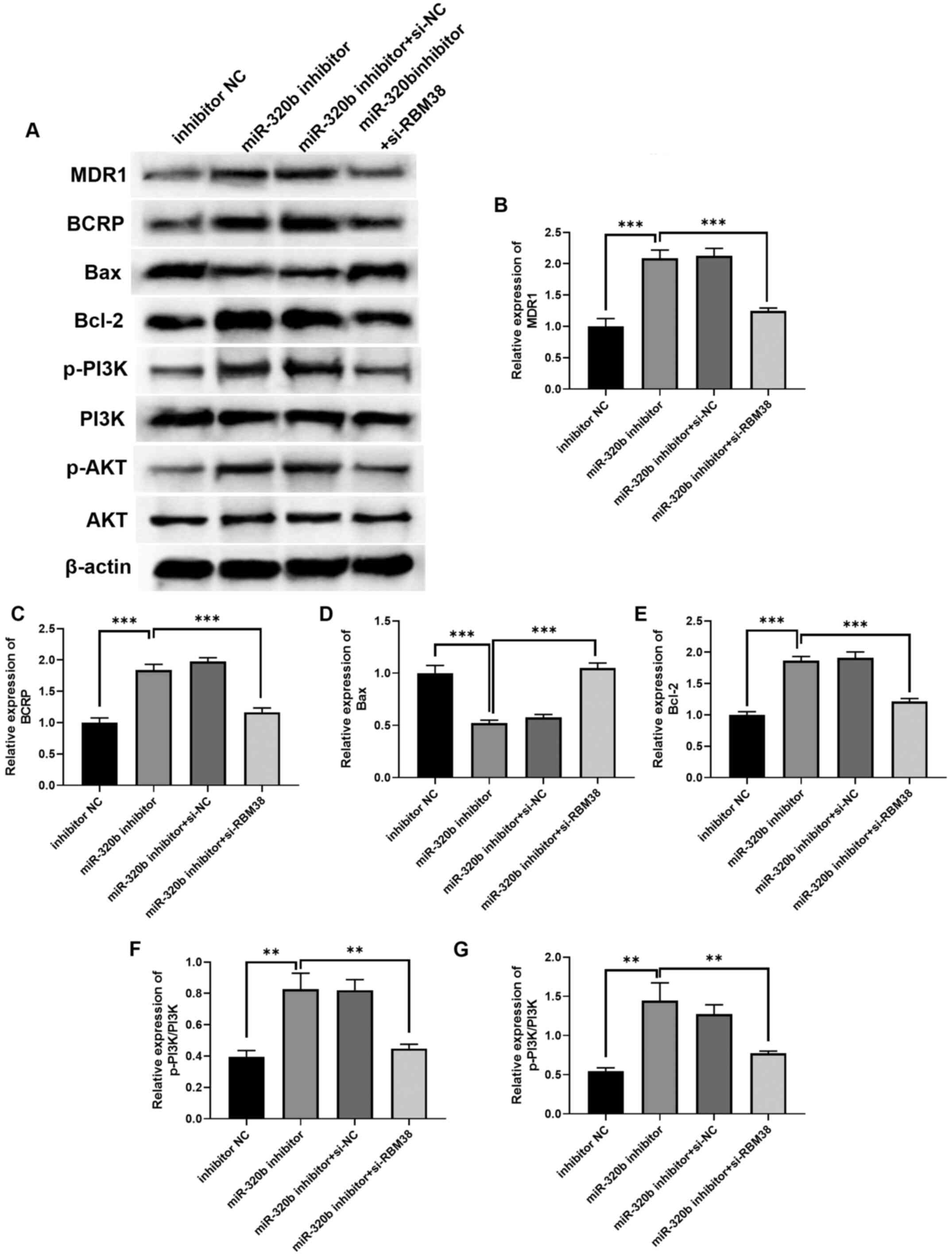 | Figure 6.RBM38 affects ADM resistance in BC by
moderating the expression levels of proteins related to apoptosis,
drug resistance and the cell cycle. (A) Western blotting
demonstrated that the RBM38/miR-320b signaling axis regulated BC
cell biological processes by altering the expression levels of
PI3K/AKT signaling pathway-, cell cycle- and drug
resistance-related proteins. The quantification of (B) MDR1, (C)
BCRP, (D) Bax, (E) Bcl-2, (F) p-PI3K/PI3K and (G) p-AKT/AKT.
**P<0.01 and ***P<0.001 as indicated. BM38, RNA binding motif
protein 38; miR, microRNA; si, small interfering RNA; MDR1,
multiple drug resistant protein 1; BCRP, breast cancer resistance
protein; p-, phosphorylated. |
Discussion
There are currently a number of effective
chemotherapeutic drugs on the market, including ADM, tamoxifen and
paclitaxel (30–32). However, the clinical therapeutic outcome is
far from satisfactory. Due to the gradual development of drug
resistance, patients with BC are only able to benefit from these
treatment options at the early stages of disease (24). The development of therapeutic
tolerance is the primary barrier to a complete cure for BC. Hence,
there is an urgent requirement for further studies to overcome the
challenge associated with drug resistance in BC treatment.
Previous studies have reported that the expression
levels of specific miRNAs, including miRNA-449, miR-140 and
miR-200a, are dysregulated in BC (11–13), and that miRNAs may
participate in the chemoresistance of BC (33,34).
miRNAs are known to inhibit gene expression by either inhibiting
transcription or inducing degradation of their target mRNAs
(35). Du et al (36) demonstrated that the overexpression
of miR-137 increased the sensitivity of BC cell-derived tumors to
ADM by targeting dual specificity phosphatase 4, both in
vitro and in vivo. The miR-202-5p/PTEN signaling axis
was also reported to mediate ADM resistance in BC cells by
regulating the PI3K/AKT signaling pathway (37). In addition, ATP binding cassette
subfamily B member 4 was found to contribute to acquired ADM
resistance in BC cells in vitro (38). However, to the best of our
knowledge, the effect of the RBM38/miR-320b signaling axis in
ADM-resistant BC remains unclear.
Although current studies have indicated that RBM38
may be a favorable factor in preventing the development of BC, the
present study revealed that this may not be the case in
drug-resistant BC. First, the results revealed that RBM38
expression was upregulated in ADM-resistant BC tissues and cells
(MCF-7/A), which indicated that RBM38 may be a key factor in the
development of ADM resistance in BC. Further investigations
demonstrated that the overexpression of RBM38 increased the
IC50 value of ADM in MCF-7/A cells, which supported that
RBM38 may decrease the sensitivity of BC cells to ADM. In addition,
the overexpression of RBM38 intensified the drug resistance of
MCF-7/A cells, by increasing the invasiveness and inhibiting the
apoptosis of cells, as well as accelerating cell cycle progression.
These findings indicated that RBM38 may play a significant role in
the drug resistance of MCF-7/A cells to ADM.
miR-320b has been reported to play an important role
in numerous types of disease, including glioma, carotid
atherosclerosis, colorectal cancer and coronary heart disease
(18,39,40).
In the present study, the results of the dual luciferase reporter
assay identified a binding site between RBM38 and miR-320b. Thus,
the association between miR-320b and RBM38 was further
investigated. The expression levels of RBM38 were found to be
negatively regulated by miR-320b. In addition, miR-320b expression
was discovered to be downregulated in ADM-resistant BC tissues and
cells. Furthermore, the overexpression of miR-320b reversed ADM
resistance, inhibited invasiveness, promoted apoptosis and arrested
the MCF-7/A cell cycle at the G0/G1 phase;
the knockdown of miR-320b exerted the opposite effect. Notably,
RBM38-knockdown rescued the effects of the miR-320b inhibitor,
indicating the presence of a negative regulatory relationship
between RBM38 and miR-320b. Investigations aiming to determine the
underlying mechanism of the miR-320b/RBM38 signaling pathway
revealed that the inhibition of miR-320b in MCF-7/A cells led to
the activation of the PI3K/AKT signaling pathway, as well as the
regulation of apoptosis-related (Bax and Bcl-2) and drug
resistance-related (MDR1 and BCRP) proteins, which was found to
contribute to ADM resistance. These results suggested that the
miR-320b/RBM38 signaling axis may play a key role in ADM resistance
in BC by regulating cellular proliferation, invasiveness, apoptosis
and the cell cycle, which provides potential novel markers for
diagnosing and overcoming drug resistance in BC. However, there are
some limitations to the current study. For example, the tissue
sample size was small. Furthermore, whether miR-320b and RBM38 can
be used to predict the disease-free survival rate of patients with
BC, and the prognostic abilities of miR-320b and RBM38 in BC, were
not investigated. These points will be addressed in future studies.
Finally, additional BC cell lines should be used to further
investigate the functions of the miR-320b/RBM38 signaling axis in
the drug resistance of BC cells to ADM.
In conclusion, the findings of the present study
indicated that RBM38, which is negatively regulated by miR-320b,
accelerated the development of ADM resistance in BC cells by
activating the PI3K/AKT signaling pathway. Thus, the RBM38/miR-320b
signaling axis may represent a novel target for alleviating drug
resistance in BC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JK and KN designed the study; HX and JL performed
the experiments; JK, KN and HX conducted the statistical analysis;
JK and KN drafted the manuscript; and JL revised and proofread the
manuscript. JK and JL confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The experimental protocol was approved by the Ethics
Committee of The Affiliated Hospital of Nantong University
(Nantong, China; approval no. 2019032174) and was performed in
accordance with the principles of the 1964 Declaration of Helsinki.
All patients provided written informed consent prior to study
commencement.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Akram M, Iqbal M, Daniyal M and Khan AU:
Awareness and current knowledge of breast cancer. Biol Res.
50:332017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zaidi Z and Dib HA: The worldwide female
breast cancer incidence and survival, 2018. Cancer Res. Jul
1–2019.(Epub ahead of print). doi:
10.1158/1538-7445.AM2019-4191.
|
|
3
|
Ellis LM and Hicklin DJ: Resistance to
targeted therapies: Refining anticancer therapy in the era of
molecular oncology. Clin Cancer Res. 15:7471–7478. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Network CGA; Cancer Genome Atlas Network,
: Comprehensive molecular portraits of human breast tumours.
Nature. 490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Daniyal A, Santoso I, Gunawan NHP,
Barliana MI and Abdulah R: Genetic Influences in Breast Cancer Drug
Resistance. Breast Cancer (Dove Med Press). 13:59–85.
2021.PubMed/NCBI
|
|
6
|
Ji X, Lu Y, Tian H, Meng X, Wei M and Cho
WC: Chemoresistance mechanisms of breast cancer and their
countermeasures. Biomed Pharmacother. 114:1088002019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu W, Tan C, He Y, Zhang G, Xu Y and Tang
J: Functional miRNAs in breast cancer drug resistance. OncoTargets
Ther. 11:1529–1541. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khordadmehr M, Shahbazi R, Ezzati H,
Jigari-Asl F, Sadreddini S and Baradaran B: Key microRNAs in the
biology of breast cancer; emerging evidence in the last decade. J
Cell Physiol. 234:8316–8326. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hanna J, Hossain GS and Kocerha J: The
potential for microRNA therapeutics and clinical research. Front
Genet. 10:4782019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Si W, Shen J, Zheng H and Fan W: The role
and mechanisms of action of microRNAs in cancer drug resistance.
Clin Epigenetics. 11:252019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tormo E, Ballester S, Adam-Artigues A,
Burgués O, Alonso E, Bermejo B, Menéndez S, Zazo S, Madoz-Gúrpide
J, Rovira A, et al: The miRNA-449 family mediates doxorubicin
resistance in triple-negative breast cancer by regulating cell
cycle factors. Sci Rep. 9:53162019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu X, Liu R, Wang M, Kumar AK, Pan F, He
L, Hu Z and Guo Z: MicroRNA-140 impedes DNA repair by targeting
FEN1 and enhances chemotherapeutic response in breast cancer.
Oncogene. 39:234–247. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu SJ, Yang L, Hong Q, Kuang XY, Di GH and
Shao ZM: MicroRNA-200a confers chemoresistance by antagonizing
TP53INP1 and YAP1 in human breast cancer. BMC Cancer. 18:742018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mulrane L, McGee SF, Gallagher WM and
O'Connor DP: miRNA dysregulation in breast cancer. Cancer Res.
73:6554–6562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang H, Cao F, Li X, Miao H, e J, Xing J
and Fu CG: miR-320b suppresses cell proliferation by targeting
c-Myc in human colorectal cancer cells. BMC Cancer. 15:7482015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Tang X, He Q, Yang X, Ren X, Wen X,
Zhang J, Wang Y, Liu N and Ma J: Overexpression of mitochondria
mediator gene TRIAP1 by miR-320b loss is associated with
progression in nasopharyngeal carcinoma. PLoS Genet.
12:e10061832016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lv GY, Miao J and Zhang XL: Long noncoding
RNA XIST promotes osteosarcoma progression by targeting Ras-related
protein RAP2B via miR-320b. Oncol Res. 26:837–846. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu X, Wang Y, Zhong W, Cheng H and Tian Z:
RNA binding protein RNPC1 suppresses the stemness of human
endometrial cancer cells via stabilizing MST1/2 mRNA. Med Sci
Monit. 26:e9213892020.PubMed/NCBI
|
|
20
|
Cho SJ, Jung YS, Zhang J and Chen X: The
RNA-binding protein RNPC1 stabilizes the mRNA encoding the
RNA-binding protein HuR and cooperates with HuR to suppress cell
proliferation. J Biol Chem. 287:14535–14544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi L, Xia T-S, Wei X-L, Zhou W, Xue J,
Cheng L, Lou P, Li C, Wang Y, Wei JF, et al: Estrogen receptor (ER)
was regulated by RNPC1 stabilizing mRNA in ER positive breast
cancer. Oncotarget. 6:12264–12278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng L, Zhang Z, Zhang S, Guo Q, Zhang F,
Gao L, Ni H, Guo X, Xiang C and Xi T: RNA binding protein RNPC1
inhibits breast cancer cell metastasis via activating
STARD13-correlated ceRNA network. Mol Pharm. 15:2123–2132. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hernandez-Aya LF and Gonzalez-Angulo AM:
Adjuvant systemic therapies in breast cancer. Surg Clin North Am.
93:473–491. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ponnusamy L, Mahalingaiah PKS and Singh
KP: Treatment schedule and estrogen receptor-status influence
acquisition of doxorubicin resistance in breast cancer cells. Eur J
Pharm Sci. 104:424–433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xue J-Q, Xia T-S, Liang X-Q, Zhou W, Cheng
L, Shi L, Wang Y and Ding Q: RNA-binding protein RNPC1: Acting as a
tumor suppressor in breast cancer. BMC Cancer. 14:3222014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhong ZF, Yu HB, Wang CM, Qiang WA, Wang
SP, Zhang JM, Yu H, Cui L, Wu T, Li DQ, et al: Furanodiene induces
extrinsic and intrinsic apoptosis in doxorubicin-resistant MCF-7
breast cancer cells via NF-κB-independent mechanism. Front
Pharmacol. 8:6482017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao HL, Xia YZ, Zhang YL, Yang L and Kong
LY: Vielanin P enhances the cytotoxicity of doxorubicin via the
inhibition of PI3K/Nrf2-stimulated MRP1 expression in MCF-7 and
K562 DOX-resistant cell lines. Phytomedicine. 58:1528852019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li YS, Zhao DS, Liu XY, Liao YX, Jin HW,
Song GP and Cui ZN: Synthesis and biological evaluation of
2,5-disubstituted furan derivatives as P-glycoprotein inhibitors
for Doxorubicin resistance in MCF-7/ADR cell. Eur J Med Chem.
151:546–556. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Muss HB, White DR, Richards F II, Cooper
MR, Stuart JJ, Jackson DV, Rhyne L and Spurr CL: Adriamycin versus
methotrexate in five-drug combination chemotherapy for advanced
breast cancer: A randomized trial. Cancer. 42:2141–2148. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Valero V, Jones SE, Von Hoff DD, Booser
DJ, Mennel RG, Ravdin PM, Holmes FA, Rahman Z, Schottstaedt MW,
Erban JK, et al: A phase II study of docetaxel in patients with
paclitaxel-resistant metastatic breast cancer. J Clin Oncol.
16:3362–3368. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Riggins RB, Schrecengost RS, Guerrero MS
and Bouton AH: Pathways to tamoxifen resistance. Cancer Lett.
256:1–24. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen X, Lu P, Wang DD, Yang SJ, Wu Y, Shen
H-Y, Zhong SL, Zhao JH and Tang JH: The role of miRNAs in drug
resistance and prognosis of breast cancer formalin-fixed
paraffin-embedded tissues. Gene. 595:221–226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Majumder S and Jacob ST: Emerging role of
microRNAs in drug-resistant breast cancer. Gene Expr. 15:141–151.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Du F, Yu L, Wu Y, Wang S, Yao J, Zheng X,
Xie S, Zhang S, Lu X, Liu Y, et al: miR-137 alleviates doxorubicin
resistance in breast cancer through inhibition of
epithelial-mesenchymal transition by targeting DUSP4. Cell Death
Dis. 10:9222019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu T, Guo J and Zhang X: MiR-202-5p/PTEN
mediates doxorubicin-resistance of breast cancer cells via PI3K/Akt
signaling pathway. Cancer Biol Ther. 20:989–998. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang JF, Wen CJ, Zhao GZ, Dai Y, Li Y, Wu
LX and Zhou HH: Overexpression of ABCB4 contributes to acquired
doxorubicin resistance in breast cancer cells in vitro. Cancer
Chemother Pharmacol. 82:199–210. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang R, Qin Y, Zhu G, Li Y and Xue J: Low
serum miR-320b expression as a novel indicator of carotid
atherosclerosis. J Clin Neurosci. 33:252–258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lv QL, Du H, Liu YL, Huang YT, Wang GH,
Zhang X, Chen SH and Zhou HH: Low expression of microRNA-320b
correlates with tumorigenesis and unfavorable prognosis in glioma.
Oncol Rep. 38:959–966. 2017. View Article : Google Scholar : PubMed/NCBI
|