Introduction
Colorectal cancer (CRC) is one of the most common
malignancies worldwide (1,2). More than 1 million new cases of CRC
were diagnosed worldwide in 2012, which accounts for ~10% of the
global cancer burden (3).
Currently, CRC is the fourth most common cause of cancer-related
mortality (4,5). Although conventional treatments such
as surgery, chemotherapy, radiotherapy and immunotherapy have
improved the survival of patients with CRC, the mortality and
relapse rates remain high (5,6).
Therefore, more specific mechanism-based treatments are greatly
needed.
Circular RNAs (circRNAs/circs) have gained attention
as a class of potential biomarkers for the early detection of CRC
(7). circRNA is a newly discovered
type of non-coding RNA, which is distinct from the traditional
linear RNA that has 5′- and 3′-ends (8). circRNAs are formed by exon skipping
or back-splicing events, during which the 5′-end of an upstream
exon is spliced together with the 3′-end of the downstream exon to
form a circular molecule of RNA (8,9).
circRNAs have been demonstrated to serve an important role in
post-transcriptional regulation by acting as microRNA (miRNA/miR)
sponges to competitively inhibit RNA/miRNA transcriptional
regulation (10,11).
The abnormal expression of circRNA is closely
associated with various diseases, including CRC (12–14).
Jiang et al (15)
identified a large set of differentially expressed circRNAs in a
primary CRC cell line (SW480) and a metastatic CRC cell line
(SW620), relative to a normal colon cell line (NCM460).
hsa_circ_000984 has been reported to promote colon cancer
growth and metastasis by sponging miR-106b (16). Furthermore, Bachmayr-Heyda et
al (17) demonstrated a global
reduction in circRNA abundance in CRC cell lines and clinical CRC
specimens compared with normal tissues. The authors described five
specific circRNAs (circ0817, circ3203, circ6229, circ7374 and
circ7780) and proposed a potentially negative association between
global circRNA abundance and cell proliferation.
Among the five circRNAs identified, the present
study focused on circ6229 (also known as circ_0000523) and
its corresponding gene/linear mRNA, methyltransferase-like 3
(METTL3). As a major RNA N6-adenosine methyltransferase,
METTL3 is widely implicated in mRNA biogenesis, decay and
translation control (18).
METTL3 has previously been demonstrated to promote the
growth, survival and invasion of numerous human cancers (19), such as lung cancer (20), hepatocellular carcinoma (21), breast cancer (22) and pancreatic cancer (23). However, the precise role of
METTL3 in the tumorigenesis and pathogenesis of CRC remains
largely unknown.
HCT116 cells are a population of malignant cells
that were isolated from the primary cell culture of a single human
colonic carcinoma (24). In
present study, the regulatory role of hsa_circ_0000523 in
HCT116 cells was explored in detail. Dual-luciferase reporter
assay, reverse transcription-quantitative PCR (RT-qPCR) and some
rescue experiments were applied to confirm the existence of the
hsa_circ_0000523/miR-let-7b/METTL3 axis. FCM
was used to count the apoptotic cells. Cell Counting Kit-8 (CCK-8)
and invasion assays were performed to monitor the activities of the
HCT116 cells. The present study suggests a potential role for the
hsa_circ_0000523/miR-let-7b/METTL3 axis in the tumorigenesis
and pathogenesis of human CRC.
Materials and methods
Cell culture
The HCT116 human CRC cell line (cat. no. CCL-247)
was obtained from American Type Culture Collection. Cells were
maintained in DMEM (Thermo Fisher Scientific, Inc.) supplemented
with 10% FBS (Thermo Fisher Scientific, Inc.), 100 units/ml
penicillin and 100 µg/ml streptomycin (Thermo Fisher Scientific,
Inc.). Cells were grown in a humidified atmosphere with 5%
CO2 at 37°C. Cells were cultured in 75-cm2
flasks or 10-cm plates (Corning, Inc.), with the confluence
maintained below 80%. After digestion with 0.25% (w/v) trypsin and
0.5 mM EDTA (Thermo Fisher Scientific, Inc.), cells were passaged
every 2–3 days with a subcultivation ratio between 1:3 and 1:8.
Plasmid constructs and small
interfering RNA (siRNA) oligos
Total RNA was extracted from the HCT116 cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.) and
further purified with two phenol-chloroform treatments, and then
treated with RQ1 DNase (Promega Corporation) to digest DNA. The
quality and quantity of the purified RNAs were measured by a Nano
Photometer spectrometer with the absorbance at 260/280 nm. The
mixture was verified by 1.2% agarose gel electrophoresis. The cDNA
was synthesized with random primers with the High-capacity cDNA
Reverse-Transcription Kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's instructions. The full-length cDNA
for human METTL3 (GenBank accession no. NM_019852.5) was
obtained from the cDNA library of HCT116 cells by PCR, using the
following protocol: 98°C for 10 min, followed by 98°C for 10 sec,
55°C for 5 sec and 72°C for 2 min for 35 cycles, and then 72°C for
10 min. The primers are listed in Table I. The vector used to generate
full-length wild-type (WT) METTL3 and
hsa_circ_0000523 was mammalian expression vector pcDNA3.1
(Thermo Fisher Scientific, Inc.). The METTL3 coding sequence
or circ_0000523 fragment was subcloned into the pcDNA3.1
vector via KpnI and BamHI double-enzymatic sites.
Positive clones were selected, and the plasmid expressing
METTL3 or hsa_circ_0000523 was verified by
sequencing. The siRNA oligos used in the present study were
designed and synthesized by Wuhan GeneCreate Biological Engineering
Co., Ltd. The sequences for these siRNA oligos were as follows:
Negative control siRNA oligo (METTL3-con) sense,
5′-GCUACUUCAGACGAGCAUUdTdT-3′; METTL3-specific siRNA oligo
(METTL3-si) sense, 5′-GCUGCACUUCAGACGAAUUdTdT-3′; negative
control oligo for hsa_circ_0000523 (circ-con) sense,
5′-CAACAGAGCAAGAAGUAGAUdTdT-3′; and circ_0000523-specific
siRNA oligo (circ-si) sense, 5′-CAACAGAGCAAGAAGAUCUAdTdT-3′.
 | Table I.Sequences of primers used for reverse
transcription-quantitative PCR in the present study. |
Table I.
Sequences of primers used for reverse
transcription-quantitative PCR in the present study.
| Gene | Primer sequence
(5′-3′) |
|---|
|
hsa_circ-0000523 | F:
CAGCATCGGAACCAGCAAAG |
|
| R:
CTGGGCTGTCACTACGGAAG |
|
miR-let-7b | F:
GGGTGAGGTAGTAGGTTGT |
|
| R:
CAGTGCGTGTCGTGGAGT |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| METTL3 | F:
GTGTCGGAGGTGATTCCAGT |
|
| R:
CTGCGCATCTCATCATCTGT |
| GAPDH | F:
GTCAGTGGTGGACCTGACCT |
|
| R:
TGCTGTAGCCAAATTCGTTG |
Transfection of plasmids and siRNA
oligos
HCT116 cells were transfected with the empty
pcDNA3.1 vector (METTL3-EV), METTL3-expressing
pcDNA3.1 vector (METTL3-OE), METTL3-con,
METTL3-si, circ-EV, circ-OE, circ-con, circ-si, mimics,
mimics NC, inhibitor and inhibitor NC using
Lipofectamine® 2000 reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. miR-let-7b mimic
sense (5′-UGAGGUAGUAGGUUGUGUGGUU-3′), its negative control
(5′-UCACAACCUCCUAGAAAGAGUAGA-3′), inhibitor
(5′-AACCACACAACCUACUACCUCA-3′) and inhibitor NC
(5′-UCUACUCUUUCUAGGAGGUUGUGA-3′) were synthesized by Wuhan
GeneCreate Biological Engineering Company. The RNA concentrations
of mimics and inhibitor were 0.32 and 0.26 µg/µl, respectively.
HCT116 cells (1×104) were seeded in 6-well plates and
maintained at a confluence of 70–80% in complete culture medium.
Then, 2.5 µg plasmid or 100 pmol siRNA oligo was added to 250 µl
Opti-MEM (Thermo Fisher Scientific, Inc.), and 5 µl Lipofectamine
2000 reagent was added to 250 µl Opti-MEM. The diluted plasmid or
siRNA oligo was mixed with diluted Lipofectamine 2000 reagent and
kept at room temperature for 20 min. After 500 µl serum-free medium
was added to each well, the Opti-MEM mixture was added. After
transfected cells were incubated at 37°C for 4 h, the culture
medium was changed to regular DMEM supplemented with 10% FBS. After
48 h, the subsequent experiments were performed.
RNA isolation and reverse
transcription
Total RNA was extracted from cells using TRIzol
reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. Briefly, after treatment, cell pellets
were resuspended in 1 ml TRIzol and mixed well. Then, 0.2 ml
chloroform was added to the cell suspension, which was shaken
vigorously for 15 sec and then incubated for 3 min at room
temperature. After centrifugation of the suspension at 12,000 × g
for 10 min at 4°C, supernatant was collected and subjected to RNA
precipitation by adding 0.5 ml isopropanol at 4°C. The RNA pellet
was obtained by centrifugation at 12,000 × g for 10 min at 4°C, and
then washed with 75% ethanol. The appropriate amount of RNase-free
distilled water was used to dissolve extracted RNA. The
concentration of RNA was measured with a NanoDrop 2000
Spectrophotometer (Thermo Fisher Scientific, Inc.). For each
sample, 1 µg RNA was reverse transcribed into cDNA using the
ReverTra Ace qPCR RT Kit (cat. no. FSQ-101; Toyobo Life Science)
according to the specifications in the product manual.
RT-qPCR
For RT-qPCR, SYBR Green Realtime PCR Master Mix
(cat. no. QPK-212; Toyobo Life Science) was used to measure the
expression of targeted genes according to the manufacturer's
protocol. For quantification of hsa_circ_0000523 and
let-7b, U6 was used as an internal reference gene.
GAPDH was used as an internal reference gene for the
quantification of METTL3. The conditions for PCR were set as
follows: Pre-denaturation at 95°C for 1 min, followed by 40 cycles
of denaturation at 95°C for 15 sec, and annealing and elongation at
60°C for 30 sec. The 7900HT Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to perform the
assay. The 2−ΔΔCq method was used to calculate
differences in target gene expression between the experimental and
control groups. The calculation formula was as follows:
ΔΔCq=ΔCqexperimentalgroup-ΔCqnormalgroup,
ΔCq=Cqtargetgene-Cqinternalreference
(25). The sequences of primers
used for RT-qPCR are presented in Table I.
Western blot (WB) assay
HCT116 human CRC cells with various treatments were
washed twice using PBS buffer. Total protein was extracted from the
treated cells using NE-PER Nuclear and Cytoplasmic Extraction
Reagents (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols and then measured using a BCA assay.
Briefly, the tube was vigorously vortexed for 15 sec. Then,
ice-cold cytoplasmic extraction reagent II was added to the tube
and the tube was vortexed for another 5 sec. The tube was
centrifuged at 16,000 × g for 5 min at 4°C, and then the
supernatant was collected. The insoluble (pellet) fraction was
suspended in ice-cold nuclear extraction reagent. Subsequently, the
collected fraction was vortexed for 15 sec and the tube was
centrifuged at 16,000 × g (15 min, 4°C). Lastly, the supernatant
fraction was transferred to a clean tube and stored at −80°C until
use. Proteins were quantified by the BCA method. The proteins (50
µg) were separated on 12% gels using SDS-PAGE (Beyotime Institute
of Biotechnology), and then quickly transferred onto a PVDF
membrane.. The membrane was blocked with 5% skimmed milk for 45 min
at 37°C, and then probed with primary antibodies for 12 h at 4°C.
The membrane was washed three times and probed with the secondary
antibodies for 1 h at 25°C. The signal was detected using an ECL
system (MilliporeSigma) and the gray level was assessed using
ImageJ 1.8.0 software (National Institutes of Health). The primary
antibodies were: Anti-METTL3 antibody (EPR18810; 1:2,000
dilution; cat. no. ab195352; Abcam) and GAPDH monoclonal Antibody
(1:10,000 dilution; catalog no. 60004-1-Ig; ProteinTech Group,
Inc.). The secondary antibody was HRP-conjugated Affinipure Goat
Anti-Rabbit IgG(H+L) (1:10,000 dilution; catalog no. SA00001-2;
ProteinTech Group, Inc.). All antibodies used in the WB assay were
diluted in 5% skimmed milk powder.
Dual-luciferase reporter assay
The binding sites among miR-let-7b,
circ_0000523 and METTL3 were predicted by Bielefeld
Bioinformatics Service online software (https://bibiserv.cebitec.uni-bielefeld.de/prevres.jsf).
The WT miR-let-7b along with the 3′-UTR of METTL3
that contained the predicted miR-let-7b binding site or a
sequence with mutations (MUT) were amplified and inserted into the
pmirGLO vector (Promega Corporation), resulting in the WT
luciferase reporter construct. The vectors (miR-let-7b-WT,
miR-let-7b-MUT, METTL3-WT or METTL3-MUT),
miR-let-7b mimics and negative control mimics, which were
synthesized by Wuhan GeneCreate Biological Engineering Company,
were transfected into CRC cells using Lipofectamine 3000 (Thermo
Fisher Scientific, Inc.). After 48 h, the luciferase activity was
measured using a luciferase reporter system (Promega Corporation),
with Renilla luciferase activity as an internal control. The
sequences of mimics miR-let-7b and mimics NC were as
follows: miR-let-7b mimic sense,
5′-UGAGGUAGUAGGUUGUGUGGUU-3′; NC sense,
5′-UCACAACCUCCUAGAAAGAGUAGA-3′.
CCK-8 assay to measure cell
proliferation
After transfection of empty pcDNA3.1 vector,
METTL3-expressing pcDNA3.1 vector, METTL3-con oligo
or METTL3-si oligo, HCT116 cells were seeded into
flat-bottomed 96-well plates at a density of
3×103−6×103 cells per well in 100 µl culture
medium. Each treatment condition was replicated in nine wells.
Cells were incubated for 24, 48 or 72 h in an incubator with 5%
CO2 at 37°C. Cell proliferation was analyzed with a
CCK-8 assay (Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. Briefly, 20 µl CCK-8 reagent (5 mg/ml) was
added to the culture medium, and cells were cultured for an
additional 4 h. The optical density values of all wells were
measured with a plate reader (Thermo Fisher Scientific, Inc.) at
450 nm.
Cell apoptosis
HCT116 cells seeded in 6-well plates were
transfected with empty pcDNA3.1 vector, METTL3-expressing
pcDNA3.1 vector, METTL3-con oligo or METTL3-si oligo,
as aforementioned. At 48 h after transfection, cells were detached
by incubation with 0.25% trypsin-EDTA solution (Thermo Fisher
Scientific, Inc.) and were harvested. Flow cytometry analysis was
performed to detect apoptosis using the Annexin V-FITC/PI apoptosis
kit (Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. Briefly, after one wash in PBS and one
wash in binding buffer, cells were stained with Annexin V-FITC/PI
for 20 min at room temperature in the dark. After another wash in
binding buffer, labelled cells were detected immediately using a
flow cytometer (CytoFLEX S; Beckman Coulter, Inc.). Data were
analyzed with CytExpert 2.0 Software (Beckman Coulter, Inc.).
Cell invasion
The cellular potential for invasion was determined
using a 24-well Transwell plate with 8.0-µm Pore Polyester Membrane
Inserts (Corning, Inc.). Matrigel (Corning, Inc.) was melted
overnight at 4°C and diluted to a final concentration of 1 mg/ml in
pre-cooled serum-free medium. Then, 100 µl diluted Matrigel was
added to the bottom of the upper chamber. The plate was then
incubated at 37°C for 4–5 h to dry the Matrigel. At 24 h after
transfection, HCT116 cells in the logarithmic growth phase were
seeded in triplicate at a density of 1×106
cells/chamber. Cells were seeded on top of the Transwell plate in
100 µl DMEM supplemented with 0.1% bovine serum albumin (Thermo
Fisher Scientific, Inc.). Then, 0.8 ml DMEM supplemented with 10%
FBS (Thermo Fisher Scientific, Inc.) was added to the lower chamber
as a chemoattractant. After 24 h of incubation at 37°C, cells on
the top surface of the insert were removed with a cotton swab.
Cells that had invaded the lower surface of the membrane were fixed
with 4% paraformaldehyde at 37°C for 15 min and stained with 800 µl
Giemsa solution at 37°C for 20 min (Beyotime Institute of
Biotechnology). Cells were visualized using a light microscope
(CKX-41; Olympus Corporation). Cell counts were obtained from three
randomly selected optical fields.
Statistical analysis
All data were analyzed using SPSS Version 21.0
software (IBM Corp.). All values are presented as the mean ± SD.
Three independent experiments were performed Quantitative data were
compared using the χ2 test. For only two comparisons,
statistically significant differences between means were determined
using an unpaired Student's t-test. For multiple comparisons, the
significance was identified by simple one-way ANOVA followed by a
Tukey's post-hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
hsa_circ_0000523 regulates METTL3
expression by modulating levels of miR-let-7b in HCT116 cells
To explore the role of hsa_circ_0000523
circRNA in the tumorigenesis and pathogenesis of CRC, an in
vitro model was established by manipulating the expression of
hsa_circ_0000523 in the HCT116 human CRC cell line. As shown
in Fig. 1A, the mRNA expression
levels of circ_0000523 were successfully increased more than
20-fold in the circ overexpression (circ-OE) group compared with
cells transfected with empty vector (circ-EV). Next, this circRNA
was knocked down in HCT116 cells. Efficient knockdown of
hsa_circ_0000523 was observed in HCT116 cells transfected
with hsa_circ_0000523-specific siRNA oligo (circ-si), in
which hsa_circ_0000523 expression was 80% less than that in
HCT116 cells transfected with control siRNA oligo (circ-con)
(Fig. 1A). Next, the effect of
hsa_circ_0000523 expression on METTL3 and
miR-let-7b, a microRNA that may be a target of
hsa_circ_0000523, was evaluated in HCT116 cells. A negative
association between the expression of hsa_circ_0000523 and
miR-let-7b was observed (Fig.
1B). Among the various treatment conditions, HCT116 cells
transfected with hsa_circ_0000523-expressing plasmid
(circ-OE) displayed the highest level of hsa_circ_0000523
expression and the lowest level of miR-let-7b
expression.
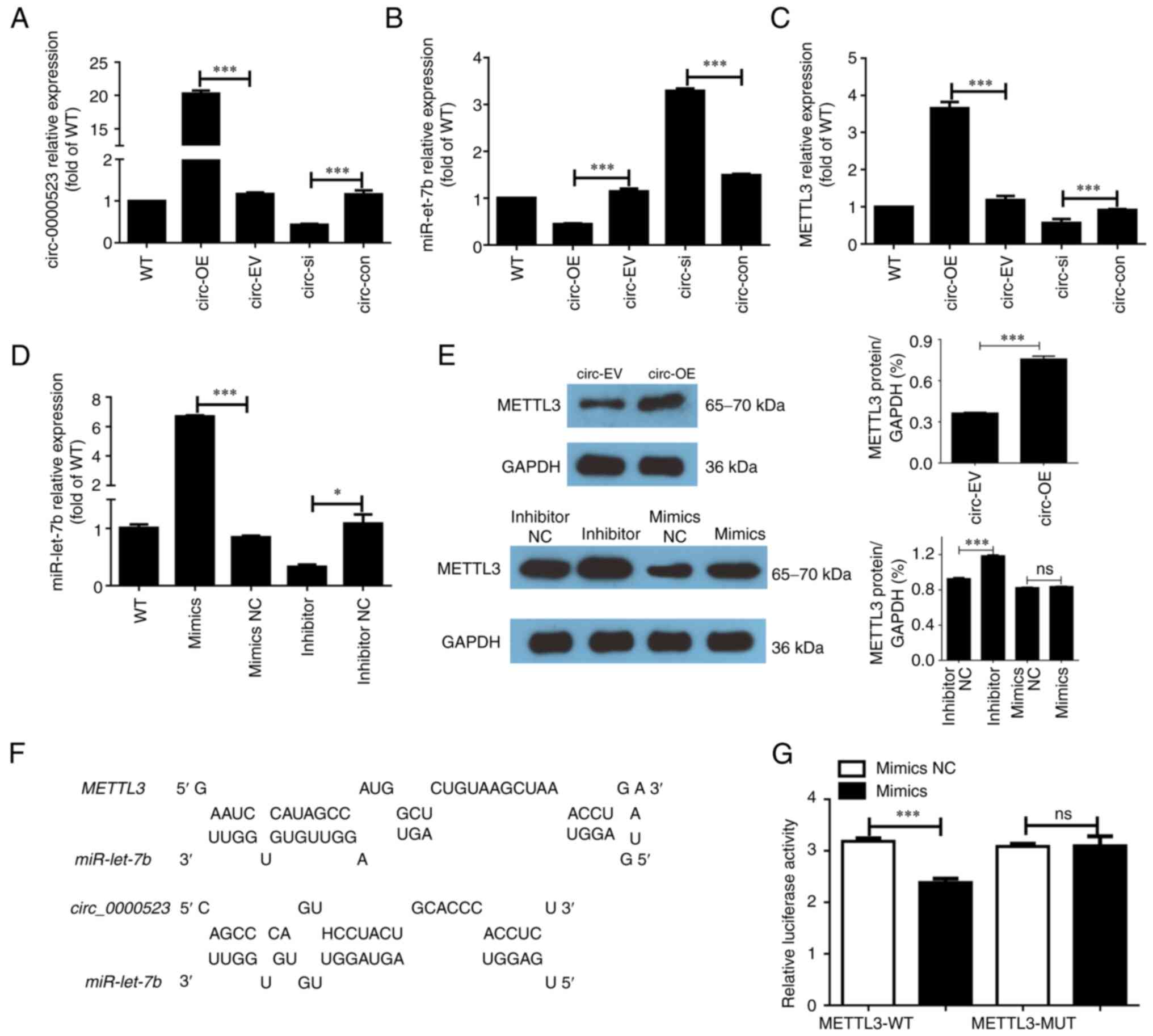 | Figure 1.hsa_circ_0000523 indirectly
regulates METTL3 expression by suppressing transcription of
miR-let-7b in HCT116 cells. (A-C) HCT116 cells are untreated
or transfected with hsa_circ_0000523-expression plasmid,
empty pcDNA3.1 vector plasmid, hsa_circ_0000523-specific
siRNA oligo or control siRNA oligo. At 48 h after transfection,
HCT116 cells were harvested and transcriptional levels of (A)
hsa_circ_0000523, (B) miR-let-7b and (C)
METTL3 are quantified by RT-qPCR. (D) RT-qPCR verifies the
successful transfection of miR-let-7b mimics and inhibitor.
(E) Protein levels of METTL3 were measured using western
blot analysis. (F) Schematic representation of miR-let-7b
and predicted target site in circ_0000523 and METTL3.
(G) Interaction between miR-let-7b and METTL3 was
detected using a luciferase reporter assay. n=3 for each group.
*P<0.05 and ***P<0.001. WT, untreated wild-type HCT116 cells;
circ-OE, HCT116 cells with overexpression of
hsa_circ_0000523; circ-EV, HCT116 cells transfected with
empty vector; circ-si, HCT116 cells transfected with
hsa_circ_0000523-specific siRNA oligo; circ-con, HCT116
cells transfected with negative control siRNA oligo; inhibitor,
inhibitor-treated groups; inhibitor NC, the control of
inhibitor-treated groups (PBS-treated groups); mimics,
miR-let-7b overexpression groups; mimics NC, the control of
mimics groups; METTL3, methyltransferase-like 3; RT-qPCR,
reverse transcription-quantitative PCR; siRNA, small interfering
RNA. |
Among all treatment groups, the circ-si group had
the highest expression of miR-let-7b (Fig. 1B). Expression of METTL3, one
of the targets of miR-let-7b, was upregulated in circ-OE
group, while knockdown of hsa_circ_0000523 exerted an
inhibitory effect (Fig. 1C). Next,
the regulatory effect of miR-let-7b on METTL3 was
verified. A mimic and an inhibitor of miR-let-7b were
synthesized and then transfected into HCT116 cells. The
transfection efficiency was validated by RT-qPCR (Fig. 1D). The reduction of METTL3
was not significant in miR-treated cells, but the miRNA inhibitor
increased the protein level of METTL3 (Fig. 1E). The binding sequence of
miR-let-7b in the 3′UTR of METTL3 mRNA and
circ_0000523 that was predicted by BiBiserv prediction
software is shown in Fig. 1F.
Lastly, a luciferase reporter assay was implemented to verify the
regulatory effect of miR-let-7b on METTL3. As shown
in Fig. 1G, luciferase activity
was reduced in the mimics and METTL3-WT groups relative to
that in the mimics NC and METTL3-WT groups. Furthermore,
transfection of mimics NC or mimics had almost no impact on
luciferase activity in the METTL3-MUT group, demonstrating
that miR-let-7b directly binds to METTL3. A
hsa_circ_0000523/miR-let-7b/METTL3 axis was
thus identified in HCT116 cells.
Positive association between METTL3
expression and proliferation in HCT116 cells
As METTL3 was the endpoint effector molecule
for the hsa_circ_0000523/miR-let-7b/METTL3
axis, it was important to elucidate the effects of METTL3
expression on proliferation, apoptosis and invasion of HCT116 cells
in the present study. Similarly, HCT116 cells with overexpression
or knockdown of METTL3 were generated by transfection of a
METTL3-expressing pcDNA3.1 plasmid or a
METTL3-specific siRNA oligo, respectively. The levels of
METTL3 mRNA were quantified using RT-qPCR and WB analysis.
The results demonstrated that METTL3 overexpression resulted
in a >200-fold increase in METTL3 transcript levels. The
WB results were consistent with the RT-qPCR findings. Compared with
the METTL3-con group, the METTL3-si group exhibited a
>70% decrease in METTL3 transcript levels (Fig. 2A).
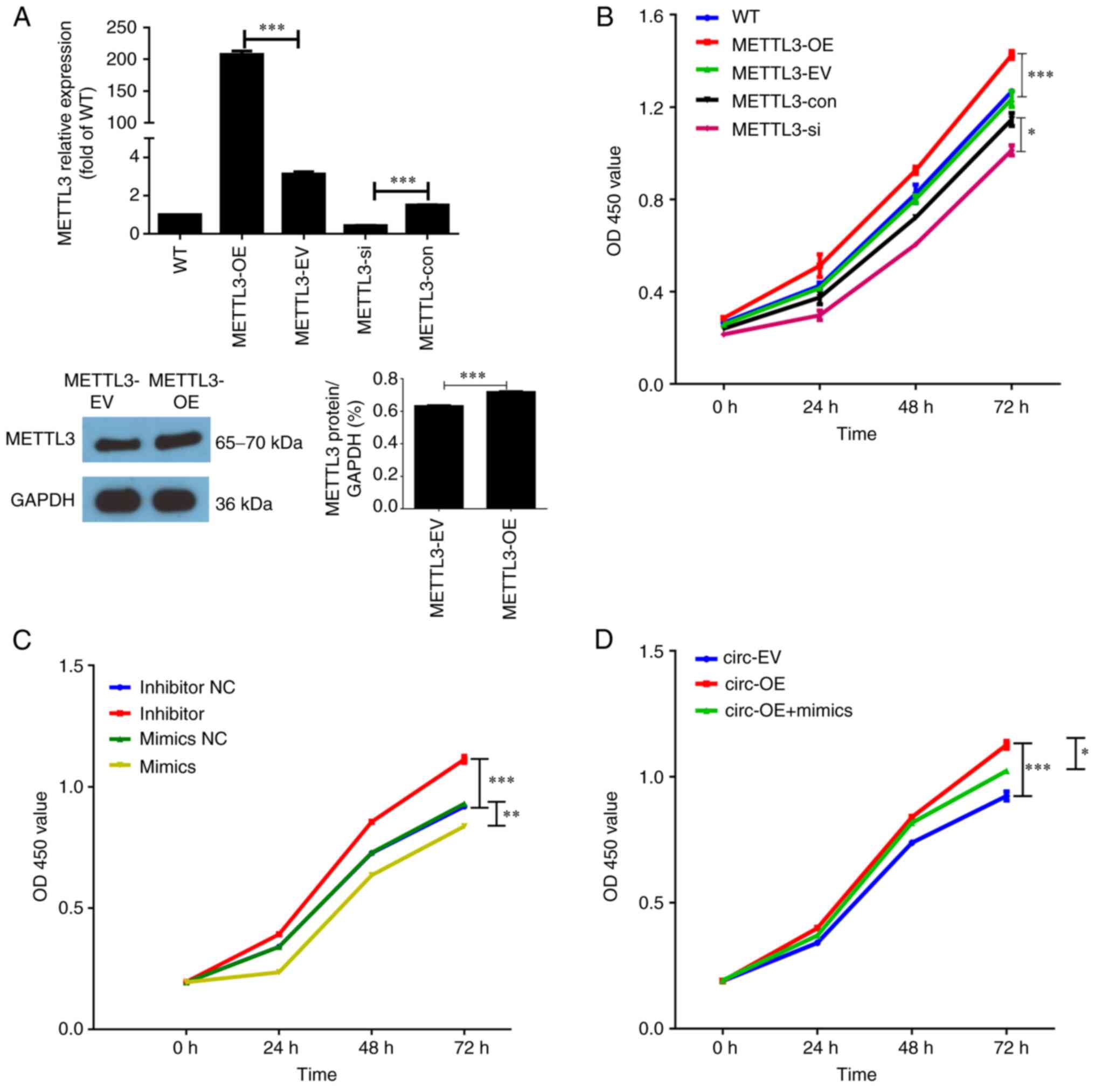 | Figure 2.METTL3, miR-let-7b and
circ_0000523 regulate proliferation of HCT116 cells. (A)
METTL3 levels were quantified by reverse
transcription-quantitative PCR or western blotting. (B-C) Cell
Counting Kit-8 assays were performed to examine the role of (B)
METTL3 and (C) miR-let-7b in cell proliferation. (D)
CCK-8 assays were used to detect the cell viability in each group.
n=3 for each group. *P<0.05, **P<0.01 and ***P<0.001. WT,
untreated wild-type HCT116 cells; METTL3-OE, HCT116 cells
with overexpression of METTL3; METTL3-EV, HCT116
cells transfected with empty vector; METTL3-si, HCT116 cells
transfected with METTL3-specific siRNA oligo;
METTL3-con, HCT116 cells transfected with negative control
siRNA oligo; inhibitor, inhibitor-treated group; inhibitor NC, the
control of inhibitor-treated group (PBS-treated group); mimics,
miR-let-7b overexpression groups; mimics NC, the control of
mimics group; circ-OE, circ_0000523 overexpression group;
circ-EV, the control of circ-OE group; circ-OE + mimics,
circ_0000523 overexpressing vector and mimics co-transfected
into HCT116 cells; METTL3, methyltransferase-like 3; siRNA,
small interfering RNA; OD, optical density. |
After transfection with plasmids or siRNA oligos,
HCT116 cells were further cultured for 24, 48 or 72 h. Cell
proliferation was measured at these time points using the CCK-8
assay. There were no significant differences in cell proliferation
among the untreated WT, empty vector-transfected and negative
control siRNA oligo-transfected HCT116 cells (Fig. 2B). Compared with the empty
vector-transfected HCT116 cells (METTL3-EV group), HCT116
cells with ectopic expression of METTL3 (METTL3-OE
group) exhibited a significantly higher rate of proliferation at
each time point. Cell proliferation was decreased in HCT116 cells
with knockdown of METTL3 compared with cells transfected
with negative control siRNA oligos (Fig. 2B). Furthermore, the effects of
miR-let-7b and circ_0000523 on HCT116 cell
proliferation were evaluated. Compared with the mimic NC groups,
proliferation was obviously inhibited in the mimic groups. By
contrast, cell viability was significantly promoted in inhibitor
groups (Fig. 2C). As shown in
Fig. 2D, overexpression of
circ_0000523 promoted HCT116 cell proliferation at all time
points. Furthermore, miR-let-7b reversed the effects of
circ_0000523 overexpression on human HCT116 cell
characteristics. Taken together, these results indicated that
hsa_circ_0000523 may regulate proliferation of CRC cells via
miR-let-7b.
METTL3 regulates apoptosis in HCT116
cells
An important question was whether and how the
expression of METTL3 impacts apoptosis in HCT116 cells;
therefore, this was evaluated using Annexin V and PI staining.
There were no significant differences in apoptotic rates among the
untreated WT, empty vector-transfected and negative control siRNA
oligo-transfected HCT116 cells, all of which exhibited an apoptotic
rate of ~15% (Fig. 3A and B). The
rate of apoptosis was significantly decreased in HCT116 cells with
ectopic expression of METTL3 compared with HCT116 cells
transfected with empty vector (12% vs. 15%, respectively). The rate
of apoptosis was higher in HCT116 cells with knockdown of
METTL3 than in cells transfected with negative control siRNA
oligos (20% vs. 15%, respectively; Fig. 3A and B). Therefore, a negative
association between METTL3 expression and the apoptotic rate
was observed, which suggests that METTL3 inhibits apoptosis
in HCT116 cells. In addition, the effect of circ_0000523 and
miR-let-7b on HCT116 cell apoptosis was investigated by flow
cytometry. Overexpression of circ_0000523 significantly
reduced apoptosis of CRC cells (Figs.
3C and S1A). Compared with
that in the mimics NC group, the number of apoptotic cells was
increased in the mimics group. By contrast, adding an inhibitor
suppressed HCT116 cell apoptosis (Figs. 3D and S1B). As shown in Figs. 3E and S1C, circ_0000523 reversed the
antitumor effects of miR-let-7b on human CRC cell line
characteristics.
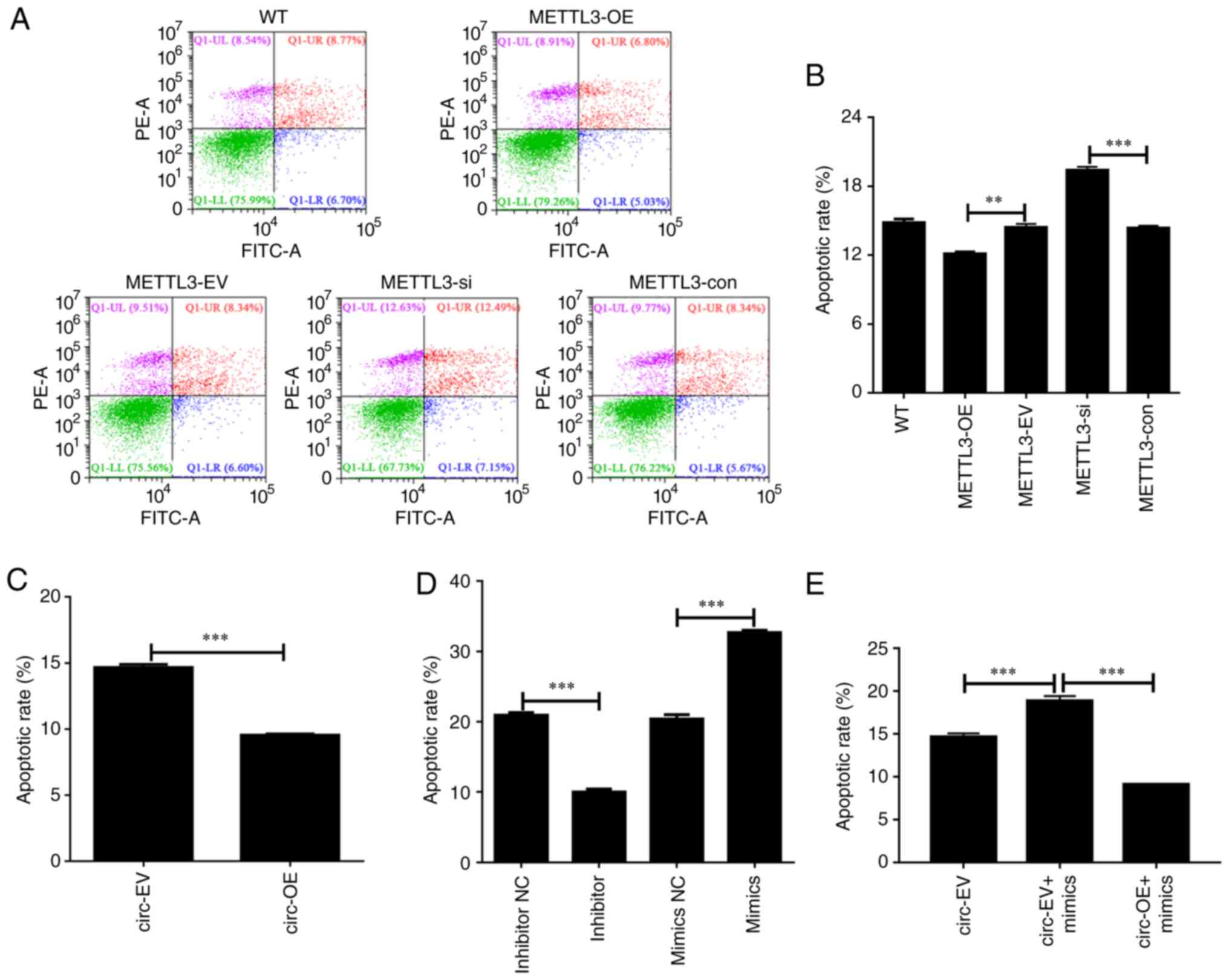 | Figure 3.METTL3, miR-let-7b and
circ_0000523 mediate apoptosis of HCT116 cells. (A) Flow
cytometry was used to detect apoptosis following knockdown or
overexpression of METTL3. (B-D) Summarized data show the
rate of apoptosis in HCT116 cells following interference with (B)
METTL3, (C) miR-let-7b and (D) circ_0000523.
(E) Representative flow cytometry apoptosis data for each group of
CRC cells are shown. Annexin V-positive cells were considered to be
apoptotic. n=3 for each group. **P<0.01 and ***P<0.001. WT,
untreated wild-type HCT116 cells; METTL3-OE, HCT116 cells
with overexpression of METTL3; METTL3-EV, HCT116
cells transfected with empty vector; METTL3-si, HCT116 cells
transfected with METTL3-specific siRNA oligo;
METTL3-OE, METTL3 overexpression group;
METTL3-con, the control of METTL3-OE group;
inhibitor, inhibitor-treated group; inhibitor NC, the control of
inhibitor-treated group (PBS-treated group); mimics,
miR-let-7b overexpression groups; mimics NC, the control of
mimics group; circ-OE, HCT116 cells with overexpression of
hsa_circ_0000523; circ-EV, HCT116 cells transfected with
empty vector; circ-EV + mimics, empty vector and mimics were
co-transfected into HCT116 cells; circ-OE + mimics,
circ_0000523 overexpressing vector and mimics co-transfected
into HCT116 cells; METTL3, methyltransferase-like 3; siRNA,
small interfering RNA. |
Higher METTL3 expression is associated
with more aggressive tumor invasion in HCT116 cells
To explore the role of METTL3 in regulating
CRC metastasis, Transwell assays were performed to examine the
invasion of HCT116 cells with various levels of METTL3
expression. After plasmid (overexpression) or siRNA oligo
(knockdown) transfection, the number of cells that had migrated to
the surface of the lower chamber (filled with DMEM containing 10%
FBS) was used as an index for the metastatic ability of HCT116
cells. As shown in Fig. 4A, a
higher number of Giemsa-positive stained cells were observed in the
group with METTL3 overexpression compared with the group
with METTL3 knockdown.
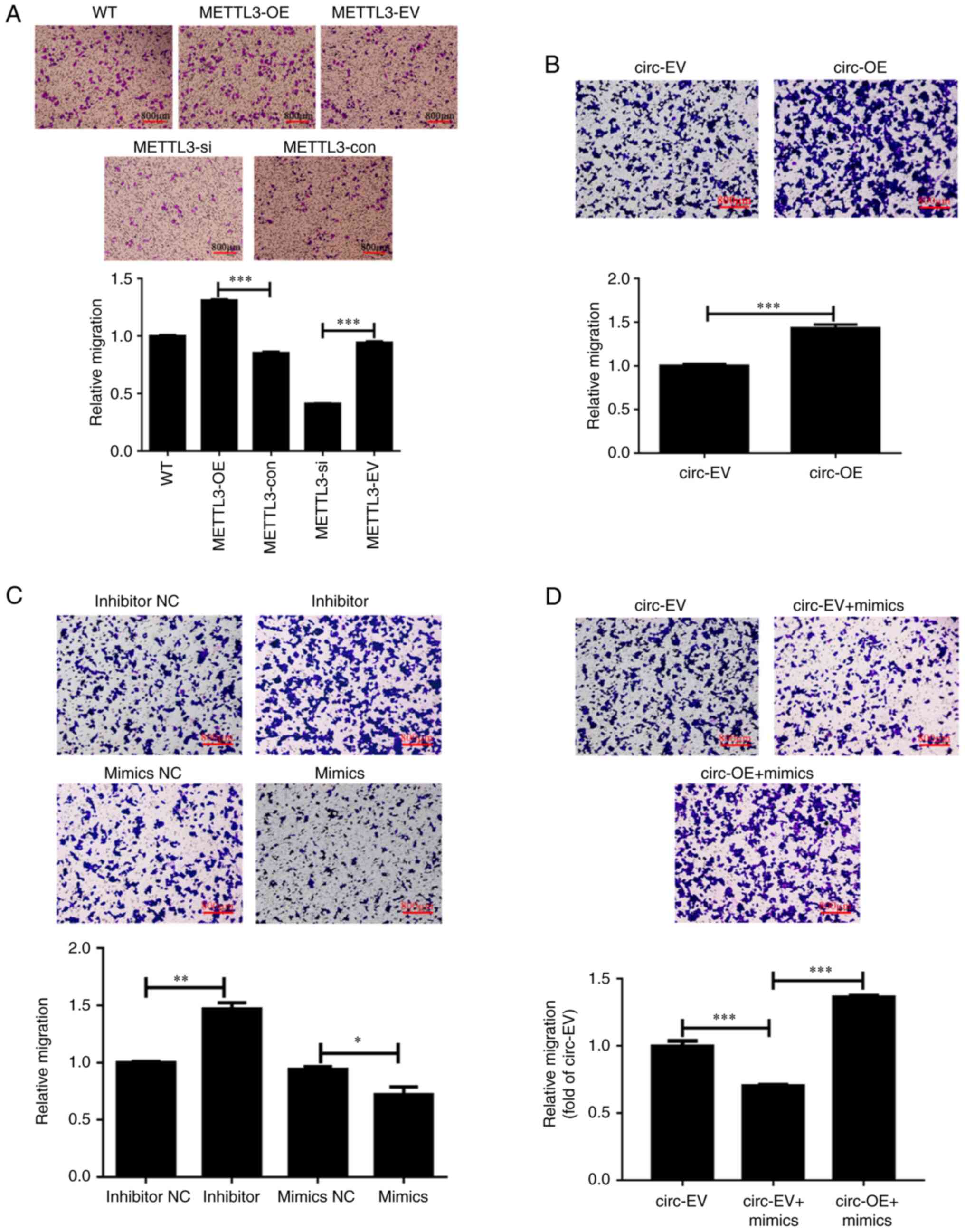 | Figure 4.METTL3, miR-let-7b and
circ_0000523 mediate invasion of HCT116 cells. (A-C)
Transwell assays reveal the invasion of HCT116 cells regulated by
(A) METTL3, (B) miR-let-7b and (C)
circ_0000523. (D) Transwell assays show the invasion
abilities in each group. Scale bar, 800 µm. n=3 for each group;
*P<0.05, **P<0.01 and ***P<0.001. WT, untreated wild-type
HCT116 cells; METTL3-OE, HCT116 cells with overexpression of
METTL3; METTL3-EV, HCT116 cells transfected with
empty vector; METTL3-si, HCT116 cells transfected with
METTL3-specific siRNA oligo; METTL3-con, the control
of METTL3-si; inhibitor, inhibitor-treated groups; inhibitor
NC, the control of inhibitor-treated groups; mimics,
miR-let-7b overexpression groups; mimics NC, the control of
mimics groups; circ-OE, HCT116 cells with overexpression of
hsa_circ_0000523; circ-EV, HCT116 cells transfected with
empty vector; circ-EV + mimics, empty vector and mimics
co-transfected into HCT116 cells; circ-OE + mimics,
circ_0000523 overexpressing vector and mimics co-transfected
into HCT116 cells; METTL3, methyltransferase-like 3; siRNA,
small interfering RNA. |
Statistical analysis confirmed the positive
association between METTL3 expression and the invasion of
HCT116 cells. There were no significant differences in invasion
among the untreated WT, empty vector-transfected and negative
control siRNA oligo-transfected HCT116 cells (Fig. 4A). Compared with empty
vector-transfected HCT116 cells (METTL3-EV group), HCT116
cells with ectopic expression of METTL3 (METTL3-OE
group) exhibited significantly higher invasion. Consistently,
invasion was decreased in HCT116 cells with METTL3 knockdown
compared with cells transfected with negative control siRNA oligos
(Fig. 4A). These results suggest
that higher expression of METTL3 is associated with more
aggressive tumor invasion in HCT116 cells. Next, it was examined
whether circ_0000523 and miR-let-7p mediate CRC cell
invasion. Compared with those in the circ-EV group, cells in the
lower chamber were increased by 43.2% in the circ-OE group,
suggesting significant levels of invasion in CRC cells (Fig. 4B). The cell invasion of
miR-let-7b-transfected cells (mimics group) obviously
decreased compared with the control group (mimics NC group), while
inhibitor treatment resulted in an obvious increase in CRC cell
invasion ability (Fig. 4C).
Furthermore, transfection of miR-let-7b mimics reduced the
invasion of CRC cells; however, this was restored by
circ_0000523 overexpression (Fig. 4D).
Discussion
circRNAs modulate the expression of parental genes
by regulating alternative splicing or transcription and acting as
competitive sponges for endogenous RNA or miRNA (26). circRNAs may provide more
comprehensive information on the key genes involved in oncogenesis
and the development of numerous cancers (27,28).
circRNAs are easily accessible and noninvasive biological markers
for the early detection of CRC (29,30).
However, few studies have reported on the specific functions of
circRNAs in the tumorigenesis and pathogenesis of human CRC. In the
present study, the expression patterns of hsa_circ_0000523
and its parental gene METTL3 were explored in the HCT116
human CRC cell line. The present results identified a potential
hsa_circ_0000523/miR-let-7b/METTL3 axis. The results of
gain-of-function and loss-of-function assays demonstrated that
expression of METTL3 promoted cell proliferation and
invasion and inhibited apoptosis in HCT116 cells. Rescue
experiments were performed to confirm the regulatory effects of
hsa_circ_0000523 on miR-let-7b. It was found that
circ_0000523 reversed the antitumor effects of
miR-let-7b on human CRC cell line characteristics.
Accumulating evidence has demonstrated the abnormal
expression and important biological functions of circRNAs in CRC
(12–14). circRNA plays various roles in CRC,
including in proliferation, metastasis and apoptosis (6). Notably, most circRNAs previously
identified by bioinformatic approaches have been found to be
downregulated in CRC cell lines and clinical CRC tissues (17). Huang et al (31). demonstrated that the expression of
circRNA itchy E3 ubiquitin ligase (circ_ITCH) was lower in
CRC tissues compared with adjacent noncancerous tissues.
circ_ITCH was found to sponge tumorigenic miR-7 and
miR-20a, which contribute to the malignancy of CRC (31). A positive association between
transcription of circ-ITCH and the parental gene ITCH
has been identified (31).
Notably, expression of circ-ITCH was found to suppress
proliferation in CRC cells (31).
Similarly, Zhu et al (32)
reported that circ-BANP was expressed in 35 CRC tissues, and
knockdown of circ-BANP decreased the proliferation of CRC
cells.
Indeed, there have been some studies reporting the
regulatory role of circRNAs in tumourigenesis and progression of
human CRC in previous years (33,34).
Jin et al (35) found a
deficiency in hsa_circ_0000523 could activate the
Wnt/β-catenin signaling pathway, which induced the development of
CRC. To the best of our knowledge, the present study found for the
first time that hsa_circ_0000523 successfully promotes CRC
via the miR-let-7b/METTL3 axis.
In addition, the expression levels of
hsa_circ_0000523, its potential target miR-let-7b,
and its parental gene METTL3 were evaluated in HCT116 cells.
Similar to the expression patterns of circ-ITCH and
ITCH, the transcript levels of hsa_circ_0000523 were
positively associated with mRNA levels of linear METTL3 in
HCT116 cells. Although the effects of hsa_circ_0000523
expression on cell proliferation were not measured, the ability of
METTL3 expression to increase HCT116 cell proliferation
suggests a similar proliferative role for hsa_circ_0000523.
However, a previous report by Jin et al (35) demonstrated that
hsa_circ_0000523 exerted anti-proliferative effects and
promoted apoptosis in two other human CRC cell lines (SW480 and
SW620). Therefore, although hsa_circ_0000523 can modulate
METTL3 expression by acting on miR-let-7b, additional
studies will be necessary to determine whether the expression of
METTL3 can impact the transcription of
hsa_circ_0000523 in HCT116 cells.
circRNAs may regulate the proliferation of CRC by
sequestering multiple miRNAs. For instance, circ-homeodomain
interacting protein kinase 3 (circ-HIPK3) has been demonstrated to
be upregulated in CRC tissue compared with normal tissue and
regulates cell proliferation by sponging nine miRNAs with 18
potential binding sites (36).
Specifically, circ-HIPK3 has been reported to bind to and inhibit
the activity of miR-124, a tumor suppressor that is typically
downregulated in CRC (37).
Therefore, circRNAs may modulate the tumorigenic proliferation of
CRC cells by diminishing the antitumor effects of certain
tumor-suppressive miRNAs.
miR-let-7b, shown here to be a potential
target of hsa_circ_0000523, is also a tumor suppressor in
multiple cancers (38). Human
miR-let-7b has been demonstrated to be downregulated in
various cancers, and the induction of tumorigenesis was revealed to
be inhibited in normal cells by ectopic expression of
miR-let-7b (39,40). Human miR-let-7 has been
reported to inhibit cancer growth by targeting various oncogenes
and inhibiting key regulators of several mitogenic pathways in
cancer (38). These results point
to a therapeutic potential for human miR-let-7 in cancer
treatment (38). Therefore, high
levels of hsa_circ_0000523 and low levels of
miR-let-7 may contribute to tumorigenesis in CRC.
On the other hand, circRNAs may regulate the
proliferation of CRC by indirectly targeting oncogenes.
METTL3, a target gene of miR-let-7b, was shown to
promote growth, survival and invasion in human lung cancer
(20). Furthermore, the oncogenic
roles of METTL3 have been demonstrated in numerous solid
tumors and hematopoietic malignancies (41,42).
For instance, it was found METTL3 depletion inhibited
tumorigenicity and sensitized lung cancer cells to
bromodomain-containing protein 4 inhibition (42). To the best of our knowledge, the
present study was the first report to describe the role of
METTL3 in CRC cells. METTL3 expression in HCT116
cells was found to be positively associated with proliferation and
cancer invasion and to be negatively associated with apoptosis.
These findings suggest that METTL3 functions as an oncogene
in CRC, potentially by promoting the translation of other
oncogenes. By contrast, METTL3 has been found to be a tumor
suppressor in renal cell carcinoma (43). METTL3 has been demonstrated
to promote cell proliferation, invasion and invasion in two human
renal cell carcinoma cell lines (CAKI-1 and CAKI-2). These effects
are mediated by modulation of the epithelial-to-mesenchymal
transition and the PI3K-Akt-mTOR signaling pathway (43). The molecular mechanisms underlying
the diverse roles of METTL3 in renal cell carcinoma and CRC
remain to be elucidated.
Through gain-of-function and siRNA oligo mediated
loss-of-function experiments, a potential
hsa_circ_0000523/miR-let-7b/METTL3 axis, which contributed
to tumorigenesis and pathogenesis in the HCT116 human CRC cell
line, was identified. METTL3 was demonstrated to promote
proliferation and invasion and to inhibit apoptosis in HCT116
cells. The present study on the
hsa_circ_0000523/miR-let-7b/METTL3 axis may facilitate
highly sensitive diagnosis of CRC and improved prognosis prediction
in patients following therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Scientific
Research Fund Project of Yunnan Education Department (grant no.
2019J1306). The funders had no role in study design, data
collection, data analysis, decision to publish or preparation of
the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
HL and YT designed and supervised the study. YW, BZ
and LL were responsible for analyzing the data, writing the
manuscript and revising it. YuZ, YoZ, LL and TS contributed to the
statistical analysis of the data and text correction. All authors
have read and approved the final manuscript. YW and BZ confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marley AR and Nan H: Epidemiology of
colorectal cancer. Int J Mol Epidemiol Genet. 7:105–114.
2016.PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Chen Z and Li J: The current
status of treatment for colorectal cancer in China: A systematic
review. Medicine (Baltimore). 96:e82422017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Favoriti P, Carbone G, Greco M, Pirozzi F,
Pirozzi RE and Corcione F: Worldwide burden of colorectal cancer: A
review. Updates Surg. 68:7–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fakih MG: Metastatic colorectal cancer:
Current state and future directions. J Clin Oncol. 33:1809–1824.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taborda MI, Ramirez S and Bernal G:
Circular RNAs in colorectal cancer: Possible roles in regulation of
cancer cells. World J Gastrointest Oncol. 9:62–69. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meng X, Li X, Zhang P, Wang J, Zhou Y and
Chen M: Circular RNA: An emerging key player in RNA world. Brief
Bioinform. 18:547–557. 2017.PubMed/NCBI
|
|
10
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang P and He X: Current research on
circular RNAs associated with colorectal cancer. Scand J
Gastroenterol. 52:1203–1210. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang P, Zuo Z, Shang W, Wu A, Bi R, Wu J,
Li S, Sun X and Jiang L: Identification of differentially expressed
circular RNAs in human colorectal cancer. Tumor Biol. Mar
28–2017.(Epub ahead of print).
|
|
14
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang W, Zhang X, Chu Q, Lu S, Zhou L, Lu
X, Liu C, Mao L, Ye C, Timko MP, et al: The circular RNA profiles
of colorectal tumor metastatic cells. Front Genet. 9:342018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu XW, Zheng BA, Hu ZM, Qian ZY, Huang CJ,
Liu XQ and Wu WD: Circular RNA hsa_circ_000984 promotes colon
cancer growth and metastasis by sponging miR-106b. Oncotarget.
8:91674–91683. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation-exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maity A and Das B: N6-methyladenosine
modification in mRNA: Machinery, function and implications for
health and diseases. FEBS J. 283:1607–1630. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Deng X, Su R, Weng H, Huang H, Li Z and
Chen J: RNA N6-methyladenosine modification in cancers: Current
status and perspectives. Cell Res. 28:507–517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m(6)A methyltransferase METTL3 promotes translation
in human cancer cells. Mol Cell. 62:335–345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen M, Wei L, Law CT, Tsang FH, Shen J,
Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al: RNA
N6-methyladenosine methyltransferase-like 3 promotes liver cancer
progression through YTHDF2-dependent posttranscriptional silencing
of SOCS2. Hepatology. 67:2254–2270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang
Z, Liu Y, Zhang X, Zhang W and Ye L: HBXIP-elevated
methyltransferase METTL3 promotes the progression of breast cancer
via inhibiting tumor suppressor let-7g. Cancer Letters. 415:11–19.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Taketo K, Konno M, Asai A, Koseki J,
Toratani M, Satoh T, Doki Y, Mori M, Ishii H and Ogawa K: The
epitranscriptome m6A writer METTL3 promotes chemo- and
radioresistance in pancreatic cancer cells. Int J Oncol.
52:621–629. 2018.PubMed/NCBI
|
|
24
|
Brattain MG, Fine WD, Khaled FM, Thompson
J and Brattain DE: Heterogeneity of malignant cells from a human
colonic carcinoma. Cancer Res. 41:1751–1756. 1981.PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shao T, Pan YH and Xiong XD: Circular RNA:
An important player with multiple facets to regulate its parental
gene expression. Mol Ther Nucleic Acids. 23:369–376. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang X, Zhang W and Shao Z: Prognostic
and diagnostic significance of circRNAs expression in lung cancer.
J Cell Physiol. 234:18459–18465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma Y, Zheng L, Gao Y, Zhang W, Zhang Q and
Xu Y: A comprehensive overview of circRNAs: Emerging biomarkers and
potential therapeutics in gynecological cancers. Front Cell Dev
Biol. 9:7095122021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tian J, Xi X, Yu J, Huang Q, Ma R, Zhang
X, Li H and Wang L: CircRNA hsa_circ_0004585 as a potential
biomarker for colorectal cancer. Cancer Manag Res. 11:5413–5423.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li RD, Guan M, Zhou Z, Dong SX and Liu Q:
The role of circRNAs in the diagnosis of colorectal cancer: A
meta-analysis. Front Med (Lausanne). 8:7662082021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang G, Zhu H, Shi Y, Wu W, Cai H and
Chen X: cir-ITCH plays an inhibitory role in colorectal cancer by
regulating the Wnt/β-catenin pathway. PLoS One. 10:e01312252015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu M, Xu Y, Chen Y and Yan F: Circular
BANP, an upregulated circular RNA that modulates cell proliferation
in colorectal cancer. Biomed Pharmacother. 88:138–144. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Peng K, Jiang P, Du Y, Zeng D, Zhao J, Li
M, Xia C, Xie Z and Wu J: Oxidized low-density lipoprotein
accelerates the injury of endothelial cells via
circ-USP36/miR-98-5p/VCAM1 axis. IUBMB Life. 73:177–187. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu D, Jia H, Zhang Z and Li S: Circ-PRMT5
promotes breast cancer by the miR-509-3p/TCF7L2 axis activating the
PI3K/AKT pathway. J Gene Med. 23:e33002021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jin Y, Yu LL, Zhang B, Liu CF and Chen Y:
Circular RNA hsa_circ_0000523 regulates the proliferation and
apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med
Biol Res. 51:e78112018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu K, Yao H, Lei S, Xiong L, Qi H, Qian
K, Liu J, Wang P and Zhao H: The miR-124-p63 feedback loop
modulates colorectal cancer growth. Oncotarget. 8:29101–29115.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Barh D, Malhotra R, Ravi B and Sindhurani
P: MicroRNA let-7: An emerging next-generation cancer therapeutic.
Curr Oncol. 17:70–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, et al: The let-7 microRNA represses cell proliferation
pathways in human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kumar MS, Erkeland SJ, Pester RE, Chen CY,
Ebert MS, Sharp PA and Jacks T: Suppression of non-small cell lung
tumor development by the let-7 microRNA family. Proc Natl Acad Sci
USA. 105:3903–3908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vu LP, Pickering BF, Cheng Y, Zaccara S,
Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al:
The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid
differentiation of normal hematopoietic and leukemia cells. Nat
Med. 23:13692017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Choe J, Lin S, Zhang W, Liu Q, Wang L,
Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al: mRNA
circularization by METTL3-eIF3h enhances translation and promotes
oncogenesis. Nature. 561:556–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li X, Tang J, Huang W, Wang F, Li P, Qin
C, Qin Z, Zou Q, Wei J, Hua L, et al: The M6A methyltransferase
METTL3: Acting as a tumor suppressor in renal cell carcinoma.
Oncotarget. 8:96103–96116. 2017. View Article : Google Scholar : PubMed/NCBI
|


















