Introduction
Lung cancer accounts for >10% of all cancer cases
worldwide and is one of the most common types of cancer (1). In 2018, 781,000 newly confirmed cases
of lung cancer and 626,000 mortalities were reported in China
(2). Small cell lung carcinoma
(SCLC), which is characterized by an unusually high proliferation
rate, a strong propensity for early metastasis and poor prognosis,
accounts for ~15% of all lung cancer cases. The 5-year survival
rate of patients with SCLC after diagnosis is only 2.8%, with a
median overall survival (OS) of ~10 months (3). In addition, the majority of patients
have metastatic disease at diagnosis, with only 1/3 of them having
earlier-stage disease, which increases the difficulty of clinical
treatment (4). Therefore, early
diagnosis of SCLC is of great importance to improve the OS of
patients with SCLC.
At present, early detection of the majority of tumor
types, including SCLC, mainly depends on the combination of
patients' timely medical consultation, as well as imaging and other
examinations. However, due to a variety of factors, including
patients' delayed medical treatment and doctors' clinical
experience, numerous patients are diagnosed at an advanced tumor
stage. Conventional methods, such as image-guided percutaneous
transthoracic puncture biopsy and bronchoscopy, are able to
accurately detect various pathological tumor types and have a role
in the diagnosis of tumors. However, biopsy and bronchoscopy are
not appropriate for patients with respiratory inadequateness or
unclear location of the tumor. With the continuous development of
molecular biology, serum tumor markers, such as neuron-specific
enolase (NSE) and progastrin-releasing peptide (ProGRP), have
gradually become diagnostic markers for malignant tumors, including
SCLC (5). However, their
specificity is not always satisfactory due to the elevated NSE and
ProGRP concentration in numerous patients. In addition, the
concentration of NSE in hemolytic samples is also increased
(6). Thus, identifying more
reliable diagnostic markers to compensate for the deficiencies of
these traditional serum markers is urgently required.
Chromosomal copy number variation is a hallmark of
cancer (7). Loss of heterozygosity
involving several chromosome 3p (chr3) regions accompanied by chr3
deletions is detected in almost 100% of SCLC cases. In addition,
these changes appear early in the pathogenesis of lung cancer. Such
3p genetic alterations suggest that the short arm of human chr3
contains several tumor suppressor genes (8). Deletions in chr3 have been identified
in lung adenocarcinoma tumors via the tumor sequencing project
initiative (9). Chr3 common
eliminated region 1 (C3CER1) on chr3p21.3 is a putative tumor
suppressor region. C3CER1 loss of heterozygosity exceeds 90% in
lung tumors compared with that of the putative tumor suppressor
gene fragile histidine triad diadenosine triphosphatase (65%) and
the tumor suppressor gene Von Hippel-Lindau (72%) (10). It is known that genetic and
epigenetic abnormalities of several genes residing in the chr3
region are important for the development of SCLC, but it remains
elusive how many they are and which of the numerous candidate tumor
suppressor genes are key factors in SCLC pathogenesis. Therefore,
exploring the diagnostic value of the chr3 gene in SCLC may
facilitate the clinical diagnosis and prognosis of SCLC.
In the present study, gene expression data obtained
from Gene Expression Omnibus (GEO) database were integrated to
perform data mining and analysis of SCLC. Next, a series of
co-differentially expressed genes were screened in SCLC. Several
analyses were carried out based on these genes, including
functional enrichment analysis, single-gene Gene Set Enrichment
Analysis (GSEA) and drug identification. Furthermore, the mRNA
levels of three diagnostic genes were detected in the SCLC cell
line NCI-H146 by reverse transcription-quantitative PCR (RT-qPCR).
The association between the genes in chr3 deletion regions and
disease progression of SCLC was analyzed and therapeutic drugs were
predicted based on these genes.
Materials and methods
Data source
The GSE40275 and GSE60052 datasets used in the
present study were downloaded from the GEO database. The GSE40275
dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40275)
contained 43 normal lung tissue samples (from 14 normal cases) and
21 SCLC tissue samples (from 8 cases of SCLC) (Table SI). The mean age of the eight
patients with SCLC (7 males and 1 female) in the GSE40275 dataset
was 67.1 years (range 54–70 years). A total of 86 samples were
included in the GSE60052 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60052),
comprising 7 normal lung tissue (from 7 normal cases) and 79 tumor
samples from patients with SCLC (Table SII). The GSE40275 dataset was
primarily used for screening of differentially expressed genes
(DEGs), weighted gene co-expression network analysis (WGCNA),
diagnostic biomarker assessment and single-sample GSEA, while the
GSE60052 dataset was employed for screening of DEGs and diagnostic
biomarker assessment.
The 505 chr3 genes (Table SIII) were obtained from the Human
Dec. 2013 (GRCh38/hg38) Assembly in the University of California
Santa Cruz (UCSC) Genome Browser Database (https://genome.ucsc.edu).
Analysis of DEGs
DEGs were identified in the GSE40275 and GSE60052
datasets individually using the ‘limma’ package in R software.
Genes that satisfied |log2 fold change (FC)|>1 and P<0.05
between normal and SCLC samples were considered as DEGs. The
overlapping genes of DEGs (co-DEGs) in the above two datasets were
obtained by Venn diagram analysis, which was performed on the jvenn
online website (http://jvenn.tou-louse.inra.fr/app/example.html).
DE-chr3 genes (overlapping genes between co-DEGs and chr3 genes)
were also identified. Furthermore, visualization and embellishment
of the Venn diagrams were achieved using the jvenn online
website.
Metascape analysis
Metascape (http://metascape.org/) is a powerful tool for
functional annotation analysis of genes (11) and was used in the present study to
perform a comprehensive functional enrichment analysis of the
DE-chr3 genes, including Gene Ontology (GO), Kyoto Encyclopedia of
Genes and Genomes (KEGG) and Reactome analyses. GO analysis was
performed in three categories: Cellular component (CC), molecular
function and biological process (BP) (12). KEGG (13) and Reactome (14) analyses were used to explore the
pathways in which genes may be involved. Terms with P<0.01 were
considered to be significantly enriched.
Ingenuity pathway analysis (IPA)
IPA (version 1–19; Qiagen Digital Insights) is a
cloud-based, graphical bioinformatics software that mines genomic
data for hidden biological significance from a biological pathway
perspective. The Ingenuity Knowledge Base (IPKB), the core
component of IPA, is a specialized biological interaction and
functional annotation database that contains information on
interactions between proteins, genes, compounds, cells, tissues,
drugs and diseases. In the present study, the DE-chr3 genes were
uploaded to IPKB and subjected to its Disease & Function
analysis.
WGCNA
Genes with mean expression values (fragments per
kilobase of exon per million mapped fragments) >1 were extracted
from all SCLC samples (GSE40275 dataset) to perform WGCNA using
clinical and expression data from the GSE40275 dataset. Clinical
trait-related modules were constructed and module genes were
identified in the R package WGCNA. In brief, a hierarchical
clustering analysis was performed on the GSE40275 dataset samples
to exclude outliers (i.e., the GSM990246 sample was an outlier and
was excluded; Fig. S1A). The
soft-threshold power was set to 11 (scale-free R2=0.85).
Modules were segmented by the dynamic tree-cutting algorithm and
MEDissThres (the dissimilarity threshold of module eigengenes) was
set to 0.5 to merge similar modules (Fig. S1B-D). Finally, Pearson
correlations between module genes and clinical features were
calculated. The genes in the modules that were most correlated with
clinical features were considered to be module genes. It should be
noted that, since the M-stage information for patients in the
GSE40275 dataset was recorded as MX for all, the M-stage was not
considered. Therefore, clinical characteristics included
pathological T and N stages (pT, pN), tumor stage (stage), sex and
age. Detailed functional annotation of module genes was performed
by GO and KEGG enrichment analysis in the clusterProfiler package
in R.
Receiver operating characteristic
(ROC) curves and area under the curve (AUC) estimation based on
diagnostic biomarkers
The overlapping genes of the module genes and
DE-chr3 genes were obtained by Venn diagram analysis and
subsequently used as candidate biomarkers. Next, ROC curves and
their corresponding AUCs were established from the GSE40275 and
GSE60052 datasets to evaluate the ability of candidate biomarkers
to correctly diagnose disease. Only candidate biomarkers with an
AUC >0.85 in both datasets were identified as diagnostic
biomarkers for SCLC. Furthermore, expression profiles were
extracted from the GSE40275 and GSE60052 databases to demonstrate
the expression patterns of the selected biomarkers.
Cell culture
Human pulmonary alveolar epithelial cells (HPAEpiC)
and the SCLC cell line NCI-H146 were purchased from the American
Type Culture Collection. HPAEpiC cells were maintained in DMEM
(HyClone; Cytiva) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Lonza Group, Ltd.). NCI-H146 cells were
cultured in RPMI-1640 medium (MilliporeSigma) supplemented with 10%
FBS and 1% penicillin-streptomycin. The cells were grown at 37°C in
a humidified atmosphere of 95% air and 5% CO2. The
experiments were carried out on cells of passage 10–25.
RNA extraction and RT-qPCR
Total RNA was extracted from HPAEpiC and NCI-H146
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The concentration and quality of RNA were
determined by spectrophotometry (Jinghua Technology) at 260 and 280
nm. Total RNA was reverse transcribed using the SureScript™
First-Strand cDNA Synthesis Kit (Genecopoeia, Inc.) according to
the manufacturer's protocol. RT-qPCR was performed using the
CFX96™ Real-Time PCR Detection System (Bio-Rad
Laboratories, Inc.) using the BlazeTaq™
SYBR®-Green qPCR Mix 2.0 kit (Genecopoeia, Inc.)
according to the manufacturer's instructions. The thermocycling
conditions were as follows: Initial denaturation at 95°C for 30
sec, followed by 40 cycles that each involved incubation at 95°C
for 10 sec, 60°C for 20 sec and 72°C for 30 sec. Relative
expression values were calculated using the 2−Δ∆Cq
method (15). The primers were
synthesized by TsingKe Biological Technology and their sequences
are listed in Table I.
 | Table I.Primer sequences for quantitative
PCR. |
Table I.
Primer sequences for quantitative
PCR.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| CDC25A |
TATGAGCAACCACTGGAGGT |
GTGACTGGGGTGTAAAAAGA |
| FYCO1 |
GTGGGGCAGGATTCGGAAAT |
TGGGGATCAGGCTGTAGGTG |
| RFTN1 |
TTCCTCCTTAGACCACCCGA |
AGTTCTCCACCATCTCCCTC |
| GAPDH |
CGCTGAGTACGTCGTGGAGTC |
GCTGATGATCTTGAGGCTGTTGTC |
Single-gene GSEA
Single-gene GSEA (in the R package clusterProfiler)
was applied to analyze the pathways enriched by each diagnostic
biomarker. In brief, samples were divided into high- and
low-expression groups using the median expression of each
diagnostic biomarker in the GSE40275 dataset. Subsequently, log2 FC
values were calculated for all genes between the high- and
low-expression groups for each diagnostic biomarker (of note, the
DEGs between the high- and low-expression groups were not defined;
only the difference between the two groups was analyzed) and were
ranked from highest to lowest according to the log2 FC value. The
sorted genes were used as the set of genes to be evaluated;
simultaneously, the KEGG pathway was used as a pre-defined set of
genes. Finally, single-gene GSEA was performed separately for each
diagnostic biomarker in the clusterProfiler package to detect the
enrichment of the pre-defined gene set in the set of genes to be
evaluated. An adjusted P<0.05 for the pathway was considered to
indicate a statistically significant difference.
Drug identification by the comparative
toxicogenomics database (CTD)
The CTD (http://ctdbase.org/) is a scientific database for
describing associations between chemicals, genes and human
diseases. For a certain gene, the CTD may provide the corresponding
target compounds in a descending order of their interactions. In
the present study, CTDs with default parameters were used to
predict the candidate drugs for the selected biomarkers.
Statistical analysis
Bioinformatics analysis was performed in R software.
The complete drug-diagnostic biomarker network was visualized with
Cytoscape software (v3.6.1; http://www.nigms.nih.gov/). For shared drugs,
intersection analysis was performed on the jvenn online website.
Subsequent shared drug-diagnostic biomarker networks were
represented in the diagrams.net online network (https://app.diagrams.net/). Histograms were
constructed using GraphPad Prism 8 software (GraphPad Software,
Inc.) to indicate the association between the tissue type (normal
and SCLC) and the mRNA levels of diagnostic biomarkers. Unless
otherwise stated, P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of the DE-chr3 genes
associated with SCLC
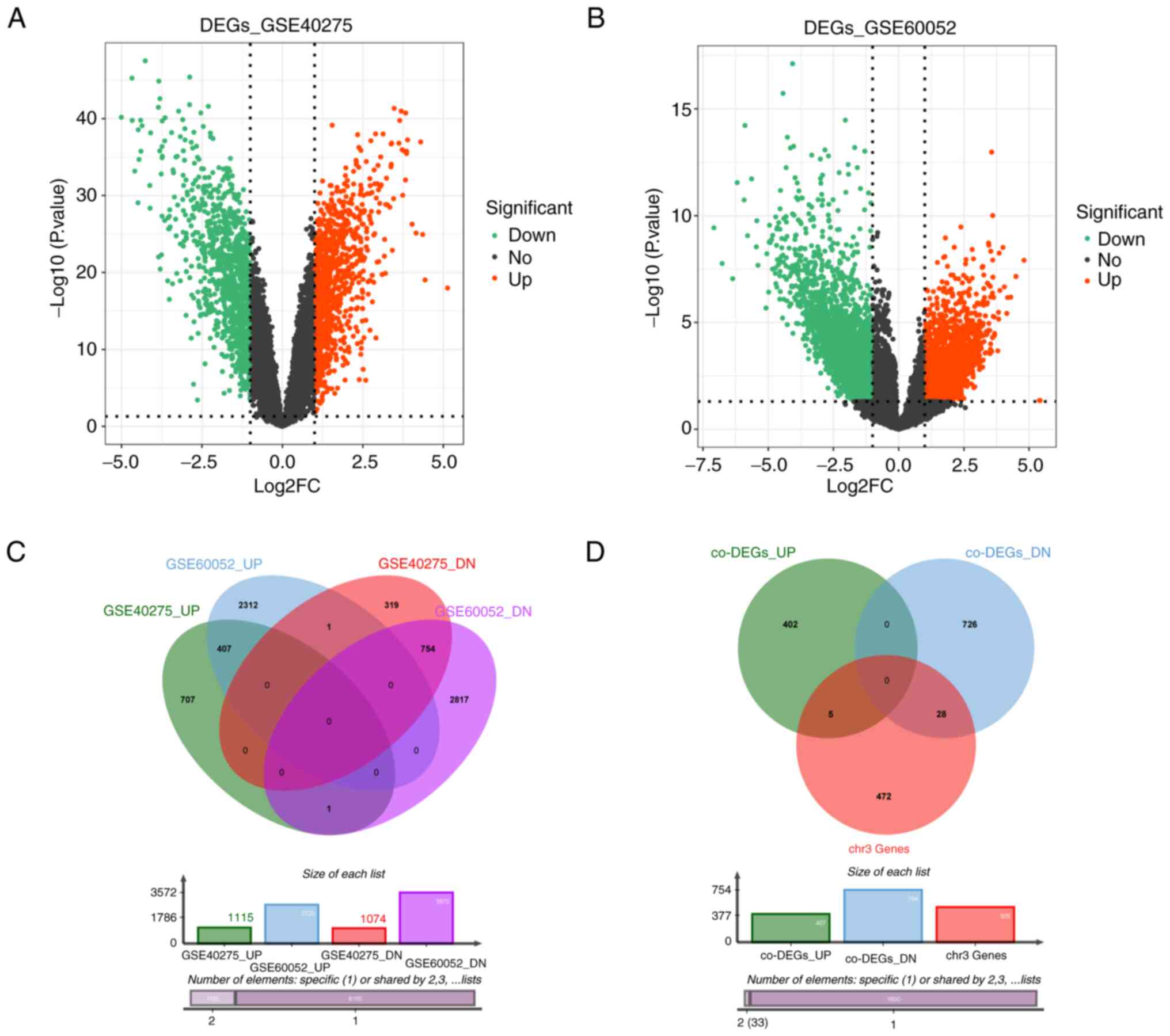
In the GSE40275 dataset, a total of 2,189 DEGs
between 21 SCLC and normal samples were identified, of which 1,115
were upregulated and 1,074 were downregulated (Fig. 1A; Table SIV). In total, 6,290 DEGs were
extracted from the GSE60052 dataset (79 SCLC vs. 7 normal),
including 2,720 upregulated and 3,572 downregulated genes (Fig. 1B; Table SV). Venn diagram analysis
indicated that a total of 1,162 co-DEGs (407 upregulated and 755
downregulated genes) were differentially expressed between SCLC and
normal samples in the above datasets (Fig. 1C; Table SVI). Subsequently, 505 genes
located in chr3:1-90000000 were obtained by Human Dec. 2013
(GRCh38/hg38) assembly in the UCSC Genome Browser Database and were
designated as chr3 genes (Table
SIII). Fig. 1D revealed that
only 33 of these 505 chr3 genes were co-DEGs, of which, 5 were
upregulated and 28 were downregulated (Fig. S2A and B; Table SVII).
Based on Metascape-GO analysis, the DE-chr3 genes
were observed to be closely associated with oxygen metabolic
process, cell motility and regulation of inflammatory responses.
Unexpectedly, regulation of supramolecular fiber organization and
supramolecular fiber organization were indicated to be
significantly enriched (Fig.
S2C-F; Table SVIII). Several
supercoiled supramolecular polymeric fibers of self-sorted
donor-acceptor molecules (supramolecular fibers) have been reported
as markers of lung cancer (16).
IPA's Disease & Function analysis revealed that these DE-chr3
genes were involved not only in cellular, tissue and organ
development, but also in cancer, tumor morphology and respiratory
diseases (Table SIX) which
suggested that the DE-chr3 genes probably have an essential role in
the development and progression of SCLC.
Search of module genes associated with
clinical features of SCLC by WGCNA
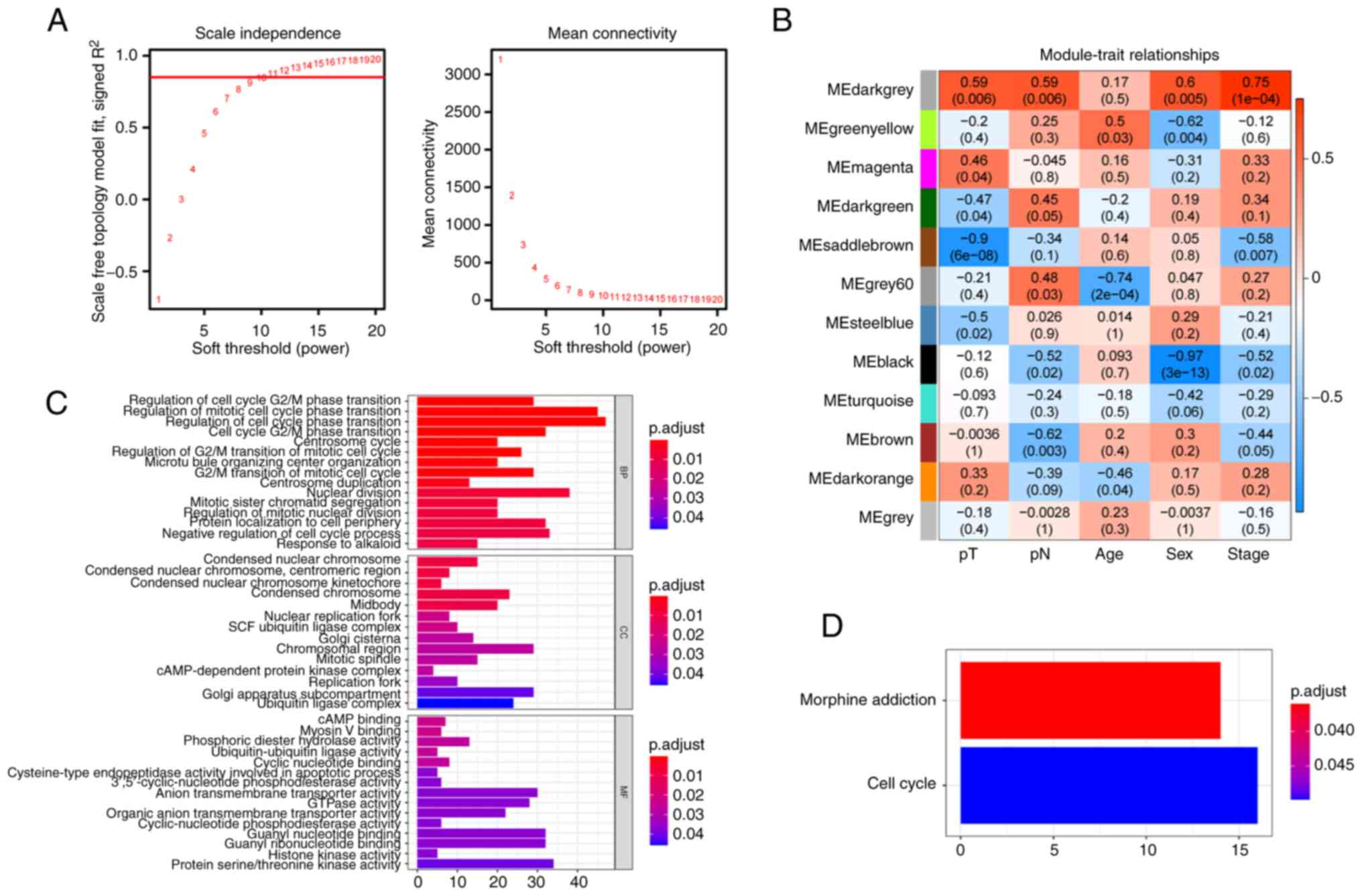
WGCNA was performed on 20 SCLC samples from the
GSE40275 dataset. With a soft threshold set to 11
(R2=0.85), the gene network infinitely approximated the
scale-free distribution (Fig. 2A).
A total of 12 co-expressed modules were then identified (Fig. 2B). Correlations between modules and
clinical characteristics were calculated, including age, sex, pT,
pN and stage. The dark grey module was strongly associated with pT
(r=0.59, P=0.006), pN (r=0.59, P=0.006), sex (r=0.6, P=0.005) and
stage (r=0.75, P=1×10−4). The dark grey module was
selected as the hub module and 1,156 genes (Table SX) of this module were
investigated in the subsequent analyses.
The potential functions of these module genes were
also explored. GO-BP analysis revealed that these genes were
significantly enriched in various cell cycle-related terms
(Fig. 2C; Table SXI). Similarly, the KEGG pathway
analysis also demonstrated that the cell cycle pathway was
significantly enriched (Fig. 2D;
Table SXII). These results
suggested that the variation in cell cycle processes may be
associated with SCLC progression.
Identification and assessment of
diagnostic biomarkers for SCLC
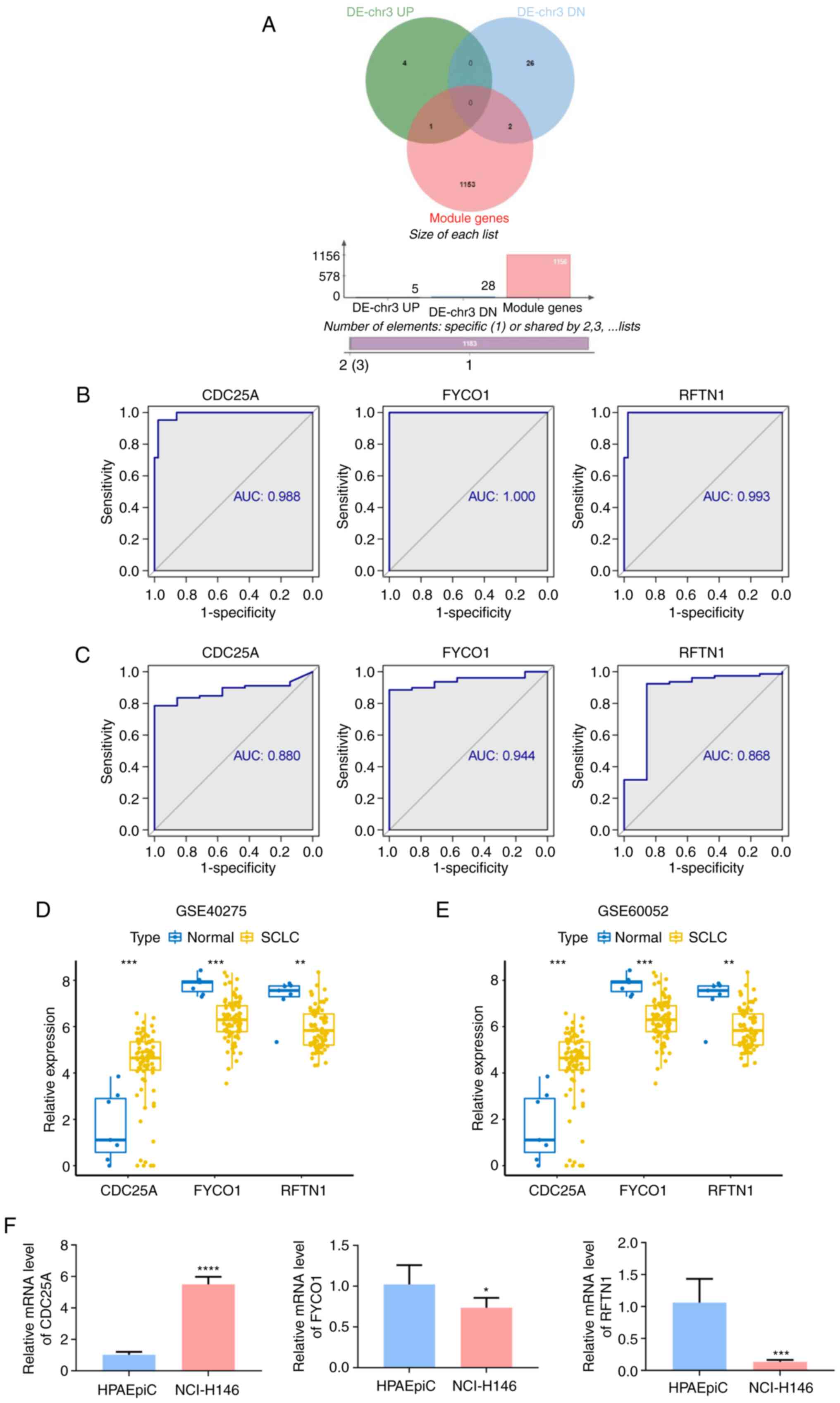
To further identify biomarkers for SCLC, three
overlapping genes for the DE-chr3 genes and the module genes were
obtained, namely cell division cycle 25 A (CDC25A), FYVE and
coiled-coil domain autophagy adaptor 1 (FYCO1) and lipid raft
linker 1 (RFTN1) (Fig. 3A). The
ability of the three overlapping genes to discriminate between
normal and SCLC samples was subsequently assessed by ROC curves
(Fig. 3B and C). CD25A had an AUC
of 0.988 in the GSE40275 dataset and of 0.880 in the GSE60052
dataset; FYCO1 had AUCs of 1 and 0.944 in the GSE40275 and GSE60052
datasets, respectively; and RFTN1 had AUCs of 0.993 and 0.868 in
the above GSE40275 and GSE60052 datasets, respectively. This
suggested that the above three overlapping genes possessed a robust
capacity to differentiate between the SCLC and normal groups and
were therefore considered as diagnostic biomarkers for SCLC in
subsequent analyses. CDC25A was upregulated in SCLC, whereas FYCO1
and RFTN1 were overexpressed in normal samples and the expression
patterns of these genes were consistent between the GSE40275
(Fig. 3D) and GSE60052 (Fig. 3E) datasets. Consistently, the
RT-qPCR results (Fig. 3F) revealed
that CDC25A exhibited a higher expression level in NCI-H146 cells
compared with that in HPAEpiC cells (P<0.0001), whereas FYCO1
and RFTN1 were significantly decreased in NCI-H146 cells (P=0.0421
and P=0.0005, respectively).
In addition, the association between the expression
of diagnostic biomarkers and the clinical characteristics of SCLC
were examined in the GSE40275 dataset. The results indicated that
the three genes were significantly associated with pT, stage and
sex. Specifically, CDC25A was markedly upregulated in pT4, stage
IIIB and male patients (Fig.
S3A); RFTN1 was markedly upregulated in pT4, stage III and male
patients (Fig. S3B); and FYCO1
was upregulated in pT2, stage I and female patients, as well as
those with pN0 (Fig. S3C).
Pathway enrichment analysis of each
diagnostic biomarker
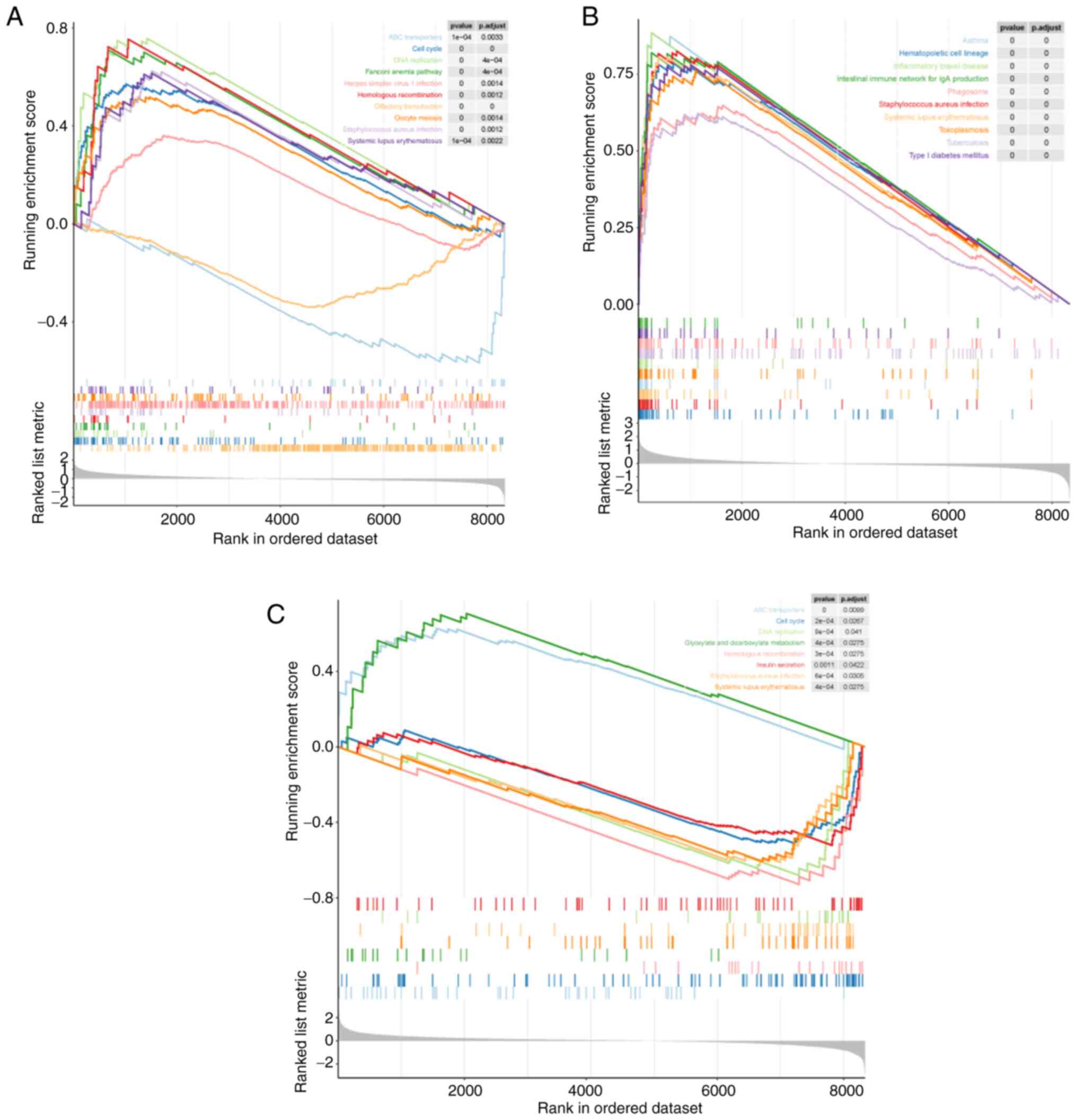
To reveal the potential pathways the diagnostic
biomarkers are involved in, single-gene GSEA was performed for each
diagnostic biomarker in the GSE40275 dataset. The results revealed
that CDC25A was enriched in a total of 22 KEGG pathways (Fig. 4A; Table SXIII); RFTN1 was significantly
associated with 52 KEGG pathways (Fig.
4B; Table SXIV); and FYCO1
was mainly involved in 8 KEGG pathways (Fig. 4C; Table SXV).
Specifically, all three diagnostic biomarkers were
involved in the KEGG pathways of ‘cell cycle’, ‘DNA replication’
and ‘homologous recombination’. This suggested that the diagnostic
biomarkers may also be involved in the proliferation process of
tumor cells. Of note, the ATP-binding cassette ‘(ABC) transporter’
pathway was significantly enriched by CDC25A and FYCO1. The ABC
transporter superfamily is known to be a family of membrane
transporter proteins. Increased intracellular drug pumping is
considered one of the mechanisms of tumor multidrug resistance and
numerous members of the ABC superfamily have also been indicated to
be involved in tumor multidrug resistance (17–19).
It was also observed that the ‘drug metabolism-other enzymes’
pathway was markedly associated with RFTN1. Such evidence indicated
that diagnostic biomarkers may aid the development of novel drugs
for SCLC. Of note, it was also observed that immune cell [T helper
‘(Th)17 cell differentiation’, ‘Th1 and Th2 cell differentiation’,
‘natural killer cell-mediated cytotoxicity’, ‘neutrophil
extracellular trap formation’] and immune response (‘antigen
processing and presentation’, ‘cell adhesion molecules’, ‘chemokine
signaling pathway’, ‘Fc e RI signaling pathway’ and ‘Rap1 signaling
pathway’)-related pathways were associated with RFTN1. The reason
why SCLC is reportedly not sensitive to immunotherapy is the
absence of a surface protein that triggers the immune response, and
this innate deficiency may also be associated with tumor immune
escape in SCLC (20). Therefore,
it was hypothesized that RFTN1 may provide a theoretical basis and
direction for identifying effective immunotherapeutic targets and
mechanisms for SCLC. In addition, several respiratory diseases were
also highlighted, including ‘asthma’ (CDC25A and RFTN1),
‘tuberculosis’ (RFTN1), ‘influenza A’ (RFTN1), ‘coronavirus
disease-COVID-19’ (RFTN1) and ‘pertussis’ (RFTN1).
Pharmaceutical prediction for
regulating the expression of diagnostic biomarkers
Based on the aforementioned results, CTD (21) was employed to predict potential
pharmaceutical agents that may modulate the expression of
diagnostic biomarkers. A total of 141 potential drugs were
identified by CTD (Table SXVI).
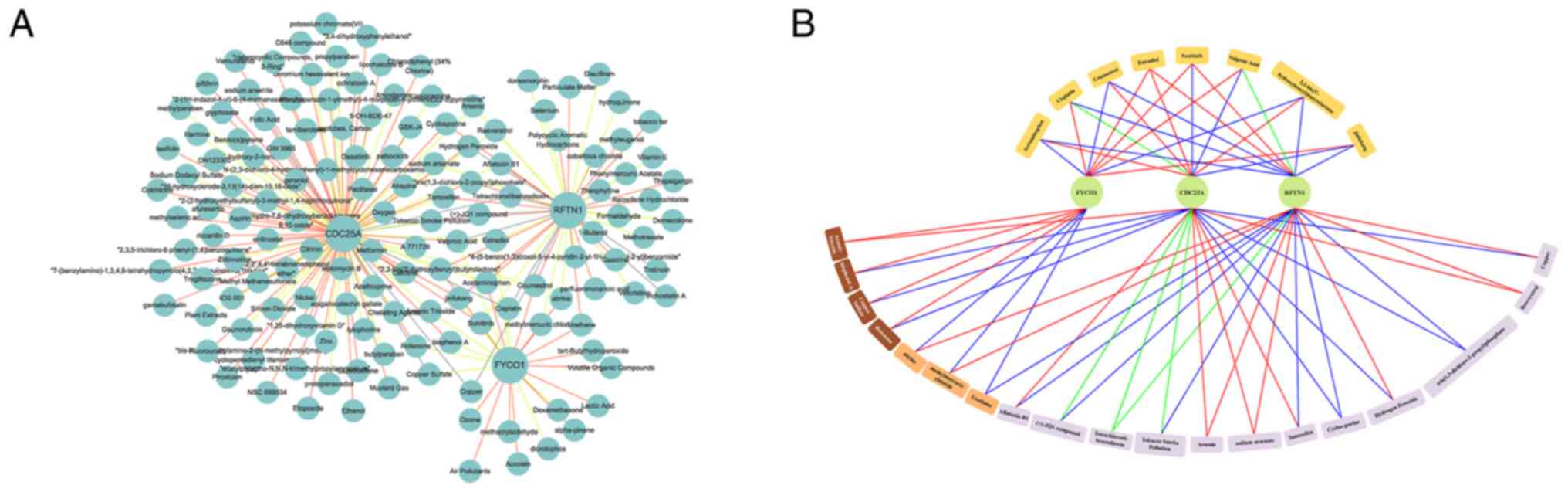
A complete drug-diagnostic biomarker network was constructed using
Cytoscape, which contained 144 nodes with 223 edges (Fig. 5A). The regulatory associations of
27 shared drugs (drugs shared by two diagnostic biomarkers and
drugs shared by all three diagnostic biomarkers) with diagnostic
biomarkers are presented in Fig.
5B. Acetaminophen, cisplatin, coumestrol, estradiol, sunitinib,
valproic acid, 2,3-bis(3′-hydroxybenzyl) butyrolactone and
Jinfukang were the shared drugs for the three diagnostic
biomarkers, which have potential to guide the development of novel
drugs for SCLC (Table II).
 | Table II.The 8 shared predicted drugs for
three diagnostic biomarkers and their interactions. |
Table II.
The 8 shared predicted drugs for
three diagnostic biomarkers and their interactions.
| A,
CDC25A∩FYCO1 |
|---|
|
|---|
|
| Interactions |
|---|
|
|
|
|---|
| Predicted drug | CDC25A | FYCO1 | RFTN1 |
|---|
| Arsenic
trioxide | UP | UP | N/A |
| Bisphenol A | DN | UP | N/A |
| Copper sulfate | DN | UP | N/A |
| Rotenone | DN | UP | N/A |
|
| B,
CDC25A∩RFTN1 |
|
|
|
Interactions |
|
|
|
| Predicted
drug | CDC25A | FYCO1 | RFTN1 |
|
| Aflatoxin B1 | UP | N/A | DN |
| (+)-JQ1
compound | UP/DN | N/A | DN |
|
Tetrachlorodi-benzodioxin | UP/DN | N/A | UP/DN |
| Tobacco smoke
pollution | UP/DN | N/A | DN |
| Arsenic | UP | N/A | UP |
| Sodium
arsenate | UP | N/A | UP |
| Tamoxifen | UP | N/A | DN |
| Cyclosporine | DN | N/A | DN |
| Hydrogen
peroxide | DN | N/A | UP |
|
Tris(1,3-dichloro-2-propyl)phosphate | DN | N/A | DN |
| Resveratrol | UP | N/A | UP |
| Copper | DN | N/A | UP |
|
| C,
FYCO1∩RFTN1 |
|
|
|
Interactions |
|
|
|
| Predicted
drug | CDC25A | FYCO1 | RFTN1 |
|
| Abrine | N/A | DN | UP |
| Methylmercuric
chloride | N/A | DN | UP |
| Urethane | N/A | DN | DN |
|
| D,
CDC25A∩FYCO1∩RFTN1 |
|
|
|
Interactions |
|
|
|
| Predicted
drug | CDC25A | FYCO1 | RFTN1 |
|
| Acetaminophen | UP | DN | DN |
| Cisplatin | UP/DN | UP | DN |
| Coumestrol | UP | DN | DN |
| Estradiol | UP | UP | UP |
| Sunitinib | DN | UP | UP |
| Valproic acid
2,3-bis | DN | UP | UP/DN |
| (3′-hydroxybenzyl)
butyrolactone | UP | DN | DN |
| Jinfukang | DN | UP | DN |
Discussion
SCLC is the most aggressive form of lung cancer.
Compared with NSCLC, SCLC is characterized by a rapid doubling time
and early, widespread metastases. The lack of early detection
modalities is one of the important barriers to progress in the
diagnosis and treatment of SCLC. Therefore, novel biomarkers with
high efficiency, sensitivity and specificity are urgently required
for the diagnosis and prognosis of SCLC. Previous studies have
indicated that chr3 alterations may be associated with the
pathogenesis of SCLC (8). Thus,
chr3 genes may become diagnostic biomarkers of SCLC.
The development of high-throughput sequencing has
facilitated the search for genes and mechanisms potentially
involved in cancer. Bioinformatics use genetic information to
improve molecular diagnoses and drug therapies (22). Integrated bioinformatics analysis
for the identification of therapeutic targets of certain cancers
based on transcriptomics, proteomics and high-throughput sequencing
may help to obtain novel information and to understand the
potential underlying molecular mechanisms. In the present study, 33
DE-chr3 genes were identified, which may have an essential role in
the development and progression of SCLC, and it was confirmed that
the variation in cell cycle processes may be associated with SCLC
progression according to GO-BP analysis. In addition, three
overlapping genes were obtained for the DE-chr3 and the module
genes (namely CDC25A, FYCO1 and RFTN1) by bioinformatics functional
assessment.
CDC25A is one of the cell cycle regulation-related
genes. CDC25A is mainly localized in the nucleus and controls
G1/S progression by dephosphorylation-dependent
inactivation of cyclin E/cyclin-dependent kinase (CDK)2 and cyclin
D/CDK4-6; G2 phase progression by dephosphorylation of
CDK1 and activation of the cyclin B1/CDK1 complex is a limiting
step (23). A previous study
suggested that CDC25A controls cell proliferation and tumorigenesis
by changing the expression of proteins involved in cyclin D1
regulation and G1/S transition (24). Maintenance of adequate CDC25A
levels is important for genomic stability and tumor suppression
(25). The activity and abundance
of CDC25A are intricately regulated and CDC25A is frequently
overexpressed in several cancer types (26–29).
Butz et al (30) revealed
that overexpression of CDK1 and CDC25A may have an important role
in promoting pituitary tumors in the G2/M transition
phase. The present study suggested that the expression of CDC25A in
SCLC was significantly increased and it was speculated that high
expression of CDC25A in SCLC may also promote the occurrence and
development of SCLC during the G2/M transition phase.
However, the specific mechanism requires to be further explored.
Rao et al (31) observed
that aberrant expression of the cell cycle regulation-related genes
cyclin D1, cyclin E, cyclin A, CDC25A and CDK4 may facilitate the
transcription and expression of genes associated with cell cycle
progression. A previous study suggested that the cell cycle could
be the pathway most closely associated with the pathogenesis of
SCLC (22). However, the functions
of the cell cycle and its regulatory proteins in SCLC have not been
fully clarified thus far.
FYCO1 is a late Rab7 effector of autophagy that is
required for the maturation of autophagosomes (32). FYCO1 has been reported to link
autophagosomes to the microtubule plus-end movement motor kinesin,
which promotes the maturation of autophagosomes and the formation
of autophagolysosomes (32).
Dionne et al (33) noticed
that FYCO1 regulated the accumulation of post-mitotic midbodies by
mediating light chain 3-dependent midbody degradation and FYCO1 was
reported as one of the key genes involved in adenoma-to-carcinoma
transition in colorectal cancer (34). Another study indicated that the
expression levels of FYCO1 in paired bladder tissue and urine
samples were significantly lower in bladder cancer than in those in
the control group (35). RFTN1 has
been studied in numerous diseases. Wang et al (36) concluded that RFTN1 may be involved
in the pathogenesis of glaucoma. Another study suggested that RFTN1
may participate in smoking behavior through modulating immune
responses or interactions with glucocorticoid receptor α and
androgen receptor (37). Zhao
et al (38) explored
molecular subtypes and core genes for lung adenocarcinoma and
screened out two core genes: Contactin 4 (CNTN4) and RFTN1. The
authors concluded that low expression of CNTN4 and RFTN1 predicted
unfavorable clinical outcomes in patients with lung adenocarcinoma.
The aforementioned studies demonstrated that FYCO1 and RFTN1 were
involved in the occurrence and development of a variety of tumors
and other diseases. However, the specific roles of CNTN4 and RFTN1
in the pathogenesis of SCLC have not been elucidated to date.
In the present study, it was determined that CDC25A
was upregulated in SCLC samples, while FYCO1 and RFTN1 were
upregulated in normal vs. SCLC samples. In addition, the three
genes were significantly associated with pT, stage and sex of
patients with SCLC. These results indicated that these three
overlapping genes may have an important role in the development of
SCLC. The results of single-gene GSEA for each diagnostic biomarker
revealed potential pathways the diagnostic biomarkers are involved
in, including ‘proliferation process of tumor cells’, ‘immune
cells’ and ‘immune response’. The specific roles of different
diagnostic biomarkers in different pathways require to be further
explored. The present results may facilitate the identification of
novel targeted drugs to improve the therapeutic effect and prolong
the survival time of patients with SCLC.
In the present study, pharmaceutical prediction for
regulating the expression of diagnostic biomarkers was performed
based on CTD. The results revealed that acetaminophen, cisplatin,
coumestrol, estradiol, sunitinib, valproic acid,
2,3-bis(3′-hydroxybenzyl) butyrolactone and Jinfukang were the
shared drugs for the aforementioned three diagnostic biomarkers,
which may potentially aid the development of novel drugs for SCLC.
A number of these drugs have been studied on lung cancer
previously. A large cohort study indicated that total non-steroidal
anti-inflammatory drug (NSAID) use was associated with a reduced
risk of lung cancer, which suggested that NSAIDs may be useful for
chemoprevention (39). Cisplatin,
a well-known chemotherapeutic drug, has been used for the treatment
of numerous human cancer types, including bladder, head and neck,
lung, ovarian and testicular cancer (40). Cisplatin exerts its anticancer
activity via multiple mechanisms, but the most accepted mechanism
involves the generation of DNA lesions by interacting with purine
bases on DNA, followed by the activation of several signal
transduction pathways, which finally lead to apoptosis (41). Coumestrol is a natural compound
exhibiting broad anticancer effects against skin melanoma, lung
cancer and colon cancer cell growth. The anticancer effect of
coumestrol is due to the direct targeting of haspin kinase
(42). Que et al (43) confirmed that Jingfukang induces
anticancer activity through oxidative stress-mediated DNA damage in
circulating human lung cancer cells. Platta et al (44) observed that the histone deacetylase
inhibitor valproic acid activates Notch1 signaling in SCLC cells,
induces changes in cell morphology and suppresses neuroendocrine
tumor markers. In addition, valproic acid profoundly inhibits SCLC
cell growth. Hubaux et al (45) demonstrated that valproic acid
improved the efficacy of a second-line regimen (vindesine,
doxorubicin and cyclophosphamide) in SCLC cells and mouse models.
The advances during the past decades in the genetics and biological
pathways driving SCLC have allowed the identification of multiple
novel therapeutic strategies. Current studies are underway to
explore combinations of immunotherapies, small molecules and
chemotherapy with immunotherapy, as well as biomarkers for the
selection of immunotherapies.
The present study has certain limitations that may
affect its conclusions. First, the conclusions were drawn based on
data from public databases rather than actual experiments,
indicating that the quality of the data cannot be guaranteed, and
that the results may be inaccurate. Furthermore, the propensity of
SCLC to metastasize extensively during the early stages of the
disease (most commonly to the brain, liver and bone) leads to a 95%
mortality rate. However, the M-stage was not considered in the
present study, as the M-stage information for all patients in the
GSE40275 dataset was recorded as MX. This may affect the accuracy
of the present results. Finally, due to a lack of public data on
the prognosis of patients with SCLC, the impact of new drugs on
survival was not further investigated in the present study.
The present study revealed three novel and powerful
diagnostic biomarkers for SCLC based on the chr3 genes CDC25A,
FYCO1 and RFTN1. These three genes were significantly associated
with pT, stage and sex. In addition, the present study provided
suggestions for the development and selection of drugs for clinical
treatment based on diagnostic biomarkers. The three aforementioned
diagnostic biomarkers may potentially guide the development of
novel drugs for SCLC. The present findings may offer novel
perspectives for patients with SCLC in future research and clinical
applications. However, additional experiments with larger sample
sizes are required in order to confirm these conclusions.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Huai Ning
(College of Physical Education, Yunnan Agricultural University,
Kunming, Yunnan, China) for providing help with the language.
Funding
This research was funded by the Natural Science Foundation of
China (grant no. 81960320), the Priority Union Foundation of Yunnan
Provincial Science and Technology Department and Kunming Medical
University (grant no. 202001AY070001-199) and the Yunnan Health
Training Project of High Level Talents (grant no. D-2019025).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW and JW conceived and designed the study. CM, JZ
and YW acquired the data. CM and JZ analyzed and interpreted the
data. HW, CM, JZ and YW were involved in the writing, review and
revision of the manuscript. HW and JW supervised the study. HW and
JW confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
SCLC
|
small-cell lung cancer
|
|
OS
|
overall survival
|
|
NSE
|
neuron-specific enolase
|
|
ProGRP
|
progastrin-releasing peptide
|
|
chr3
|
chromosome 3p
|
|
GEO
|
Gene Expression Omnibus
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
DEGs
|
Differential expression genes
|
|
WGCNA
|
weighted gene co-expression network
analysis
|
|
UCSC
|
University of California Santa
Cruz
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
IPKB
|
Ingenuity Knowledge Base
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under curve
|
|
HPAEpiC
|
human pulmonary alveolar epithelial
cells
|
|
ATCC
|
American Type Culture Collection
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
References
|
1
|
Semenova EA, Nagel R and Berns A: Origins,
genetic landscape, and emerging therapies of small cell lung
cancer. Genes Dev. 29:1447–1462. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rudin CM, Brambilla E, Faivre-Finn C and
Sage J: Small-cell lung cancer. Nat Rev Dis Primers. 7:32021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nicholson AG, Chansky K, Crowley J,
Beyruti R, Kubota K, Turrisi A, Eberhardt WE, van Meerbeeck J,
Rami-Porta R; Staging and Prognostic Factors Committee, ; et al:
The international association for the study of lung cancer lung
cancer staging project: Proposals for the revision of the clinical
and pathologic staging of small cell lung cancer in the forthcoming
eighth edition of the TNM classification for lung cancer. J Thorac
Oncol. 11:300–311. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zamay TN, Zamay GS, Kolovskaya OS, Zukov
RA, Petrova MM, Gargaun A, Berezovski MV and Kichkailo AS: Current
and prospective protein biomarkers of lung cancer. Cancers (Basel).
9:1552017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu L, Teng J, Zhang L, Cong P, Yao Y, Sun
G, Liu Z, Yu T and Liu M: The combination of the tumor markers
suggests the histological diagnosis of lung cancer. Biomed Res Int.
2017:20139892017.PubMed/NCBI
|
|
7
|
Li YL, Roberts ND, Wala JA, Shapira O,
Schumacher SE, Kumar K, Khurana E, Waszak S, Korbel JO, Haber JE,
et al: Patterns of somatic structural variation in human cancer
genomes. Nature. 578:112–121. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zabarovsky ER, Lerman MI and Minna JD:
Tumor suppressor genes on chromosome 3p involved in the
pathogenesis of lung and other cancers. Oncogene. 21:6915–6935.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weir BA, Woo MS, Getz G, Perner S, Ding L,
Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al:
Characterizing the cancer genome in lung adenocarcinoma. Nature.
450:893–898. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Petursdottir TE, Thorsteinsdottir U,
Jonasson JG, Moller PH, Huiping C, Bjornsson J, Egilsson V, Imreh S
and Ingvarsson S: Interstitial deletions including chromosome 3
common eliminated region 1 (C3CER1) prevail in human solid tumors
from 10 different tissues. Genes Chromosomes Cancer. 41:232–242.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
The Gene Ontology Consortium, . Expansion
of the gene ontology knowledgebase and resources. Nucleic Acids
Res. 45:D331–D338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fabregat A, Jupe S, Matthews L,
Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger
F, May B, et al: The reactome pathway knowledgebase. Nucleic Acids
Res. 46:D649–D655. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sandeep A, Praveen VK, Kartha KK,
Karunakaran V and Ajayaghosh A: Supercoiled fibres of self-sorted
donor-acceptor stacks: A turn-off/turn-on platform for sensing
volatile aromatic compounds. Chem Sci. 7:4460–4467. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li W, Zhang H, Assaraf YG, Zhao K, Xu XJ,
Xie JB, Yang DH and Chen ZS: Overcoming ABC transporter-mediated
multidrug resistance: Molecular mechanisms and novel therapeutic
drug strategies. Drug Resist Updat. 27:14–29. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chauncey TR: Drug resistance mechanisms in
acute leukemia. Curr Opin Oncol. 13:21–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pérez-Tomás R: Multidrug resistance:
Retrospect and prospects in anti-cancer drug treatment. Curr Med
Chem. 13:1859–1876. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu MR, Huang Y, Bender ME, Girard L,
Kollipara R, Eglenen-Polat B, Naito Y, Savage TK, Huffman KE,
Koyama S, et al: Evasion of innate immunity contributes to small
cell lung cancer progression and metastasis. Cancer Res.
81:1813–1826. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Davis AP, Grondin CJ, Johnson RJ, Sciaky
D, Wiegers J, Wiegers TC and Mattingly CJ: Comparative
toxicogenomics database (CTD): Update 2021. Nucleic Acids Res.
49:D1138–D1143. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liao Y, Yin GF, Wang X, Zhong P, Fan XM
and Huang CL: Identification of candidate genes associated with the
pathogenesis of small cell lung cancer via integrated
bioinformatics analysis. Oncol Lett. 18:3723–3733. 2019.PubMed/NCBI
|
|
23
|
Shen T and Huang S: The role of Cdc25A in
the regulation of cell proliferation and apoptosis. Anticancer
Agents Med Chem. 12:631–639. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sadeghi H, Golalipour M, Yamchi A,
Farazmandfar T and Shahbazi M: CDC25A pathway toward tumorigenesis:
Molecular targets of CDC25A in cell-cycle regulation. J Cell
Biochem. 120:2919–2928. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ray D, Terao Y, Fuhrken PG, Ma ZQ, DeMayo
FJ, Christov K, Heerema NA, Franks R, Tsai SY, Papoutsakis ET and
Kiyokawa H: Deregulated CDC25A expression promotes mammary
tumorigenesis with genomic instability. Cancer Res. 67:984–991.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sengupta S, Jana S and Bhattacharyya A:
TGF-β-Smad2 dependent activation of CDC 25A plays an important role
in cell proliferation through NFAT activation in metastatic breast
cancer cells. Cell Signal. 26:240–252. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brunetto E, Ferrara AM, Rampoldi F,
Talarico A, Cin ED, Grassini G, Spagnuolo L, Sassi I, Ferro A,
Cuorvo LV, et al: CDC25A protein stability represents a previously
unrecognized target of HER2 signaling in human breast cancer:
Implication for a potential clinical relevance in trastuzumab
treatment. Neoplasia. 15:579–590. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Albert H, Santos S, Battaglia E, Brito M,
Monteiro C and Bagrel D: Differential expression of CDC25
phosphatases splice variants in human breast cancer cells. Clin
Chem Lab Med. 49:1707–1714. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang P, Zou F, Zhang X, Li H, Dulak A,
Tomko RJ Jr, Lazo JS, Wang Z, Zhang L and Yu J: microRNA-21
negatively regulates Cdc25A and cell cycle progression in colon
cancer cells. Cancer Res. 69:8157–8165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Butz H, Németh K, Czenke D, Likó I,
Czirják S, Zivkovic V, Baghy K, Korbonits M, Kovalszky I, Igaz P,
et al: Systematic investigation of expression of G2/M transition
genes reveals CDC25 alteration in nonfunctioning pituitary
adenomas. Pathol Oncol Res. 23:633–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rao PC, Begum S, Sahai M and Sriram DS:
Coptisine-induced cell cycle arrest at G2/M phase and reactive
oxygen species-dependent mitochondria-mediated apoptosis in
non-small-cell lung cancer A549 cells. Tumour Biol.
39:10104283176945652017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Olsvik HL, Lamark T, Takagi K, Larsen KB,
Evjen G, Øvervatn A, Mizushima T and Johansen T: FYCO1 contains a
C-terminally extended, LC3A/B-preferring LC3-interacting Region
(LIR) motif required for efficient maturation of autophagosomes
during basal autophagy. J Biol Chem. 290:29361–29374. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dionne LK, Peterman E, Schiel J, Gibieža
P, Skeberdis VA, Jimeno A, Wang XJ and Prekeris R: FYCO1 regulates
accumulation of post-mitotic midbodies by mediating LC3-dependent
midbody degradation. J Cell Sci. 130:4051–4062. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sillars-Hardebol AH, Carvalho B, de Wit M,
Postma C, Delis-van Diemen PM, Mongera S, Ylstra B, van de Wiel MA,
Meijer GA and Fijneman RJ: Identification of key genes for
carcinogenic pathways associated with colorectal
adenoma-to-carcinoma progression. Tumour Biol. 31:89–96. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Eissa S, Matboli M, Awad N and Kotb Y:
Identification and validation of a novel autophagy gene expression
signature for human bladder cancer patients. Tumor Biology. Apr
5–2017.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang J, Qu D, An J, Yuan G and Liu Y:
Integrated microarray analysis provided novel insights to the
pathogenesis of glaucoma. Mol Med Rep. 16:8735–8746. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li M, Chen Y, Yao J, Lu S, Guan Y, Xu Y,
Liu Q, Sun S, Mi Q, Mei J, et al: Genome-wide association study of
smoking behavior traits in a Chinese Han population. Front
Psychiatry. 11:5642392020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao Y, Gao Y, Xu X, Zhou J and Wang H:
Multi-omics analysis of genomics, epigenomics and transcriptomics
for molecular subtypes and core genes for lung adenocarcinoma. BMC
Cancer. 21:2572021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Slatore CG, Au DH, Littman AJ, Satia JA
and White E: Association of nonsteroidal anti-inflammatory drugs
with lung cancer: Results from a large cohort study. Cancer
Epidemiol Biomarkers Prev. 18:1203–1207. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ghosh S: Cisplatin: The first metal based
anticancer drug. Bioorg Chem. 88:1029252019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim JE, Lee SY, Jang M, Choi HK, Kim JH,
Chen H, Lim TG, Dong Z and Lee KW: Coumestrol epigenetically
suppresses cancer cell proliferation: Coumestrol is a natural
haspin kinase inhibitor. Int J Mol Sci. 18:22282017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Que Z, Zhou Z, Luo B, Dong C, Jiang Y, Li
H and Tian J: Jingfukang induces anti-cancer activity through
oxidative stress-mediated DNA damage in circulating human lung
cancer cells. BMC Complement Altern Med. 19:2042019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Platta CS, Greenblatt DY, Kunnimalaiyaan M
and Chen H: Valproic acid induces Notch1 signaling in small cell
lung cancer cells. J Surg Res. 148:31–37. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hubaux R, Vandermeers F, Cosse JP,
Crisanti C, Kapoor V, Albelda SM, Mascaux C, Delvenne P, Hubert P
and Willems L: Valproic acid improves second-line regimen of small
cell lung carcinoma in preclinical models. ERJ Open Res.
1:00028–2015. 2015. View Article : Google Scholar : PubMed/NCBI
|