Introduction
Primary liver cancer remains one of the commonest
types of cancers and hepatocellular carcinoma (HCC) is its major
histologic subtype (1). Incidence
and mortality of liver cancer are associated with the infection of
viral hepatitis, which is a disease with significant geographic
distributions worldwide (2).
Although multiple novel techniques are now available for treatment
of this heterogeneous disease, identification and validation of
molecular factors that hold prognostic and therapeutic promise are
urgently needed.
Through past efforts in finding novel molecular
markers associated with survival outcomes of patients with breast
and gastric cancers by in silico data-mining analysis, it
was found that the zinc-fingers and homeoboxes (ZHX) family members
may be among the targets (3,4). ZHX
factors, including ZHX1, ZHX2 and ZHX3, have been reported as a
group of transcription factors with two zinc-finger motifs and five
homeobox DNA-binding domains existing in the cell nucleus (5–10).
Evidence has indicated that ZHX factors are important
transcriptional regulators in downstream signaling that is involved
in the osteogenic differentiation of mesenchymal stem cells,
development and differentiation of hematopoietic cells and
maintenance of neural progenitors (5,11,12).
Misexpression of ZHX factors has been associated with development
of various diseases, such as neurological, hematological and kidney
diseases (5,13,14).
Moreover, results from relevant studies suggest that ZHX family
members are involved in initiation and development of a variety of
types of cancer (3–5). The crucial roles of ZHX factors
provide reason enough for them as candidate biomarkers for cancer
surveillance, diagnosis and survival prediction. Nevertheless, to
the best of the authors' knowledge, the prognostic values of
individual ZHX factors in liver cancer remain to be elucidated. The
present study examined the expression patterns of ZHX factors and
the corresponding prognostic implications in liver cancer, using
integrative bioinformatics analyses with a set of online available
databases, including the Oncomine (http://www.oncomine.org/) (15), Tumor IMmune Estimation Resource
(TIMER) 2.0 (16), Cancer Cell
Line Encyclopedia (CCLE) database (http://sites.broadinstitute.org/ccle//) (3,4,17),
Kaplan-Meier Plotter (http://kmplot.com/analysis/) (18,19)
and cBioPortal (http://www.cbioportal.org/) (20,21).
Further, immunohistochemistry was performed to confirm ZHX3 protein
expression in liver cancer, as well as its association with
clinicopathologic variables and survival outcomes.
Materials and methods
Oncomine database analysis
The present study analyzed the expression of
distinct ZHX factors in cancers through the Oncomine database
(15). When the transcriptional
expression of ZHX factors in tumor tissues were compared to those
in noncancerous tissues, P<0.01 with a fold-change = 2 was
considered as statistically significant. Paired Student's t-test
was used.
Tumor IMmune Estimation Resource
(TIMER) database analysis
TIMER web server is an integrated online database
for comprehensive analysis of immune infiltrates through multiple
types of cancer (16). In the
current study, the gene expression profile of ZHX factors in
multiple types of cancer were evaluated via TIMER database analysis
(https://cistrome.shinyapps.io/timer/).
CCLE database analysis
The mRNA expression levels of specific ZHX factors
in diverse types of cancer cell lines were determined using the
CCLE database (http://portals.broadinstitute.org/ccle/), as described
previously (3,4,17).
Kaplan-Meier Plotter survival
analysis
The prognostic impacts of ZHX mRNA levels were
analyzed using the Kaplan-Meier Plotter online database, which
includes the information of 54,675 genes on survival using 10,461
clinical cancer samples, including 364 from patients with liver
cancer for outcome prediction analysis (18,19).
Data sources contain those from the Gene Expression Omnibus (GEO),
the European Genome-phenome Archive (EGA) and the Cancer Genome
Atlas (TCGA). To investigate the overall survival (OS) and
relapse-free survival (RFS) rates, patients were separated into
high and low-expression groups according to the median mRNA
expression levels so that survival analyses were conducted to
produce Kaplan-Meier plots. Hazard ratio with 95% confidence
interval and log-rank P-values were calculated.
cBioPortal cancer genomics database
analysis
The effects of genomic alterations of ZHX genes
containing mutations and copy-number variance on OS and
disease-free survival (DFS) rates in patients with liver cancer
were analyzed using the cBioPortal online database (20,21).
The raw data used prior to bioinformatic analysis are derived from
GEO and TCGA. In the present study, OncoPrint in cBioPortal were
employed to demonstrate the proportion and distribution of samples
with genetic alterations in ZHX genes.
Immunohistochemistry and
evaluation
The immunohistochemical staining for ZHX3 protein
expression was performed using a standard EnVision complex method
previously described (3,4,22,23).
One tissue microarray chip containing 94 primary HCC tissues and 86
adjacent noncancerous tissues was purchased from Outdo Biotech Co.,
Ltd. Following deparaffinization, rehydration and antigen
retrieval, 4-µm sections of tissue samples were incubated with a
rabbit polyclonal anti-ZHX3 antibody (catalog no. ab84677;
dilution, 1:500; Abcam) overnight at 4°C. ZHX3 protein staining was
visualized using an EnVision antibody complex (anti-Mouse/Rabbit)
method with an Envision Detection kit (OriGene Technologies, Inc.)
and 3,3′-diaminobenzidine as the chromogen substrate. Nuclei were
counterstained with 0.5% hematoxylin for 2 min at room
temperature.
A total of 10 random microscopic fields per slide
(magnification, ×400) were evaluated by two independent observers
who were unaware of the clinical information. Immunostaining was
graded semi-quantitatively by multiplication of staining intensity
and percentage of positive cells. The mean percentage of positively
stained cells was scored as follows: 0–5% (0); 5–25% (1); 26–50% (2); 51–75% (3); and 76–100% (4). The staining intensity was categorized
as follows: Absent (0); weak (1);
moderate (2); and strong (3). Tumor samples exhibiting a final
staining score of <2 were defined as low ZHX3 expression and
those with scores ≥2 as high ZHX3 expression.
Statistical analysis
Statistical analyses were conducted using the SPSS
17.0 statistical software package (SPSS Inc.). Associations between
the expression levels of ZHX factors and clinicopathological
variables were assessed using the Pearson's χ2 test or
Fisher's exact test. Survival curves were produced using the
Kaplan-Meier method and compared with the log-rank test. The
prognostic significance of the clinicopathological variables was
determined using a univariate Cox regression analysis. A Cox
proportional hazards regression model for multivariate analysis was
employed for factors that achieved significance in the univariate
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
mRNA expression profile of ZHX factors
in human cancers
Hitherto, three ZHX factors were characterized in a
variety of types of human cancer. Our previous study revealed that
the Oncomine database provided a total of 308, 434 and 416 unique
analyses for ZHX1, ZHX2 and ZHX3, respectively (3). However, the mRNA levels of ZHX
factors were not found in liver cancer datasets. The present study
thus examined the mRNA expression of ZHX factors in multiple types
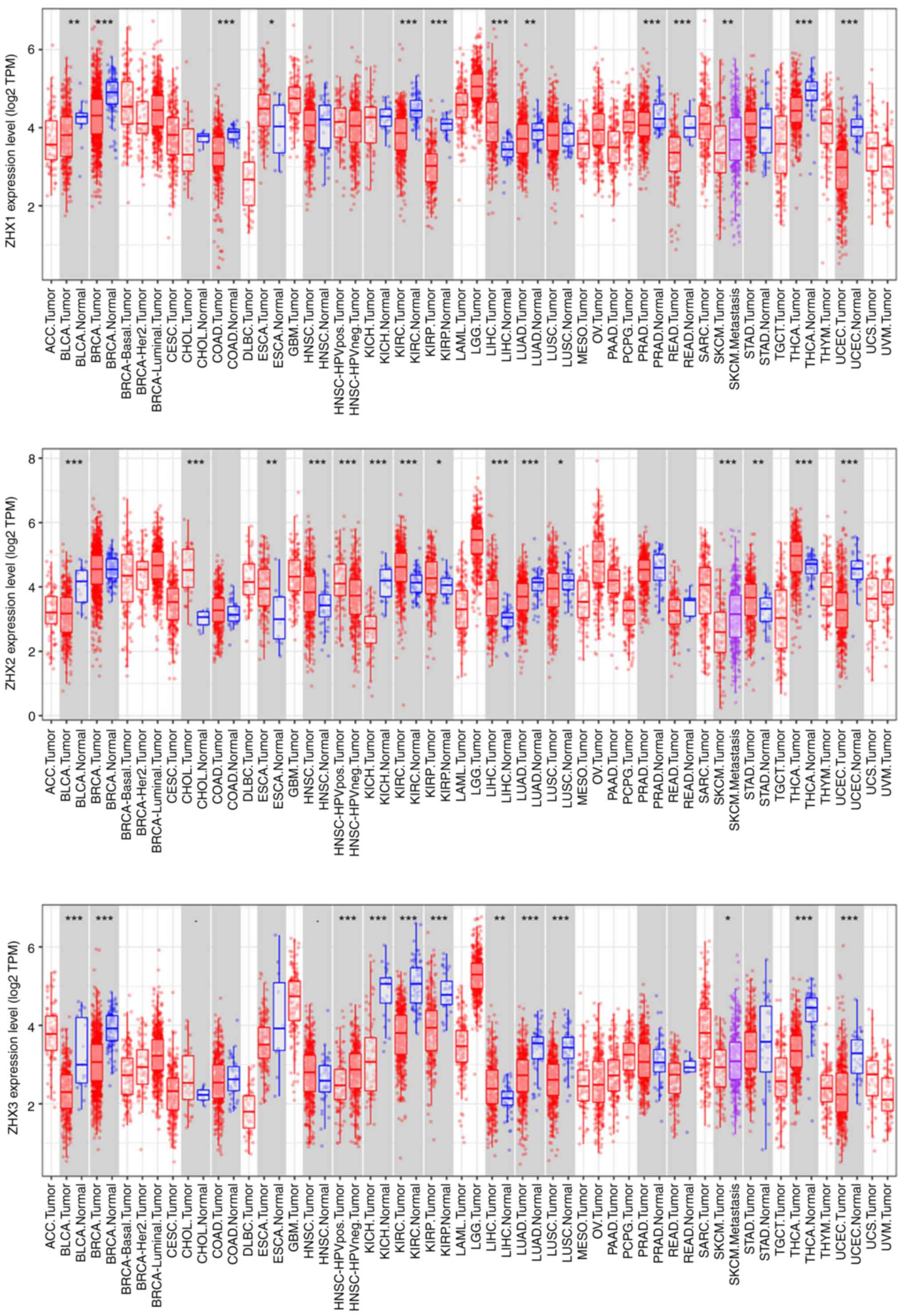
of cancer using the TIMER online database. The expression of all
three ZHX factors was significantly higher in liver hepatocellular
carcinoma (LIHC) tissues than in normal tissues (Fig. 1). Additionally, analyses from the
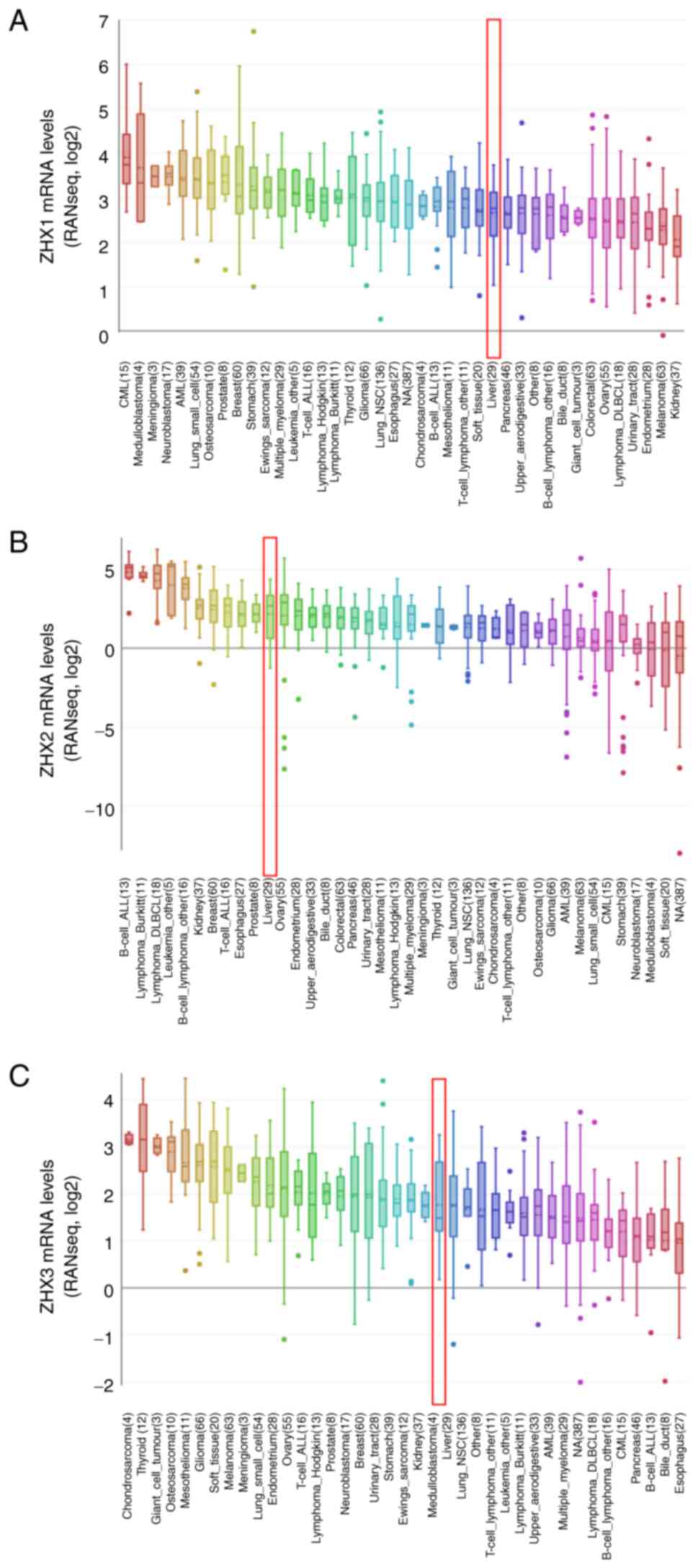
CCLE database revealed that the mRNA levels of ZHX1, ZHX2 and ZHX3
in liver cancer cells ranked the 27th, 11th and 23rd highest across
all types of cancer, respectively (Fig. 2).
Association between the expression of
ZHX factors and survival outcomes
The present study next identified the prognostic
impacts of ZHX family members on patient outcome via Kaplan-Meier
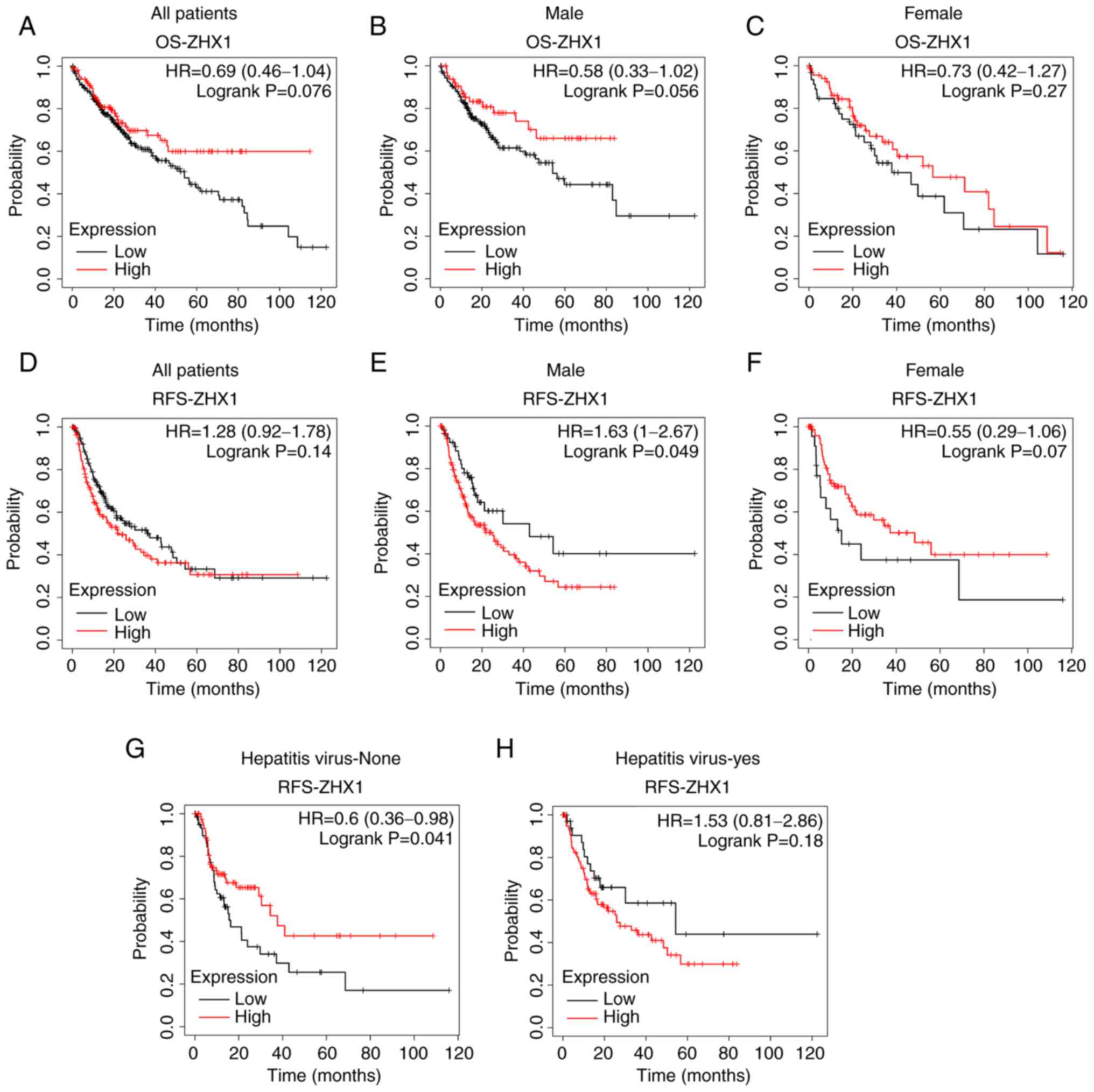
plotter survival analysis. ZHX1 mRNA level was not significantly
associated with OS in patients with liver cancer (Fig. 3A). Subgroup analyses showed no
significant association between ZHX1 mRNA expression and male
patients or female patients (Fig. 3B
and C). Similarly, ZHX1 mRNA expression was not correlated with
RFS in patients with liver cancer. Low expression of ZHX1 predicted
a longer RFS rate in male patients, but not in female patients
(Fig. 3E and F). Increased ZHX1
expression also displayed a longer RFS rate in patients without
hepatitis virus infection (Fig. 3G and
H).
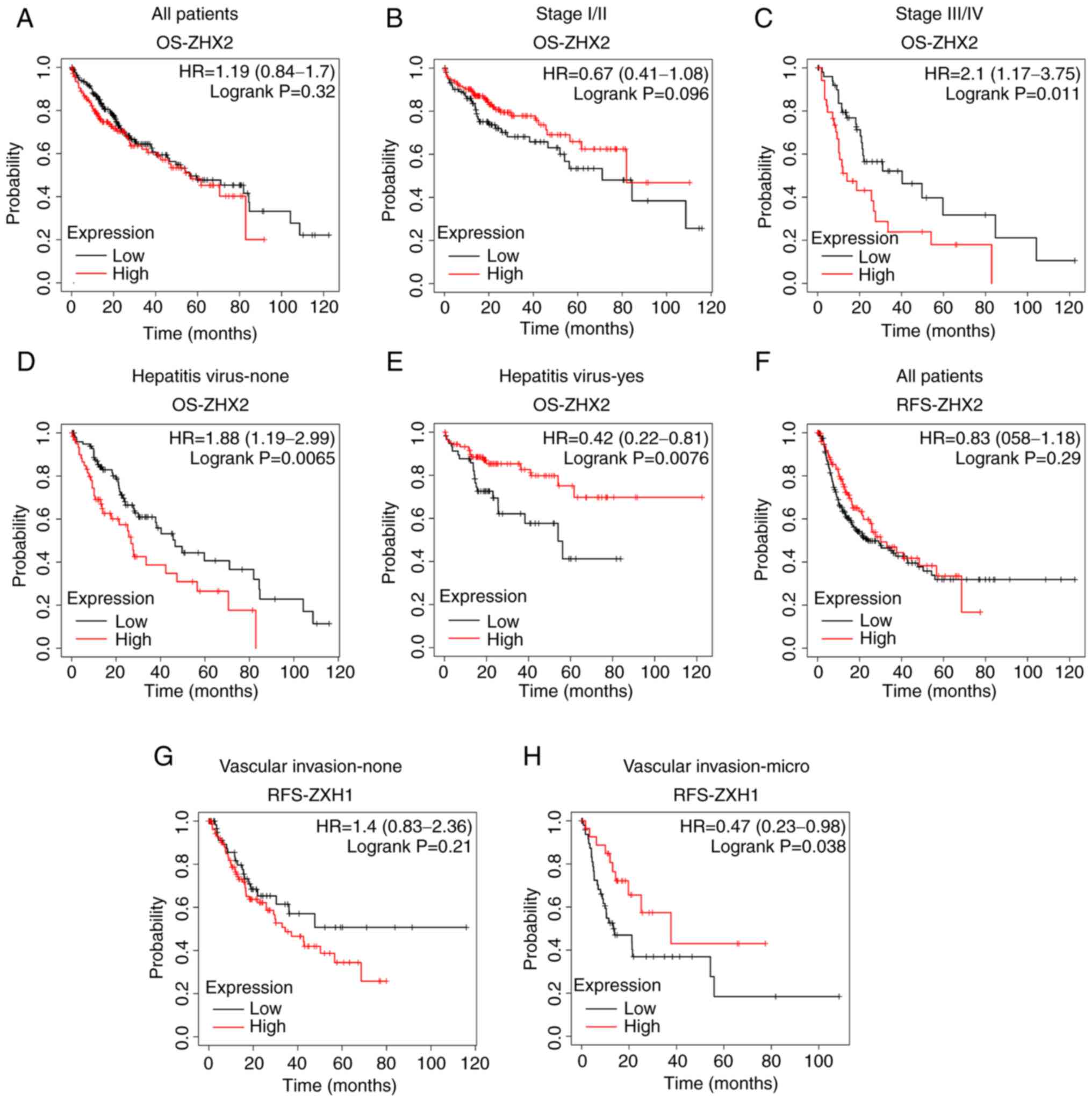
No significant association was observed between ZHX2
mRNA levels and OS in patients with liver cancer (Fig. 4A). Subgroup analyses suggested that
decreased ZHX2 expression indicated a longer OS rate in patients
with stage III/IV tumors but not in patients with stage I/II tumors
(Fig. 4B and C). Decreased ZHX2
mRNA level was associated with an improved OS in patients without
hepatitis virus infection (Fig.
4D), whereas increased ZHX2 expression was associated a
favorable OS in patients with hepatitis virus infection (Fig. 4E). Similarly, ZHX2 expression was
not significantly associated with RFS in patients with liver cancer
(Fig. 4F). High expression of ZHX2
implied longer RFS times in patients with micro vascular invasion,
but not in those without vascular invasion (Fig. 3G and H).
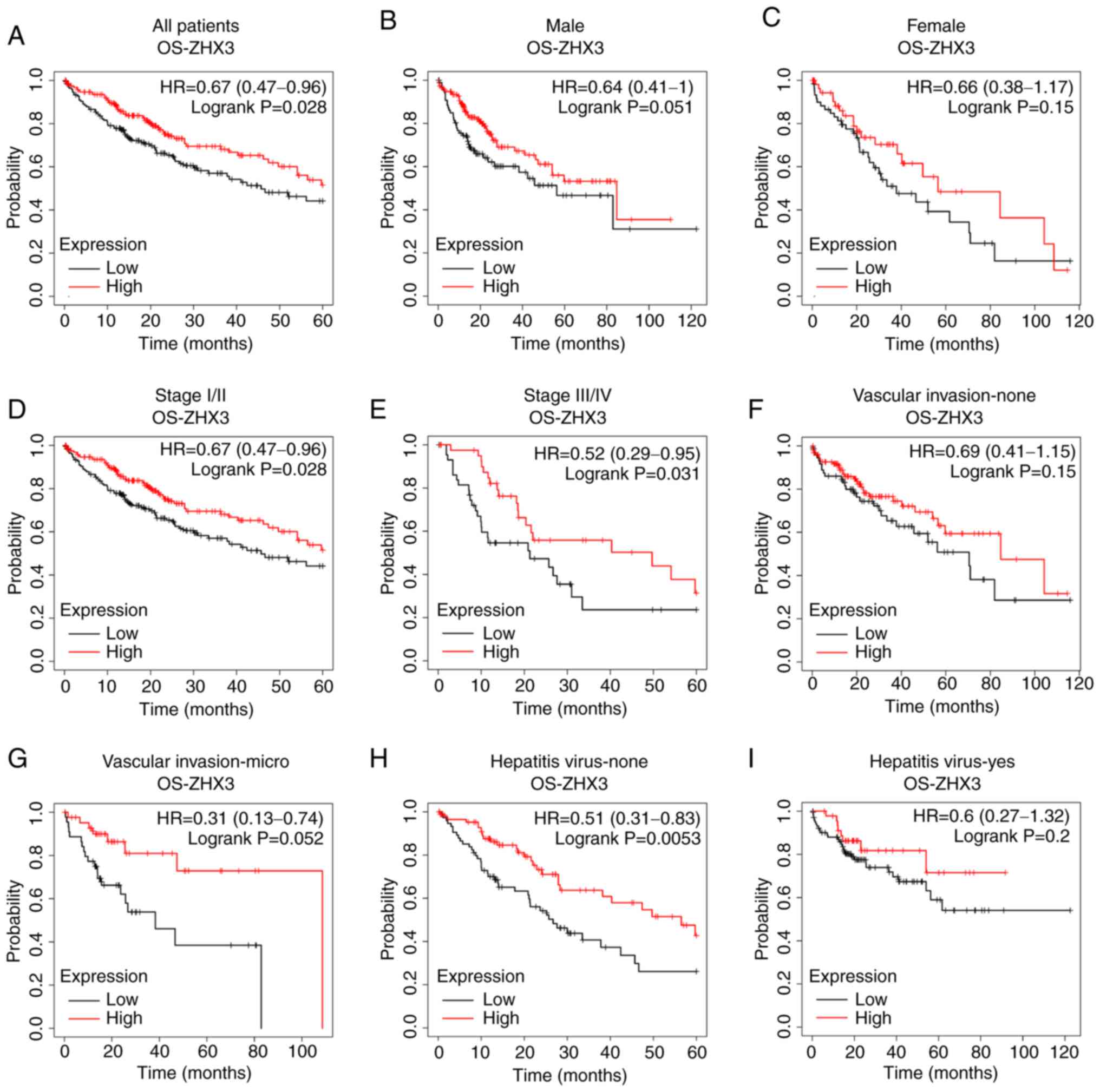
Regarding ZHX3, its upregulation was found to be
associated with a prolonged OS rate in patients with liver cancer
(Fig. 5A). Subgroup analyses
showed that no significant correlation between ZHX2 expression and
OS either in male patients or in female patients (Fig. 5B and C). Increased ZHX3 expression
exhibited longer OS times in patients with Stage I/II tumors and
Stage III/IV tumors (Fig. 5D and
E). High ZHX3 mRNA level represented an improved OS rate in
patients with micro vascular invasion, but not in those without
micro vascular invasion (Fig. 5F and
G). In addition, elevated ZHX3 expression illustrated a longer
OS in patients without hepatitis virus infection (Fig. 5H), but not in those with hepatitis
virus infection (Fig. 5I).
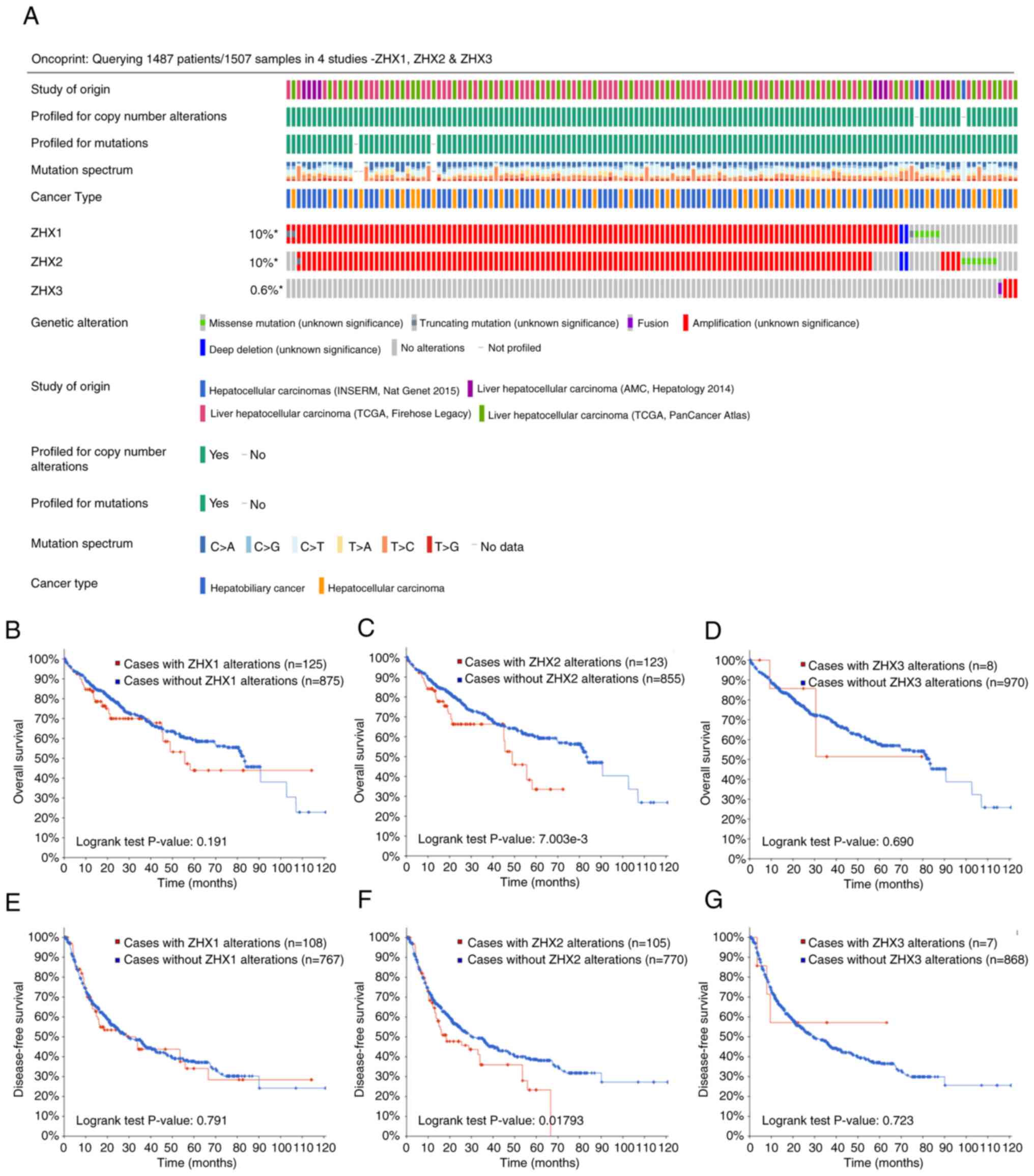
Correlation between genetic
alterations of ZHX factors and survival outcomes
The prognostic association between genetic
alterations of ZHX factors and outcomes in patients with liver
cancer was further characterized using the CbioPORTAL online
database. The genetic alteration rates for ZHX1, ZHX 2 and ZHX3
were 10, 10 and 0.6%, respectively (Fig. 3). The genetic alteration of ZHX2
was found to be associated with OS in patient with liver cancer
(Fig. 6C). Nevertheless, no other
significant relationship was observed between genetic alterations
of ZHX factors and patient survival, as regarding either OS or DFS
(Fig. 6B and D-G).
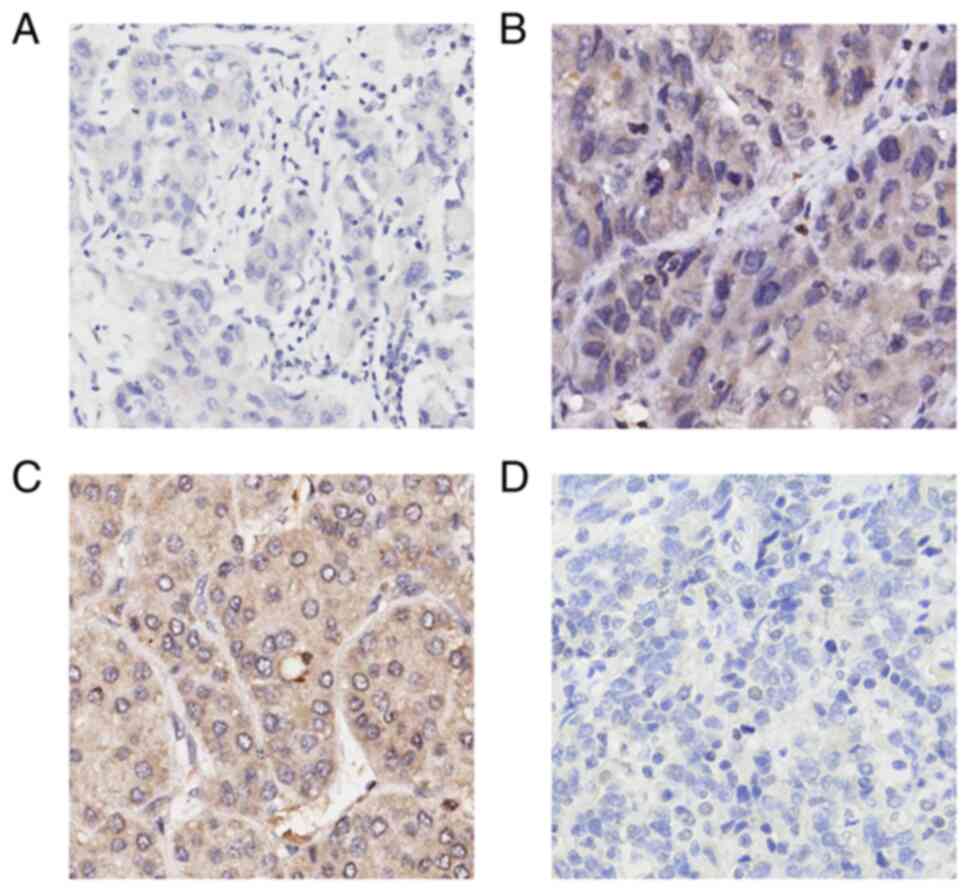
ZHX3 expression is an independent
prognostic factor in liver cancer
To support the above results, the expression status
of ZHX3 protein was thus examined using one tissue microarray chip
containing total 94 primary HCC specimens. A high level of ZHX3
protein expression primarily in the cytoplasm of cancer cells in
48.9% (46/94) of the HCC specimens tested was observed (Fig. 7). Low ZHX3 expression was found to
be associated with larger tumor size, advanced TNM staging and T
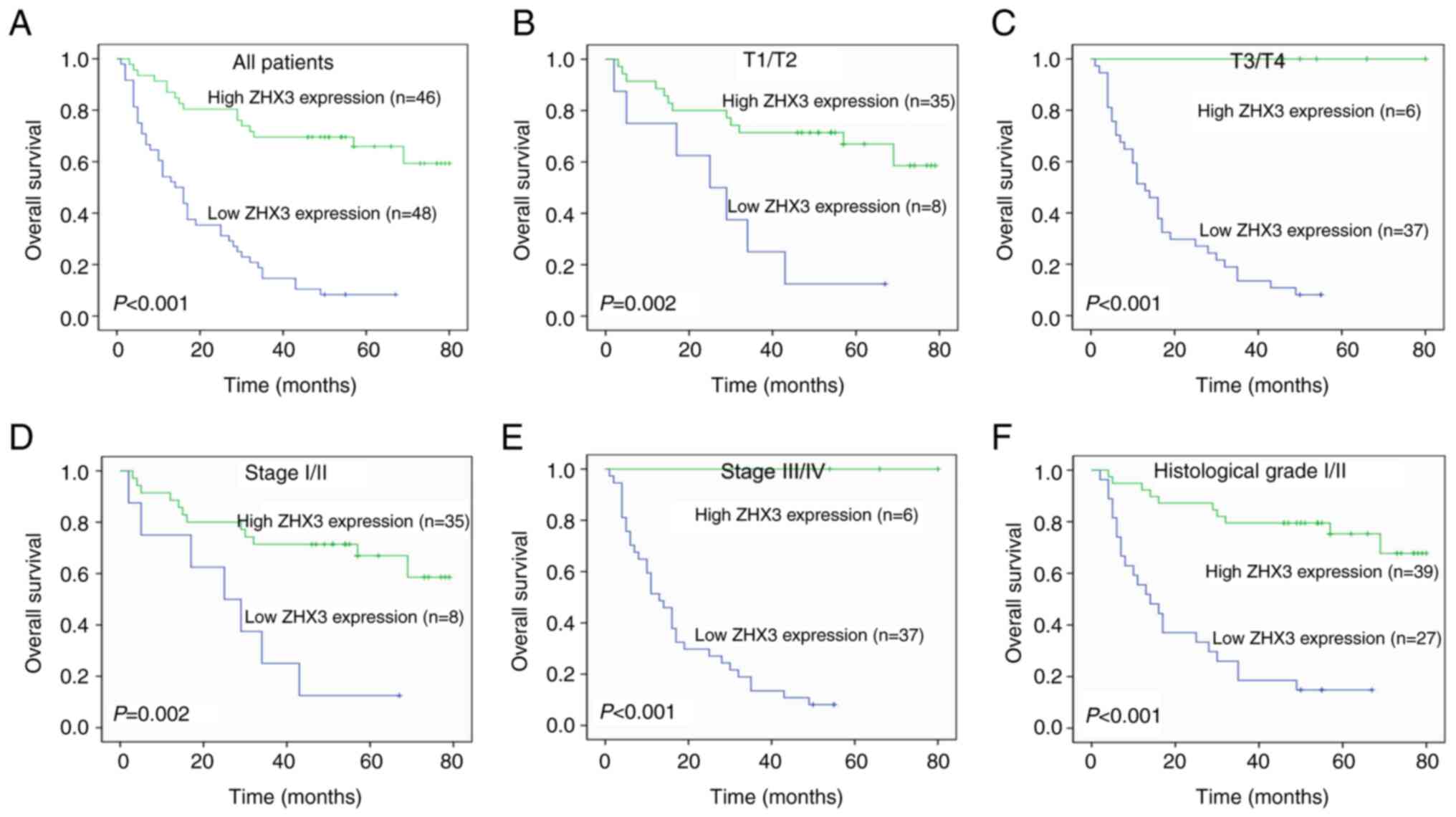
stage, positive thrombus status and TP53 expression (Table I). Kaplan-Meier survival analyses
demonstrated that patients with high ZHX3 expression had an
improved OS compared with those with low ZHX3 expression (Fig. 8A). Subgroup analyses showed that
high ZHX3 expression indicated an improved OS in patients both with
T1/T2 tumors and T3/T4 tumors (Fig. 8B
and C). ZHX3 overexpression also exhibited a longer OS in
patients both with Stage I/II and Stage III/IV tumors (Fig. 8D and E). In addition, elevated ZHX3
suggested an improved OS in patients with histological grade I/II
tumors (Fig. 8F). In the
univariate analysis, larger tumor size, advanced TNM stage, higher
histological grade, positive thrombus status and ZHX3 expression
were determined to be associated with an unfavorable OS (Table II). After correcting the
prognostic variables obtained in the univariate analysis, only
histological grade and ZHX3 expression kept the independent
implication in the multivariate analysis (Table II).
 | Table I.Correlation between ZHX3 expression
and clinicopathological variables in liver cancer. |
Table I.
Correlation between ZHX3 expression
and clinicopathological variables in liver cancer.
|
|
| ZHX3
expression |
|
|---|
|
|
|
|
|
|---|
| Parameters | No. of
patients | Low, n
(%) | High, n
(%) | P-value |
|---|
| Age |
|
|
|
|
| ≤60
years | 53 | 29 (54.7) | 24 (45.3) | 0.491 |
| >60
years | 40 | 19 (47.5) | 21 (52.5) |
|
| NA | 1 |
|
|
|
| Sex |
|
|
|
|
|
Male | 10 | 3 (30.0) | 7 (70.0) | 0.194 |
|
Female | 84 | 45 (53.6) | 39 (46.4) |
|
| Tumor size |
|
|
|
|
| ≤5
cm | 39 | 9 (23.1) | 30 (76.9) | <0.001 |
| >5
cm | 54 | 38 (70.4) | 16 (29.6) |
|
| NA | 1 |
|
|
|
| Histological
grade |
|
|
|
|
|
I/II | 66 | 27 (40.9) | 39 (59.1) | 0.932 |
|
III | 28 | 21 (75.0) | 7 (25.0) |
|
| TNM Stage |
|
|
|
|
|
I/II | 43 | 8 (18.6) | 35 (81.4) | <0.001 |
|
III/IV | 43 | 37 (86.0) | 6 (14.0) |
|
| NA | 8 |
|
|
|
| T Stage |
|
|
|
|
|
T1/T2 | 43 | 8 (18.6) | 35 (81.4) | <0.001 |
|
T3/T4 | 43 | 37 (86.0) | 6 (14.0) |
|
| NA | 8 |
|
|
|
| Thrombus |
|
|
|
|
|
Negative | 75 | 36 (48.0) | 39 (52.0) | 0.013 |
|
Positive | 7 | 7 (100.0) | 0 (0.0) |
|
| NA | 12 |
|
|
|
| Cirrhosis |
|
|
|
|
|
Negative | 58 | 31 (53.4) | 27 (46.6) | 0.557 |
|
Positive | 36 | 17 (47.2) | 19 (52.8) |
|
| AFP |
|
|
|
|
|
Negative | 39 | 23 (59.0) | 26(41.0) | 0.167 |
|
Positive | 54 | 24 (44.4) | 30(55.6) |
|
| NA | 1 |
|
|
|
| CD34 |
|
|
|
|
|
Negative | 37 | 17 (45.9) | 20 (54.1) | 0.524 |
|
Positive | 55 | 29 (52.7) | 26 (47.3) |
|
| NA | 1 |
|
|
|
| Ki67 |
|
|
|
|
|
Negative | 44 | 19 (43.2) | 25 (56.8) | 0.179 |
|
Positive | 49 | 28 (57.1) | 21 (42.9) |
|
| NA | 1 |
|
|
|
| TP53 |
|
|
|
|
|
Negative | 44 | 17 (38.6) | 27 (61.4) | 0.030 |
|
Positive | 49 | 30 (61.2) | 19 (28.8) |
|
| PDL-1 |
|
|
|
|
|
Negative | 43 | 22 (51.2) | 21 (48.8) | 0.911 |
|
Positive | 42 | 22 (52.4) | 20 (47.6) |
|
| NA | 9 |
|
|
|
| CD8 |
|
|
|
|
|
Negative | 42 | 20 (47.6) | 22 (52.4) | 0.528 |
|
Positive | 46 | 25 (54.3) | 21 (45.6) |
|
| NA | 6 |
|
|
|
 | Table II.Univariate and multivariate analyses
of the factors correlated with overall survival of liver carcinoma
patients. |
Table II.
Univariate and multivariate analyses
of the factors correlated with overall survival of liver carcinoma
patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Tumor size |
|
|
|
|
| >5
cm vs. ≤5 cm | 2.397
(1.365-4.210) | 0.002 | 1.002
(0.445-2.256) | 0.996 |
| TNM Stage |
|
|
|
|
| III/IV
vs. I/II | 2.860
(1.611-5.077) | <0.001 | 0.824
(0.321-2.115) | 0.687 |
| Histological
grade |
|
|
|
|
| III/IV
vs. I/II | 3.401
(2.002-5.779) | <0.001 | 2.067
(1.07-3.995) | 0.031 |
| Thrombus |
|
|
|
|
|
Positive vs.
Negative | 2.644
(1.117-6.259) | 0.027 | 1.732
(0.580-5.170) | 0.325 |
| ZHX3
expression |
|
|
|
|
| Low vs.
high | 0.179
(0.098-0.329) | <0.001 | 0.173
(0.066-0.453) | <0.001 |
Discussion
The present study is part of a continuing effort to
explore molecular targets of liver cancer behaviors with
reliability to predict outcome and promise as targets for directed
therapy. Identification of this issue may be important to improve
clinical management of liver cancer in the future. Consequently,
the results of the present study using data-mining analyses as well
as immunohistochemistry provided an in-depth investigation into the
prognostic values of ZHX family members in patients with liver
cancer.
ZHX1 has been identified as a tumor suppressor in
several types of cancer (24–28).
On the contrary, two reports show that ZHX1 might act as an
oncogene in cholangiocarcinoma and glioblastoma (29,30).
To the best of the authors' knowledge, except for our two reports
(3,4), no other study has unraveled the
association between ZHX1 expression and outcomes of patients with
cancer. Of note, its prognostic impact on different cancers appears
to be contradictory. The present authors previously reported that
high ZHX1 expression predicts worse OS for breast cancer but
present better OS for gastric cancer, suggesting its diverse roles
in development of different types of cancer (3,4). It
was inferred that different sample sources, histological types and
intrinsic differences in each type of cancers may be possible to
explain this disparity. Although there no relevance was found
between ZHX1 expression and OS in patients with liver cancer in the
present study, a prognostic value for ZHX1 was identified in
subgroup analyses, i.e., a significant association between low ZHX1
mRNA levels and longer RFS in male patients as well as in patient
without hepatitis virus infection.
Several studies have reported tumor-suppressor roles
of ZHX2 in multiple types of cancer, including liver cancer
(31–38). However, no significant association
was observed between ZHX2 expression and OS or RFS in patients with
liver cancer. Decreased ZHX2 expression was only observed to be
correlated with an improved OS in patients with Stage III/IV tumors
or an improved RFS in patients with micro vascular invasion.
Dysregulation of ZHX2 has been described to function in the
transcriptional inhibition of cancer markers in normal hepatocytes
(31). It has been noted that gene
promoter methylation-medicated silencing of ZHX2 frequent occurs in
HCC and overexpression of ZHX2 suppresses proliferation and
augments the chemo-sensitivity of HCC cells (32–35).
It has been also reported that HBV inhibits ZHX2 expression and
accelerates the proliferation of HCC cells through the activation
of miR-155 and, conversely, ZHX2 represses HBV replication through
epigenetic and non-epigenetic manners (35,36).
These observations seem consistent with the findings of the present
study, i.e., ZHX2 expression predicted better OS in patients with
hepatitis infection, suggesting that ZHX2 may exert different
functions according the different microenvironment during
development of liver cancer.
Consistent with our previous study in breast cancer
(3), attenuated ZHX3 expression
was observed to be correlated with unfavorable OS in patients with
liver cancer. The data also demonstrated that elevated ZHX3 was
associated with an improved OS in patients with both Stage I/II and
Stage III/IV tumors, suggesting that ZHX3 might be valuable in
predicting the outcomes of patients with early-stage malignancy.
This conclusion is contrary to the oncogene function of ZHX3 in
gastric cancer in another study (4). To support the observation by in
silico analyses, protein expression of ZHX3 was also examined
by immunohistochemistry in cancer tissues. The data of the present
study characterized that decreased ZHX3 levels were significantly
associated with malignant properties and suggested that ZHX3
expression is an independent prognostic factor in liver cancer.
Notably, the genetic alteration rate of ZHX3 was lower than that of
ZHX1 and ZHX2 in liver cancer, which is similar to our previous
studies in breast and gastric cancers (3,4).
This lower frequency of ZHX3 gene alteration in the types of cancer
that we observed suggest that ZHX3 may exert more important
biological functions as an tumor suppressor gene.
In summary, the present study systematically
examined the expression pattern of ZHX factors and the
corresponding prognostic significance in liver cancer, based on
in silico analysis and immunohistochemistry analyses. The
results suggested that ZHX family members are distinct prognostic
biomarkers for this disease. Future research should be performed to
discover the exact functions of ZHX family members in liver cancer,
which may support that ZHX factors could serve as prognostic
predicators and promising therapeutic targets for precision
medicine.
Acknowledgements
Not applicable.
Funding
This work was supported by the Natural Science Foundation of
Ningxia Hui Autonomous Region, China (grant no. 2021AAC03318) and
in part by the National Natural Science Foundation of China (grant
no. 81860426).
Availability of data and materials
The dataset used and/or analyzed in the current
study is available from the corresponding authors on reasonable
request.
Authors' contributions
YY, FH and SH conceived the study, designed and
performed the experiments, analyzed and interpret the data and
drafted the manuscript. YY and SH confirm the authenticity of all
raw data. All authors read and approved the final manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy and integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
The study protocol conformed to the ethical
guidelines outlined in the Declaration of Helsinki and was approved
by the Institutional Review Board (approval no. 07-170) of Ningxia
Hui Autonomous Region People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu J, Tao Q, Cheung KF, Jin H, Poon FF,
Wang X, Li H, Cheng YY, Röcken C, Ebert MP, et al: Epigenetic
identification of ubiquitin carboxyl-terminal hydrolase L1 as a
functional tumor suppressor and biomarker for hepatocellular
carcinoma and other digestive tumors. Hepatology. 48:508–518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
You Y, Ma Y, Wang Q, Ye Z, Deng Y and Bai
F: Attenuated ZHX3 expression serves as a potential biomarker that
predicts poor clinical outcomes in breast cancer patients. Cancer
Manag Res. 11:1199–1210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
You Y, Bai F, Li H, Ma Y, Yao L, Hu J and
Tian Y: Prognostic value and therapeutic implications of ZHX family
member expression in human gastric cancer. Am J Transl Res.
12:3376–3388. 2020.PubMed/NCBI
|
|
5
|
Liu Y, Ma D and Ji C: Zinc fingers and
homeoboxes family in human diseases. Cancer Gene Ther. 22:223–226.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barthelemy I, Carramolino L, Gutiérrez J,
Barbero JL, Márquez G and Zaballos A: A novel mouse homeodomain
protein containing two zinc-fingers and five homeodomains. Biochem
Biophys Res Commun. 224:870–876. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hirano S, Yamada K, Kawata H, Shou Z,
Mizutani T, Yazawa T, Kajitani T, Sekiguchi T, Yoshino M,
Shigematsu Y, et al: Rat zinc-fingers and homeoboxes 1 (ZHX1), a
nuclear factor-YA-interacting nuclear protein, forms a homodimer.
Gene. 290:107–114. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kawata H, Yamada K, Shou Z, Mizutani T,
Yazawa T, Yoshino M, Sekiguchi T, Kajitani T and Miyamoto K:
Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX
family, functions as a transcriptional repressor. Biochem J.
373:747–757. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamada K, Kawata H, Shou Z, Hirano S,
Mizutani T, Yazawa T, Sekiguchi T, Yoshino M, Kajitani T and
Miyamoto K: Analysis of zinc-fingers and homeoboxes
(ZHX)-1-interacting proteins: Molecular cloning and
characterization of a member of the ZHX family, ZHX3. Biochem J.
373:167–178. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kawata H, Yamada K, Shou Z, Mizutani T and
Miyamoto K: The mouse zinc-fingers and homeoboxes (ZHX) family;
ZHX2 forms a heterodimer with ZHX3. Gene. 323:133–140. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suehiro F, Nishimura M, Kawamoto T, Kanawa
M, Yoshizawa Y, Murata H and Kato Y: Impact of zinc fingers and
homeoboxes 3 on the regulation of mesenchymal stem cell osteogenic
differentiation. Stem Cells Dev. 20:1539–1547. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu G, Clement LC, Kanwar YS, Avila-Casado
C and Chugh SS: ZHX proteins regulate podocyte gene expression
during the development of nephrotic syndrome. J Biol Chem.
281:39681–39692. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Clement LC, Liu G, Perez-Torres I, Kanwar
YS, Avila-Casado C and Chugh SS: Early changes in gene expression
that influence the course of primary glomerular disease. Kidney
Int. 72:337–347. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nagel S, Ehrentraut S, Meyer C, Kaufmann
M, Drexler HG and MacLeod RA: Aberrantly expressed OTX homeobox
genes deregulate B-cell differentiation in hodgkin lymphoma. PLoS
One. 10:e01384162015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancerdrug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lanczky A, Nagy A, Bottai G, Munkacsy G,
Szabo A, Santarpia L and Gyorffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2,178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
You Y, Li H, Qin X, Zhang Y, Song W, Ran Y
and Gao F: Decreased CDK10 expression correlates with lymph node
metastasis and predicts poor outcome in breast cancer patients - a
short report. Cell Oncol (Dordr). 38:485–491. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
You Y, Li H, Qin X, Ran Y and Wang F.:
Down-regulated ECRG4 expression in breast cancer and its
correlation with tumor progression and poor prognosis-A short
report. Cell Oncol (Dordr). 39:89–95. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Liu D, Liang X, Gao L, Yue X, Yang
Y, Ma C and Liu J: Construction of a recombinant eukaryotic human
ZHX1 gene expression plasmid and the role of ZHX1 in hepatocellular
carcinoma. Mol Med Rep. 8:1531–1536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Ma X, Cai Q, Wang X, Yu B, Cai Q,
liu B, Zhu Z and Li C: MiR-199a-3p promotes gastric cancer
progression by targeting ZHX1. FEBS Lett. 588:4504–4512. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma X, Huang M, Wang Z, Liu B, Zhu Z and Li
C: ZHX1 inhibits gastric cancer cell growth through inducing
cell-cycle arrest and apoptosis. J Cancer. 7:60–68. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kwon RJ, Kim YH, Jeong DC, Han ME, Kim JY,
Liu L, Jung JS and Oh SO: Expression and prognostic significance of
zinc fingers and homeoboxes family members in renal cell carcinoma.
PLoS One. 12:e01710362017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guan J, Liu Z, Xiao M, Hao F, Wang C, Chen
Y, Lu Y and Liang J: MicroRNA-199a-3p inhibits tumorigenesis of
hepatocellular carcinoma cells by targeting ZHX1/PUMA signal. Am J
Transl Res. 9:2457–2465. 2017.PubMed/NCBI
|
|
29
|
Kwon RJ, Han ME, Kim JY, Liu L, Kim YH,
Jung JS and Oh SO: ZHX1 promotes the proliferation, migration and
invasion of cholangiocarcinoma cells. PLoS One. 11:e01655162016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kwon RJ, Han ME, Kim YJ, Kim YH, Kim JY,
Liu L, Heo W and Oh SO: Roles of zinc-fingers and homeoboxes 1
during the proliferation, migration, and invasion of glioblastoma
cells. Tumour Biol. 39:10104283176945752017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yamada K, Ogata-Kawata H, Matsuura K,
Kagawa N, Takagi K, Asano K, Haneishi A and Miyamoto K: ZHX2 and
ZHX3 repress cancer markers in normal hepatocytes. Front Biosci
(Landmark Ed). 14:3724–3732. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lv Z, Zhang M, Bi J, Xu F, Hu S and Wen J:
Promoter hypermethylation of a novel gene, ZHX2, in hepatocellular
carcinoma. Am J Clin Pathol. 125:740–746. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yue X, Zhang Z, Liang X, Gao L, Zhang X,
Zhao D, Liu X, Ma H, Guo M, Spear BT, et al: Zinc fingers and
homeoboxes 2 inhibits hepatocellular carcinoma cell proliferation
and represses expression of cyclins A and E. Gastroenterology.
142:1559–1570.e2. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luan F, Liu P, Ma H, Yue X, Liu J, Gao L,
Liang X and Ma C: Reduced nucleic ZHX2 involves in oncogenic
activation of glypican 3 in human hepatocellular carcinoma. Int J
Biochem Cell Biol. 55:129–135. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Song X, Tan S, Wu Z, Xu L, Wang Z, Xu Y,
Wang T, Gao C, Gong Y, Liang X, et al: HBV suppresses ZHX2
expression to promote proliferation of HCC through miR-155
activation. Int J Cancer. 143:3120–3130. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu L, Wu Z, Tan S, Wang Z, Lin Q, Li X,
Song X, Liu Y, Song Y, Zhang J, et al: Tumor suppressor ZHX2
restricts hepatitis B virus replication via epigenetic and
non-epigenetic manners. Antiviral Res. 153:114–123. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu S, Zhang M, Lv Z, Bi J, Dong Y and Wen
J: Expression of zinc-fingers and homeoboxes 2 in hepatocellular
carcinogenesis: A tissue microarray and clinicopathological
analysis. Neoplasma. 54:207–211. 2007.PubMed/NCBI
|
|
38
|
Armellini A, Sarasquete ME, García-Sanz R,
Chillón MC, Balanzategui A, Alcoceba M, Fuertes M, López R,
Hernández JM, Fernández-Calvo J, et al: Low expression of ZHX2, but
not RCBTB2 or RAN, is associated with poor outcome in multiple
myeloma. Br J Haematol. 141:212–215. 2008. View Article : Google Scholar : PubMed/NCBI
|