Introduction
Esophageal cancer (EC) is one of the most serious
types of cancer worldwide, and it presents a high mortality rate
and poor prognosis (1). Esophageal
squamous cell carcinoma (ESCC), which is the most common type of
esophageal cancer, is often diagnosed at an advanced stage
(2), and ESCC patients have a high
recurrence rate and low 5-year survival rate (3). Various factors are involved in the
development, growth, and invasion of ESCC. For example,
insulin-like growth factor-binding protein-7 (IGFBP7), which is
also known as insulin-like growth factor binding protein related
protein 1 (IGFBPrP1), is a secretory protein with a molecular mass
of approximately 30 kDa (4), and
it has distinct characteristics and participates in cell
proliferation, senescence, and apoptosis in numerous cancers
(5). IGFBP7 expression has been
reported in gastrointestinal cancers, including esophageal
adenocarcinoma (EAC) (6).
Cancer invasion and metastasis are not only
determined by cancer cells but also by the tumor microenvironment
(TME), with cancer-associated fibroblasts (CAFs) and the
extracellular matrix (ECM) playing important roles during the
progression of cancer (7). The TME
also plays a critical role in ESCC (8). α-smooth muscle actin (α-SMA) is a
marker of CAF activation, and the collagen I content in the stroma
cells of the TME is correlated with the aggressiveness and outcome
of tumors (9). Transforming growth
factor-β1 (TGFβ1) has been recognized as a key signal mediator
involved in oncogenesis (10,11),
and it plays an important role in the TME by accelerating invasion,
metastasis, angiogenesis, and immunosuppression. TGFβ1 upregulates
α-SMA and collagen I, thus creating an important proinvasion and
proangiogenesis niche for cancer development (12,13).
The TGFβ signaling pathway is a critical pathway for generating a
fibrotic TME, and the TGFβ1/SMAD signaling pathway is a classical
pathway associated with cancer. Recent studies have shown that
TGFβ1 cannot activate p-SMAD2/3-deficient hepatic stellate cells
(HSCs) to synthesize ECM (14,15).
Therefore, the TGFβ1/SMAD signaling pathway plays an important role
in ECM deposition.
IGFBP7 can upregulate the expression of TGFβ1 and
α-SMA and act as an upstream factor for TGFβ1 in the activation of
the SMAD signaling pathway, thus leading to increases in collagen I
in HSCs (16). However, whether
IGFBP7 expression activates the SMAD signaling pathway to
upregulate the expression of TGFβ1, α-SMA, and ECM to remodel the
TME during esophageal squamous cell carcinoma (ESCC) progression
remains unknown. The present study investigated the expression of
IGFBP7 and its effect on TGFβ1 and the TME in ESCC and analyzed the
associated changes in the expression of TGFβ1, α-SMA, collagen I,
p-SMAD2/3, and SMAD2/3.
Materials and methods
Tissue samples
A total of 45 patients aged 18 to 80 years who were
diagnosed with ESCC via biopsy at the Affiliated Lianyungang
Oriental Hospital of Xuzhou Medical University (Lianyungang, China)
between April 2017 and June 2019 were eligible for inclusion and
enrolled in this study. Early esophageal cancer refers to the
cancer that only invades the esophageal mucosa or submucosa without
lymph node metastasis. When the tumor has involved the muscularis
or adventitia or outside the adventitia, and has local or distant
lymph node metastasis, the cancer has developed to the advanced
stage. The samples were divided into three groups: early tumor
group (n=15), advanced tumor group (n=15), and paracancer control
group (n=15). Normal paracancer tissues adjacent to the tumor were
sampled. The present study was formally approved by the Medical
Ethics Committee of the Affiliated Lianyungang Oriental Hospital of
Xuzhou Medical University (2017-006-01), and written informed
consent was obtained from each patient.
Cell culture and treatment with
AdIGFBP7 and LvshTGFβ1
The EC109 cell line was purchased from the Aiyan
Biological Co. Cells were maintained in Dulbecco's modified Eagle's
medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin, and 100 µg/ml streptomycin. The cell
lines were grown in a humidified 37°C incubator with 5%
CO2. AdIGFBP7 was purchased from GenePharma Co.
Adenoviral vectors carrying no cDNA (CAd) were used as the negative
control. LvshTGFβ1 was designed and synthesized by Sangon Biotech
Co. the Lentiviral vectors carrying negative shRNA (LvshNC) served
as negative control. The TGFβ1 shRNA sequence was as follows:
5′-GCAGCTGTACATTGACTTTAG-3′. The negative shRNA sequence was
5′-TTCTCCGAACGTGTCACGT-3′. The shRNA and LV10 vector were
constructed by Sangon Biotech Co. at the final concentration of 200
nM and 50 ng/µl, respectively. The 293T cell line was purchased
from the Cell Bank of the Chinese Academy of Sciences and used to
generate the virions. RNAi-Mate (A615555; bbi-biotech GmbH) was
used as the transfection reagent according to the manufacturer's
instructions. Transfection was performed at 37°C for 6 h and then
the transfection medium was replaced with full culture medium for
72 h. Viral supernatant was collected and centrifuged at 1,800 × g
at 4°C for 4 min, and then at 26,000 × g at 4°C for 2 h. The
transfection efficiency was assessed by the percentage of the
number of RFP-positive cells to the total cells. Further
experiments with EC109 cells were followed. In the treatment group,
EC109 cells were transfected with AdIGFBP7 and LvshTGFβ1 at a
multiplicity of infection (MOI) of 40 and 100, respectively. Cells
were harvested at 24, 48 and 72 h.
Immunohistochemistry
Immunohistochemistry was performed to examine the
expression of IGFBP7. The patient samples were washed with PBS,
fixed in formalin, embedded in paraffin, and then sliced. The
tissue slices were then deparaffinized, hydrated, and incubated
with hydrogen peroxide (3%) for 20 min to block endogenous
peroxidase activities. Antigen retrieval was performed using 10 mM
sodium citrate buffer (pH=6.0) and a microwave histoprocessor for
10 min. Primary antibodies against IGFBP7 (1:100; cat. no.
ab171085; Abcam) were then added and incubated overnight at 4°C,
followed by biotinylated secondary antibodies (1:500; cat. no.
ab207995; Abcam) at 37°C for 20 min. In this experiment, PBS was
used as the negative control instead of the primary antibody.
Slices were then incubated with 3,3-diaminobenzidine (DAB) solution
for 5 min. Integrated optical density (IOD) values were measured
using the Image Pro Plus software (version 6.0; Media Cybernetics,
Inc.).
Western blot analysis
The total protein from the tissue specimens or cells
were extracted using a kit following the manufacturer's protocol
(cat. no. 3100; KeyGEN BioTECH Co., Ltd.). The protein
concentration was determined using a BCA protein concentration
assay kit following the manufacturer's instructions (cat. no.
23227; Thermo Fisher Scientific, Inc.). Proteins (40 µg) from the
tissue samples and cell homogenates were separated by sodium
dodecyl sulfate polyacrylamide gel electrophoresis (10% SDS-PAGE)
and subjected to western blotting. The separated proteins were
subsequently transferred onto PVDF membranes and then incubated in
a blocking solution with 3% skimmed milk for 2 h at room
temperature and with primary antibodies against IGFBP7 (1:1,000;
cat. no. ab171085; Abcam), TGFβ1 (1:1,000; cat. no. ab92486;
Abcam), α-SMA (1:1,000; cat. no. ab5694; Abcam), collagen I
(1:1,000; cat. no. ab34710; Abcam), p-SMAD2/3 (1:1,000; cat. no.
ab272332; Abcam), SMAD2/3 (1:1,000; cat. no. ab217553; Abcam), and
β-actin (1:1,000; cat. no. ab8227; Abcam) overnight at 4°C. After
washing, horseradish peroxidase-conjugated goat anti-rabbit
secondary antibody (1:5,000; cat. no. D110066; Sangon Biotech, Co.,
Ltd.) was added and incubated for 2 h at room temperature. An
enhanced chemiluminescence system (Bio-Rad Laboratories, Inc.) was
used to visualize the protein bands, and the relative protein
expression was normalized to that of β-actin. Protein expression
was quantified using Bio-Rad Quantity One software (version 4.6.2;
Bio-Rad Laboratories, Inc.).
Statistical analysis
All statistical analyses were performed using SPSS
16.0 software (SPSS, Inc.) Data are presented as mean ± SD. An
analysis of variance (ANOVA) and SNK-q test were performed
to determine whether the differences among the groups were
significant, with P≤0.05 considered to indicate a statistically
significant difference.
Results
Expression of IGFBP7, TGFβ1, α-SMA,
and collagen I are increased in esophageal squamous cell
carcinoma
Tissue samples from the patients in each
experimental group were examined to identify changes in IGFBP7,
TGFβ1, α-SMA, and collagen I expression. Immunohistochemistry
staining showed that the IGFBP7 protein was upregulated in both the
early tumor group and advanced tumor group compared with the
control group, with IGFBP7 expression markedly upregulated in the
advanced tumor group compared with the early tumor group (Fig. 1A). A semi-quantitative evaluation
of IGFBP7, as indicated by the integrated optical density (IOD),
showed that the expression of IGFBP7 was gradually increased in the
early tumor group and advanced tumor group relative to the control
group (P<0.05; Fig. 1B), with
the highest IOD observed in the advanced tumor group. As shown in
Fig. 1C, the western blot analysis
showed that IGFBP7, TGFβ1, α-SMA, and collagen I were significantly
upregulated in both the early tumor group and advanced tumor group
compared with the control group. In the advanced tumor group, the
expression of IGFBP7, TGFβ1, α-SMA, and collagen I was markedly
upregulated compared with that of the early tumor group
(P<0.05). These results suggest that IGFBP7 is positively
correlated with TGFβ1 and participates in the process of esophageal
squamous cell carcinoma.
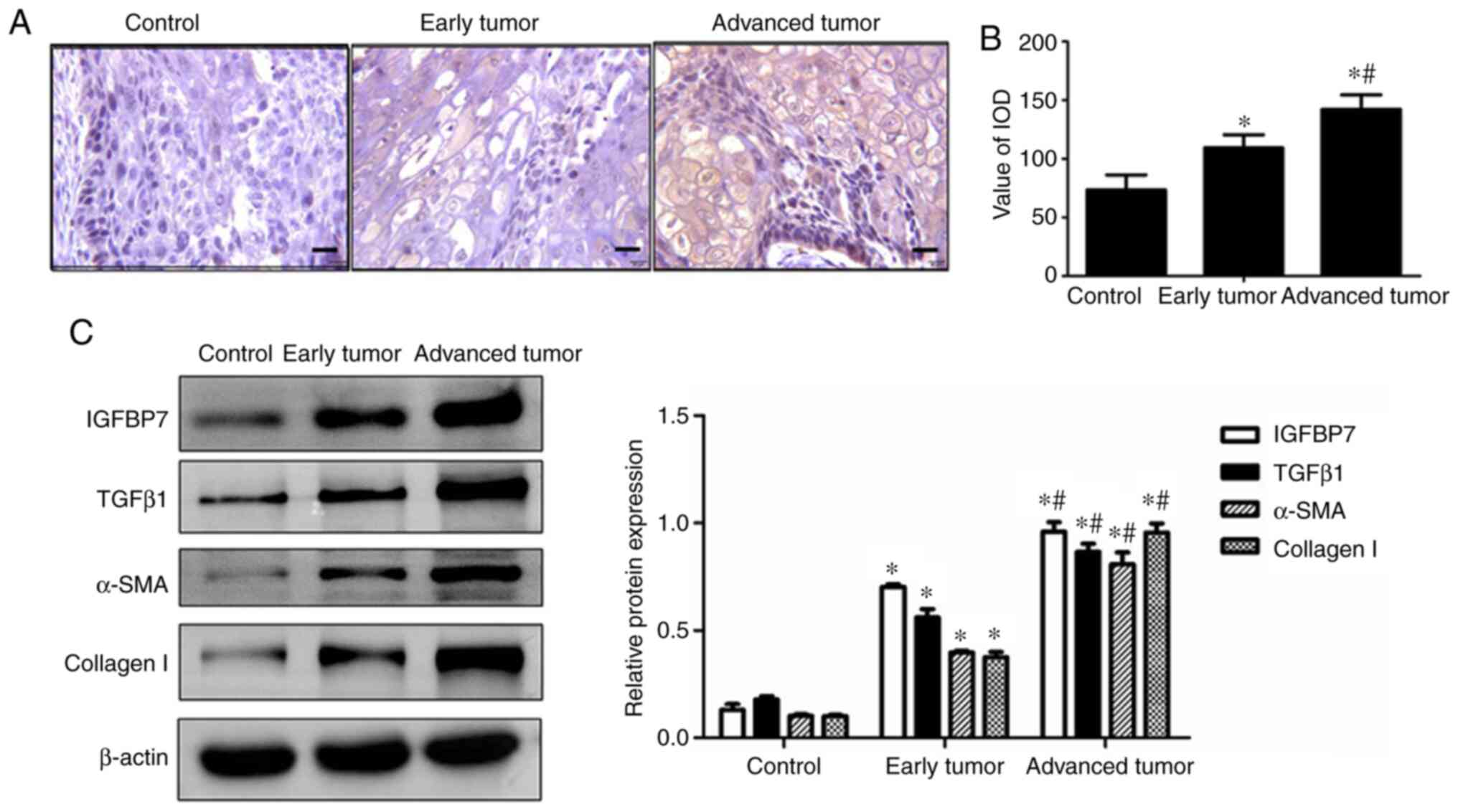 | Figure 1.Expression of IGFBP7, TGFβ1, α-SMA,
and collagen I in esophageal squamous cell carcinoma. (A) IGFBP7
expression was examined by immunohistochemistry staining (scale
bar, 50 µm). (B) IOD value of the positive-brown particles was
calculated. (C) Expression of IGFBP7, TGFβ1, α-SMA, and collagen I
was examined by western blotting. β-actin served as an internal
control (n=15). *P<0.05 vs. the control group; #P<0.05 vs.
the early tumor group. IGFBP7, insulin-like growth factor-binding
protein-7; TGFβ1, transforming growth factor-β1; α-SMA, α-smooth
muscle actin. |
Effect of IGFBP7 on the expression of
TGFβ1 and α-SMA in the EC109 cell line
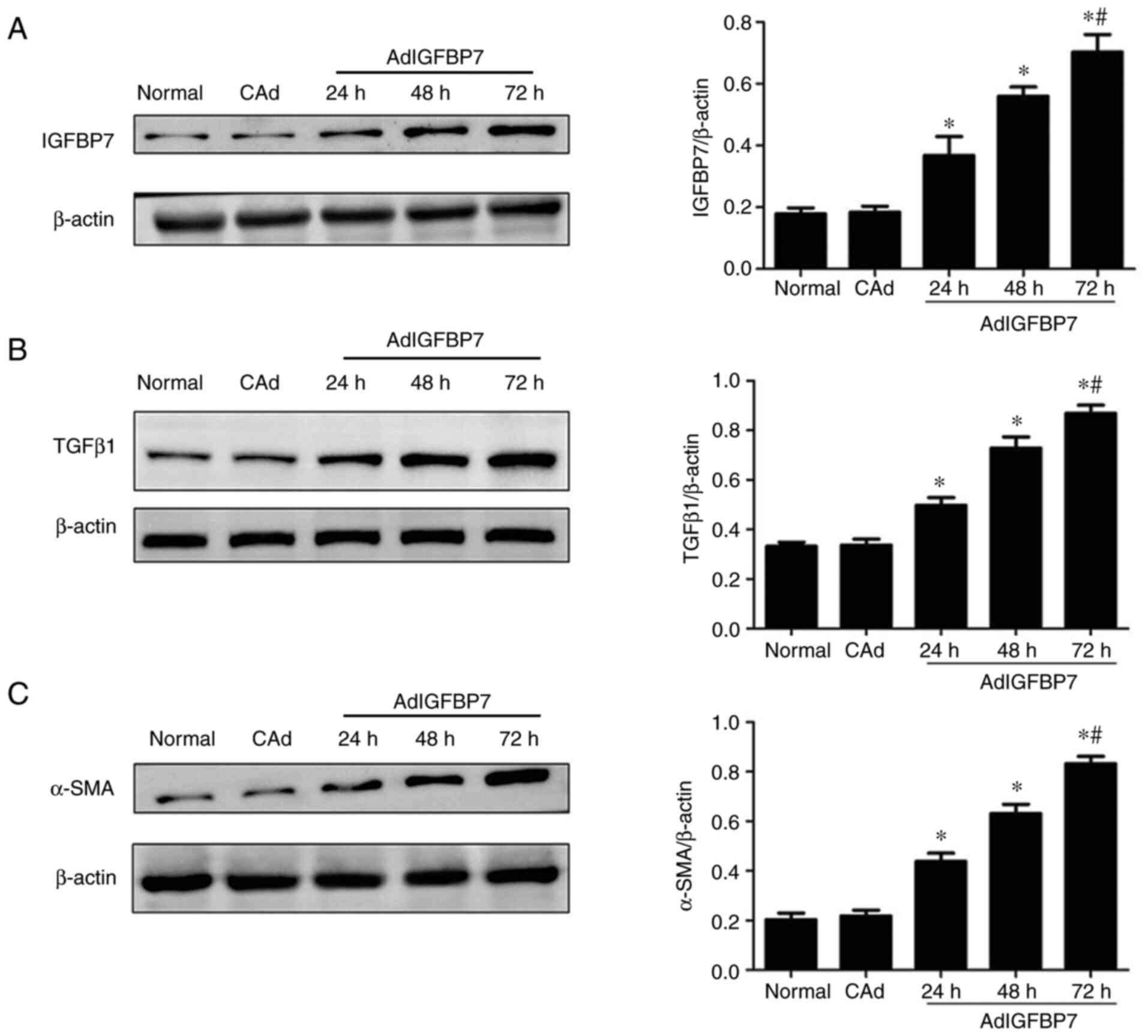
After the EC109 cells were treated with AdIGFBP7,
western blotting was performed to assess the expression of IGFBP7,
TGFβ1, and α-SMA in each experimental group. As shown in Fig. 2A, the IGFBP7 protein level
gradually increased from 24 to 72 h, thus demonstrating that
AdIGFBP7 transfection was effective. The western blot analysis
showed that the TGFβ1 and α-SMA proteins were upregulated at 24,
48, and 72 h in the treatment groups compared with the control
group (P<0.05); moreover, the levels gradually increased in a
time-dependent manner and peaked at 72 h. There were no changes in
the normal and CAd groups (P<0.05; Fig. 2B and C). These results suggest that
IGFBP7 upregulates the expression of TGFβ1 and α-SMA in esophageal
squamous cell carcinoma.
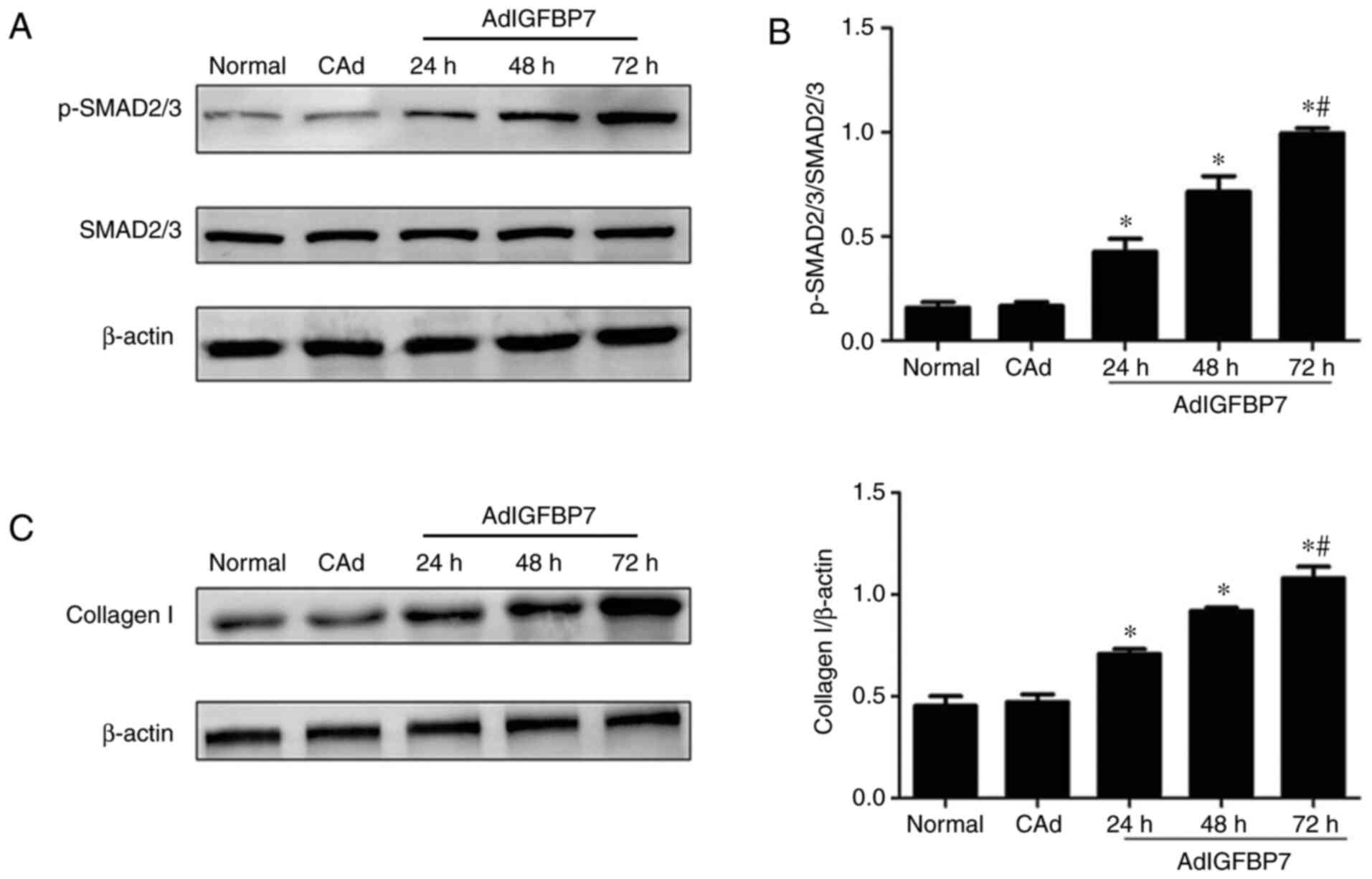
Effect of IGFBP7 on the expression of
p-SMAD2/3, SMAD2/3, and collagen I in the EC109 cell line
Following treatment with AdIGFBP7, the expression of
p-SMAD2/3, SMAD2/3, and collagen I was examined via western
blotting, and the results showed that the p-SMAD2/3 protein was
upregulated at 24, 48, and 72 h and peaked at 72 h (P<0.05;
Fig. 3A). No changes in SMAD2/3
protein expression were observed in the experimental groups
(Fig. 3A). The ratio of p-SMAD2/3
to SMAD2/3 was also significantly upregulated from 24 to 72 h in a
time-dependent manner (P<0.05; Fig.
3B). As shown in Fig. 3C,
collagen I expression gradually increased from 24 to 72 h, with a
peak observed at 72 h (P<0.05). These results suggest that
IGFBP7 may promote collagen I expression by activating the
TGFβ1/SMAD signaling pathway in esophageal squamous cell
carcinoma.
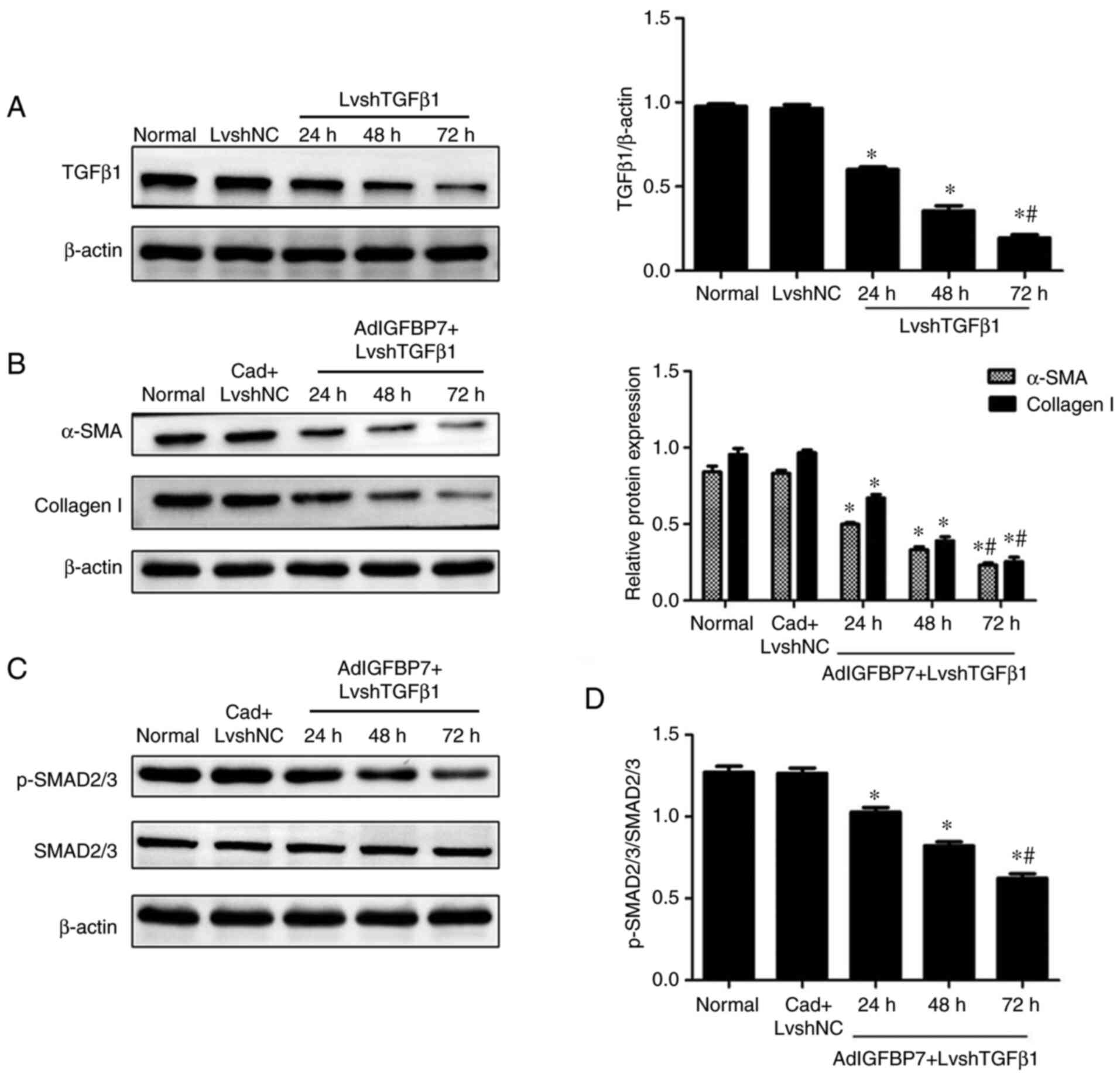
Inhibition of TGFβ1 expression reduces
the expression of α-SMA, collagen I, and p-SMAD2/3 in
IGFBP7-treated EC109 cell line
Following treatment of the EC109 cell line with
LvshTGFβ1, the TGFβ1 protein level was gradually and significantly
decreased from 24 to 72 h, which showed that LvshTGFβ1 transfection
was effective (Fig. 4A). After the
EC109 cell line was treated with both AdIGFBP7 and LvshTGFβ1, the
western blotting analysis showed that the expression of α-SMA,
collagen I, and p-SMAD2/3 and the ratio of p-SMAD2/3 to SMAD2/3
were gradually decreased from 24 to 72 h compared with that of the
control group. There was no changes in the CAd plus LvshNC compared
with the normal group. Changes in the expression of SMAD2/3 protein
were not observed among the groups (Fig. 4B-D). These results suggest that
TGFβ1 possibly plays an important role in IGFBP7-induced α-SMA and
ECM production in esophageal squamous cell carcinoma.
Discussion
Esophageal carcinoma can be divided into two
different histological subtypes, i.e., squamous cell carcinoma and
adenocarcinoma (EAC), of which squamous cell carcinoma accounts
(ESCC) for the majority of EC cases (3). Many factors are involved in the
occurrence and development of ESCC. For example, insulin-like
growth factor-binding protein-7 (IGFBP7) participates in the
progression of many tumors (17),
and previous research has demonstrated that the protein level of
IGFBP7 is significantly increased in EAC (18). In this study, we observed that
IGFBP7 was significantly upregulated in ESCC; thus, IGFBP7 is
increased not only in EAC but also in ESCC. Furthermore, we found
that the expression of IGFBP7 was markedly increased in the
advanced tumor group compared with the early tumor group. These
data suggest that IGFBP7 possibly performs an important function in
the process of esophageal squamous cell carcinoma.
Transforming growth factor-β1 (TGFβ1) is an
important cytokine that regulates the tumor microenvironment (TME)
(19) and affects the formation
and development of tumors in many ways (20). A previous study showed that
TGF-β-induced cell proliferation and apoptosis play important roles
in the progression of ESCC (21).
In addition, TGFβ1 expression in cancer-associated fibroblasts
(CAFs) was significantly associated with the overall survival of
patients with ESCC (22). In the
present study, we found that the TGFβ1 protein was significantly
upregulated in ESCC. α-smooth muscle actin (α-SMA) and collagen I
represent the most important cell components of the TME, and they
are upregulated in ESCC. In addition, TGFβ1, α-SMA, and collagen I
were markedly upregulated in the advanced tumor group compared with
the early tumor group. Furthermore, the changes in TGFβ1, α-SMA,
and collagen I were consistent with those of IGFBP7. These data
suggest that IGFBP7 is positively correlated with the expression of
TGFβ1, α-SMA, and collagen I; thus, IGFBP7 may promote the
development of esophageal cancer.
TGFβ1 expression was induced by IGFBP7 in the liver
of rats in vivo (23) and
hepatic stellate cells (HSCs) in vitro (24). However, whether IGFBP7 directly
induces TGFβ1 expression in ESCC has not been clarified. Therefore,
we transfected the EC109 cell line with AdIGFBP7, and subsequent
changes in TGFβ1 expression were observed. Our experiments showed
that the expression of TGFβ1 was gradually increased in a
time-dependent manner, which indicates that IGFBP7 can upregulate
TGFβ1 expression in other cell lines (as indicated above) as well
as the EC109 cell line. Previous studies have demonstrated that the
expression of α-SMA is induced by IGFBP7 in fibroblasts (25) and HSCs (26). In this study, we also found that
the expression of α-SMA was gradually increased in the EC109 cell
line treated with AdIGFBP7. Considering the above results, we
suggest that IGFBP7 is likely to participate in ESCC through
upregulating the expression of TGFβ1 and α-SMA, which may result in
TME remodeling.
The TGFβ1/SMAD signaling pathway performs an
important function in the regulation of ECM production. Previous
studies revealed that IGFBP7 can activate and induce ECM production
in HSCs via the TGFβ1/SMAD signaling pathway (27,28).
Thus, we tested the effects of IGFBP7 on p-SMAD2/3 and collagen I
in the EC109 cell line and found that the protein expression levels
of p-SMAD2/3 and collagen I were significantly increased following
treatment with AdIGFBP7, which was consistent with previous
reports. Endogenous TGFβ1 in fibroblasts might be involved in the
induction of α-SMA by IGFBP7. We observed that the expression of
α-SMA was significantly decreased by LvshTGFβ1 in the
AdIGFBP7-treated EC109 cell line, which suggests that α-SMA
expression was induced by IGFBP7 in a TGFβ1-dependent manner. Our
data also showed that the expression of p-SMAD2/3 and collagen I
were significantly decreased by LvshTGFβ1 in the AdIGFBP7-treated
EC109 cell line. These results demonstrated that the inhibition of
TGFβ1 expression possibly affected the IGFBP7-induced SMAD pathway
and collagen I expression in the EC109 cell line. Taken together,
our results indicate that IGFBP7 has an important effect on the
TGFβ1/SMAD signaling pathway and induces α-SMA upregulation and ECM
production, which may remodel the TME in ESCC.
In conclusion, the present study suggests that
increased IGFBP7 may accelerate ESCC progression by promoting the
expression of TGFβ1, α-SMA, and collagen I by activating the
TGFβ1/SMAD signaling pathway, which could remodel the TME. However,
the effects of IGFBP7 knockdown on TGFβ1/SMAD signaling and IGFBP7
on TME cell components in ESCC were not analyzed in this study,
which will be used as a future research perspective. Further
investigations are required to elucidate additional mechanisms
underlying the effect of IGFBP7 on ESCC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL designed the experiments and prepared the
manuscript. JZ, YW, CM, DW, and LP performed the experiments. LC
analyzed the data. All authors read and approved the final
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved. XL and LC confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
The present study strictly conformed to the ethical
rules of the Medical Ethics Committee of the Affiliated Lianyungang
Oriental Hospital of Xuzhou Medical University (2017-006-01), and
written informed consent was obtained from each patient (Jiangsu,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shi C, Pan B, Shi F, Xie ZH, Jiang YY,
Shang L, Zhang Y, Xu X, Cai Y, Hao JJ and Wang MR: Sequestosome 1
protects esophageal squamous carcinoma cells from apoptosis via
stabilizing SKP2 under serum starvation condition. Oncogene.
37:3260–3274. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Golyan F, Druley T and Abbaszadegan M:
Whole-exome sequencing of familial esophageal squamous cell
carcinoma identified rare pathogenic variants in new predisposition
genes. Clin Transl Oncol. 22:681–693. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fatehi Hassanabad A, Chehade R, Breadner D
and Raphael J: Esophageal carcinoma: Towards targeted therapies.
Cell Oncol (Dordr). 43:195–209. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Komiya E, Sato H, Watanabe N, Ise M,
Higashi S, Miyagi Y and Miyazaki K: Angiomodulin, a marker of
cancer vasculature, is upregulated by vascular endothelial growth
factor and increases vascular permeability as a ligand of integrin
αvβ3. Cancer Med. 3:537–549. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fang P, Hwa V and Rosenfeld R: IGFBPs and
cancer. Novartis Found Symp. 262:215–268. 2004.PubMed/NCBI
|
|
6
|
Nancarrow DJ, Clouston AD, Smithers BM,
Gotley DC, Drew PA, Watson DI, Tyagi S, Hayward NK and Whiteman DC;
Australian Cancer Study; Study of Digestive Health, : Whole genome
expression array profiling highlights differences in mucosal
defense genes in Barrett's esophagus and esophageal adenocarcinoma.
PLoS One. 6:e225132011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zou MX, Zheng BW, Liu FS, Wang XB, Hu JR,
Huang W, Dai ZH, Zhang QS, Liu FB, Zhong H, et al: The Relationship
between tumor-stroma ratio, the immune microenvironment, and
survival in patients with spinal chordoma. Neurosurgery.
85:E1095–E1110. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang L, Han H, Wang Z, Shi L, Yang M and
Qin Y: Targeting the microenvironment in esophageal cancer. Front
Cell Dev Biol. 9:6849662021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bayer SV, Grither WR, Brenot A, Hwang PY,
Barcus CE, Ernst M, Pence P, Walter C, Pathak A and Longmore GD:
DDR2 controls breast tumor stiffness and metastasis by regulating
integrin mediated mechanotransduction in CAFs. Elife. 8:e455082019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao HC, Huang YZ, Liu YQ, Chen Y, Wang ZH
and Yin GH: Role of TG2 and TGF-β1 in the pathogenesis of human
breast cancer. Oncol Lett. 20:2212020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chung JY, Chan MK, Li JS, Chan AS, Tang
PC, Leung KT, To KF, Lan HY and Tang PM: TGF-β Signaling: From
tissue fibrosis to tumor microenvironment. Int J Sci.
22:75752021.
|
|
12
|
Nie Y, Yang Y, Zhang J, Cai G, Chang Y,
Chai G and Guo C: Shikonin suppresses pulmonary fibroblasts
proliferation and activation by regulating Akt and p38 MAPK
signaling pathways. Biomed. Pharmacother. 95:1119–1128. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Angioni R, Sánchez-Rodríguez R, Viola A
and Molon B: TGF-β in Cancer: Metabolic driver of the tolerogenic
crosstalk in the tumor microenvironment. Cancers (Basel).
13:4012021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gwon MG, An HJ, Kim JY, Kim WH, Gu H, Kim
HJ, Leem J, Jung HJ and Park KK: Anti-fibrotic effects of synthetic
TGF-β1 and Smad oligodeoxynucleotide on kidney fibrosis in vivo and
in vitro through inhibition of both epithelial dedifferentiation
and endothelial-mesenchymal transitions. FASEB J. 34:333–349. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu HH, Chen DQ, Wang YN, Feng YL, Cao G,
Vaziri ND and Zhao YY: New insights into TGF-β/Smad signaling in
tissue fibrosis. Chem Biol Interact. 292:76–83. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li X, Zhang Q, Zhang H, Guo X, Fan H and
Liu L: Interaction between insulin-like growth factor binding
protein-related protein 1 and transforming growth factor beta 1 in
primary hepatic stellate cells. Hepatobiliary Pancreat Dis Int.
16:395–404. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu CT, Xu YW, Guo H, Hong CQ, Huang XY,
Luo YH, Yang SH, Chu LY, Li EM and Peng YH: Serum insulin-like
growth factor binding protein 7 as a potential biomarker in the
diagnosis and prognosis of esophagogastric junction adenocarcinoma.
Gut Liver. 14:727–734. 2020. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Creemers A, Ebbing EA, Pelgrim TC, Lagarde
SM, van Etten-Jamaludin FS, van Berge Henegouwen MI, Hulshof MCCM,
Krishnadath KK, Meijer SL, Bijlsma MF, et al: A systematic review
and meta-analysis of prognostic biomarkers in resectable esophageal
adenocarcinomas. Sci Rep. 8:132812018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vitiello GAF, Amarante MK, Oda JMM, Hirata
BKB, de Oliveira CEC, Campos CZ, de Oliveira KB, Guembarovski RL
and Watanabe MAE: Transforming growth factor beta 1 (TGFβ1)
plasmatic levels in breast cancer and neoplasia-free women:
Association with patients' characteristics and TGFB1 haplotypes.
Cytokine. 130:1550792020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang C, Wang T, Lv D, Li L, Yue J, Chen HZ
and Xu L: Acquired resistance to EGFR TKIs mediated by
TGFβ1/Integrin β3 signaling in EGFR-Mutant lung cancer. Mol Cancer
Ther. 18:2357–2367. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen D, Zhang Z, Mao C, Zhou Y, Yu L, Yin
Y, Wu S, Mou X and Zhu Y: ANRIL inhibits p15(INK4b) through the
TGFβ1 signaling pathway in human esophageal squamous cell
carcinoma. Cell Immunol. 289:91–96. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang H, Xie C, Yue J, Jiang Z, Zhou R,
Xie R, Wang Y and Wu S: Cancer-associated fibroblasts mediated
chemoresistance by a FOXO1/TGFβ1 signaling loop in esophageal
squamous cell carcinoma. Mol Carcinog. 56:1150–1163. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo Y, Zhang Y, Zhang Q, Guo X, Zhang H,
Zheng G and Liu L: Insulin-like growth factor binding
protein-related protein 1 (IGFBPrP1) contributes to liver
inflammation and fibrosis via activation of the ERK1/2 pathway.
Hepatol Int. 9:130–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Zhang QQ, Guo XH, Zhang HY and
Liu LX: IGFBPrP1 induces liver fibrosis by inducing hepatic
stellate cell activation and hepatocyte apoptosis via Smad2/3
signaling. World J Gastroenterol. 20:6523–6533. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rao C, Lin SL, Ruan WJ, Wen H, Wu DJ and
Deng H: High expression of IGFBP7 in fibroblasts induced by
colorectal cancer cells is co-regulated by TGF-β and Wnt signaling
in a Smad2/3-Dvl2/3-dependent manner. PLoS One. 9:e853402014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kong Y, Huang T, Zhang H, Zhang Q, Ren J,
Guo X, Fan H and Liu L: The lncRNA NEAT1/miR-29b/Atg9a axis
regulates IGFBPrP1-induced autophagy and activation of mouse
hepatic stellate cells. Life Sci. 237:1169022019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ren JJ, Huang TJ, Zhang QQ, Zhang HY, Guo
XH, Fan HQ, Li RK and Liu LX: Insulin-like growth factor binding
protein related protein 1 knockdown attenuates hepatic fibrosis via
the regulation of MMPs/TIMPs in mice. Hepatobiliary Pancreat Dis
Int. 18:38–47. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo X, Zhang H, Zhang Q, Li X and Liu L:
Screening for and validation of a hepatic fibrosis-related pathway
induced by insulin-like growth factor-binding protein-related
protein 1. Eur J Gastroenterol Hepatol. 28:762–772. 2016.
View Article : Google Scholar : PubMed/NCBI
|