Introduction
The Wnt/β-catenin signaling pathway (also known as
the canonical Wnt pathway) is responsible for embryonic development
and tissue homeostasis (1).
Aberrant activation of this pathway by genetic and epigenetic
alterations is involved in human diseases such as cancer (2–4). In
colorectal cancer (CRC), frequent activation of Wnt/β-catenin
signaling pathway by somatic mutations in APC regulator of WNT
signaling pathway (APC) or the β-catenin gene
(CTNNB1) has been reported. In the cBioPortal for Cancer
Genomics (https://www.cbioportal.org/), a
public database of cancer genomes, mutations of APC and
CTNNB1 were found in 64 and 6%, respectively, of 3,051 CRC
tissues. Loss of function mutations in APC or activating
mutations in CTNNB1 result in the stabilization and
accumulation of β-catenin protein in the cells. The accumulated
β-catenin interacts with T cell factor (TCF)/lymphoid
enhancer-binding factor (LEF) transcription factors in the nucleus,
and induces the expression of their target genes (Wnt target genes)
(5). To date, more than one
hundred Wnt target genes have been identified, and a list of the
genes is shown on the Wnt homepage at https://web.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes.
Studies of their function have helped to further understand the
molecular mechanisms of carcinogenesis and the complex regulatory
mechanisms underlying this signaling pathway. Representative
examples of the aberrant activation of this pathway contributing to
carcinogenesis include MYC proto-oncogene (MYC) and cyclin
D1 (CCND1). MYC was identified by serial analysis of
gene expression using HT29 cells containing a zinc-inducible APC,
and affects a wide variety of functions such as cell proliferation,
angiogenesis, and promotion of anaerobic metabolism (6). Cyclin D1 is known to regulate
G1-S cell cycle progression, and it was identified through the
analysis of human genes involved in controlling cell growth, the
promoter regions of which contain the core TCF/LEF-binding sites
(7).
It is of note that chromatin immunoprecipitation
coupled with high-throughput sequencing (ChIP-seq) analysis using
six different cell lines and anti-TCF7L2 antibody identified
116,000 non-redundant TCF7L2-binding sites, with 1,864 sites common
to the cell lines tested, suggesting the existence of as yet
unidentified Wnt target genes in human cells (8). To understand the precise molecular
mechanism underlying the development of Wnt-driven cancer, we
previously searched for new target genes by microarray using
β-catenin-depleted CRC cells and ChIP-seq of TCF7L2. Integrated
analysis of these data identified 11 candidate genes that are
directly regulated by the β-catenin/TCF7L2 complex (9). Among these candidates, we focused in
this study on motile sperm domain containing 1 (MOSPD1), and
revealed that MOSPD1 is a novel direct target of the Wnt
signaling pathway. Furthermore, we identified three Wnt responsive
elements in the 3′-flanking region of MOSPD1, and showed
that the elements are involved in the transcriptional activation.
These data will help deepen our understanding of colorectal
carcinogenesis, as well as the regulatory mechanism of MOSPD1.
Materials and methods
Cell culture
Human CRC cell lines, HCT116 (CCL-247) and SW480
(CCL-228), and a human endocervical adenocarcinoma cell line, HeLa
(CCL-2) were purchased from the American Type Culture Collection.
All cell lines were grown in appropriate media (McCoy's 5a Modified
Medium for HCT116, Leibovitz's L-15 Medium for SW480, and Eagle's
Minimum Essential Medium for HeLa) supplemented with 10% fetal
bovine serum (BioSera), and antibiotic/antimycotic solution
(Fujifilm Wako Pure Chemical). HCT116 and HeLa cells were
maintained in 5% CO2 at 37°C, and SW480 cells were
maintained without CO2 supplementation at 37°C.
Reporter plasmids and luciferase
assay
Two genomic regions of 5′-putative (GRCh38-chrX:
134,932,384-134,933,013) and 3′-putative enhancers (GRCh38-chrX:
134,885,255-134,886,704) were amplified by PCR using
region-specific primer sets and genomic DNA extracted from the
peripheral blood of healthy volunteers as a template. After
digestion with XhoI and BglII restriction enzymes,
the PCR products were cloned into pGL4.23 vector (Promega) to
generate pGL4.23-MOSPD1-5′E and pGL4.23-MOSPD1-3′E. The primer
sequences are shown in Table SI.
Construction of pCAGGS-dominant negative TCF7L2 (dnTCF7L2) was
described previously (10).
Significant suppression of β-catenin/TCF-dependent transcriptional
activity using pCAGGS-dnTCF7L2 has been confirmed in the previous
studies (11–13). HCT116 cells were transfected with
these reporter plasmids together with β-catenin siRNAs or
pCAGGS-dnTCF7L2 using Lipofectamine 2000 (Thermo Fisher
Scientific). pRL-null plasmids were co-transfected with the
reporter plasmids for normalization. 48 h after the transfection,
the cells were lysed and reporter activities were measured using
dual luciferase kit (TOYO B-Net) and Lumat LB9507 Luminometer
(Berthold Technologies). Firefly luciferase activity was normalized
to Renilla luciferase activity (pRL-null).
Site-directed mutagenesis
TCF-binding motifs were searched by JASPAR, a
database for transcription factor binding profiles (14). Mutant reporter plasmids containing
substitutions in the consensus sequence of the TCF-binding motifs
were prepared by site-directed mutagenesis. Wild type-plasmid DNA
of pGL4.23-MOSPD1-3′E was amplified using KOD-Plus-Neo (Toyobo) and
a set of mutagenic primers (Table
SII). The PCR products were digested with DpnI
restriction enzyme (Takara Bio) to cleave the methylated template
DNA, followed by transformation into Escherichia coli.
Insertion of mutations in the plasmids was confirmed by Sanger
sequencing (data not shown; 3500×l DNA Analyzer; Thermo Fisher
Scientific).
Gene silencing
For the knockdown of β-catenin, two β-catenin siRNAs
(siβ-catenin#9: 5′-GAUCCUAGCUAUCGUUCUU-3′ and siβ-catenin#10:
5′-UAAUGAGGACCUAUACUUA-3′; Merck) were used. Control siRNA
(siControl, ON-TARGET plus non-targeting Pool, #D-001810-10-20) was
purchased from Horizon Discovery. Cells were transfected with 10 nM
of the indicated siRNA using Lipofectamine RNAiMAX or Lipofectamine
2000 (Thermo Fisher Scientific) for 48 h.
Western blotting
Total protein was extracted from cultured cells
using SDS sample buffer (25 mM Tris-HCl, pH 6.8, 0.8% sodium
dodecyl sulfate, 4% glycerol). After boiling the samples for 10
min, the protein was separated by SDS-PAGE, and transferred onto a
nitrocellulose membrane (GE Healthcare). The membranes were blocked
with 5% milk in TBS-T (Tris-buffered saline-Tween-20), and then
incubated with primary antibody; anti-MOSPD1 (GTX32111; GeneTex),
anti-β-catenin (9582; Cell Signaling Technology), or anti-β-actin
(A5441; Merck). Horseradish peroxidase-conjugated goat anti-mouse
or anti-rabbit IgG (GE Healthcare) served as the secondary antibody
for the ECL Detection System (GE Healthcare).
Chromatin immunoprecipitation
assay
Chromatin immunoprecipitation followed by qPCR
(ChIP-qPCR) was performed as described previously (15). Briefly, HCT116 cells were
cross-linked with 1% formaldehyde for 10 min at room temperature,
and 0.1 M glycine was added to quench the formaldehyde. Chromatin
was extracted and sheared by micrococcal nuclease digestion (New
England Biolabs). Subsequently, protein-DNA complexes were
immunoprecipitated with 10 µg of anti-TCF7L2 antibody (05-511;
Merck) bound to Dynabeads Protein G (Thermo Fisher Scientific).
Normal mouse IgG (Santa Cruz Biotechnology) was used as a negative
control. The precipitated protein-DNA complexes were purified by
conventional DNA extraction methods, and the purified DNA was
subjected to qPCR analysis using KAPA SYBR FAST ABI prism Kit (Kapa
Biosystems) and a set of primers encompassing the TCF-binding
motifs located in the 3′-flanking region of MOSPD1.
Amplification of a region upstream of the GAPDH gene was
used as a negative control. Sequences of the primers are shown in
Table SIII.
Chromatin conformation capture (3C)
assay
3C was performed as described previously, with minor
modifications (16). Briefly,
SW480 cells were cross-linked with 1% formaldehyde for 10 min at
room temperature, and then treated with 0.125 M glycine. The
cross-linked chromatin was digested at 37°C overnight with 400
units of HindIII (Takara Bio), and subsequently
heat-inactivated for 25 min at 65°C in the presence of SDS (1.6%)
prior to ligation. DNA fragments were ligated with 2,000 U of T4
DNA ligase (New England Biolabs) for 8 h at 16°C. Samples were
treated with Proteinase K (300 µg; Merck) at 37°C overnight to
reverse the cross-links. After treatment with RNase A (300 µg;
Merck), the DNA was purified by phenol/chloroform extraction and
ethanol precipitation. Nested PCR (KOD One, Toyobo) was performed
to investigate a possible interaction between the promoter and
enhancer regions of MOSPD1. The sequences of 1st and nested
primers are shown in Table
SIV.
Immunohistochemical staining
All colorectal tumor tissues and corresponding
non-cancerous tissues were obtained with written informed consent
from resected specimens of 11 patients who underwent surgery. The
clinical and histological information of the 11 CRCs is shown in
Supplementary Table SV. Tissue
sections were deparaffinized with xylene and rehydrated in a graded
series of ethanol. Antigen retrieval was performed using 0.01 M
citrate buffer (pH 6.0) and autoclave heating at 110°C for 10 min.
After blocking endogenous peroxidase activity in 0.3%
H2O2 (Fujifilm Wako Pure Chemical) for 5 min,
slides were incubated with 5% goat serum (ab7481; Abcam) for 8 min,
followed by the incubation with anti-MOSPD1 (GeneTex, 1:200) or
anti-β-catenin antibody (RB-1491; NeoMarkers, 1:300) at 4°C
overnight. Secondary antibody, Dako EnVision™+ Dual Link System-HRP
(Dako), and ImmPACT DAB Substrate Kit (Vecter Laboratories) were
then used to visualize the immunoreactivity. Tissue sections were
counterstained with hematoxylin (Merck).
Statistical analysis
Gene expression values of human colorectal tumors
(GSE21510) (17) were obtained
from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). An unpaired t-test
with Benjamini-Hochberg correction was applied to evaluate the
differential expression of MOSPD1 between 104 tumors and 25
matched non-cancerous controls. Correlation between the expression
values of MOSPD1 and known Wnt target genes was determined
using Pearson's correlation coefficient (r). Experiments
were performed in biological triplicate and data are presented as
the mean ± standard deviation (SD). To compare the means between
two groups in ChIP-qPCR, the unpaired t-test was used. Statistical
analysis of data from reporter assays was performed using one-way
analysis of variance (ANOVA) followed by Tukey's or Dunnett's
multiple comparisons test. We used the BellCurve for Excel software
for the analyses (Social Survey Research Information). P<0.05
was considered to indicate a statistically significant
difference.
Results
The expression of MOSPD1 is regulated
by Wnt/β-catenin signaling in colorectal cancer cells
In the previous study, we identified a total of 11
target genes whose expression was commonly down-regulated by the
introduction of β-catenin siRNAs and a dominant-negative form of
TCF7L2 (dnTCF7L2) in HCT116, SW480, and LS174T cells (9). Subsequent qPCR analysis revealed that
the expression of PDE4D, PHLDB2, OXR1, FRMD5, and
MOSPD1 was significantly decreased by the knockdown of
β-catenin. To verify the association of MOSPD1 with the
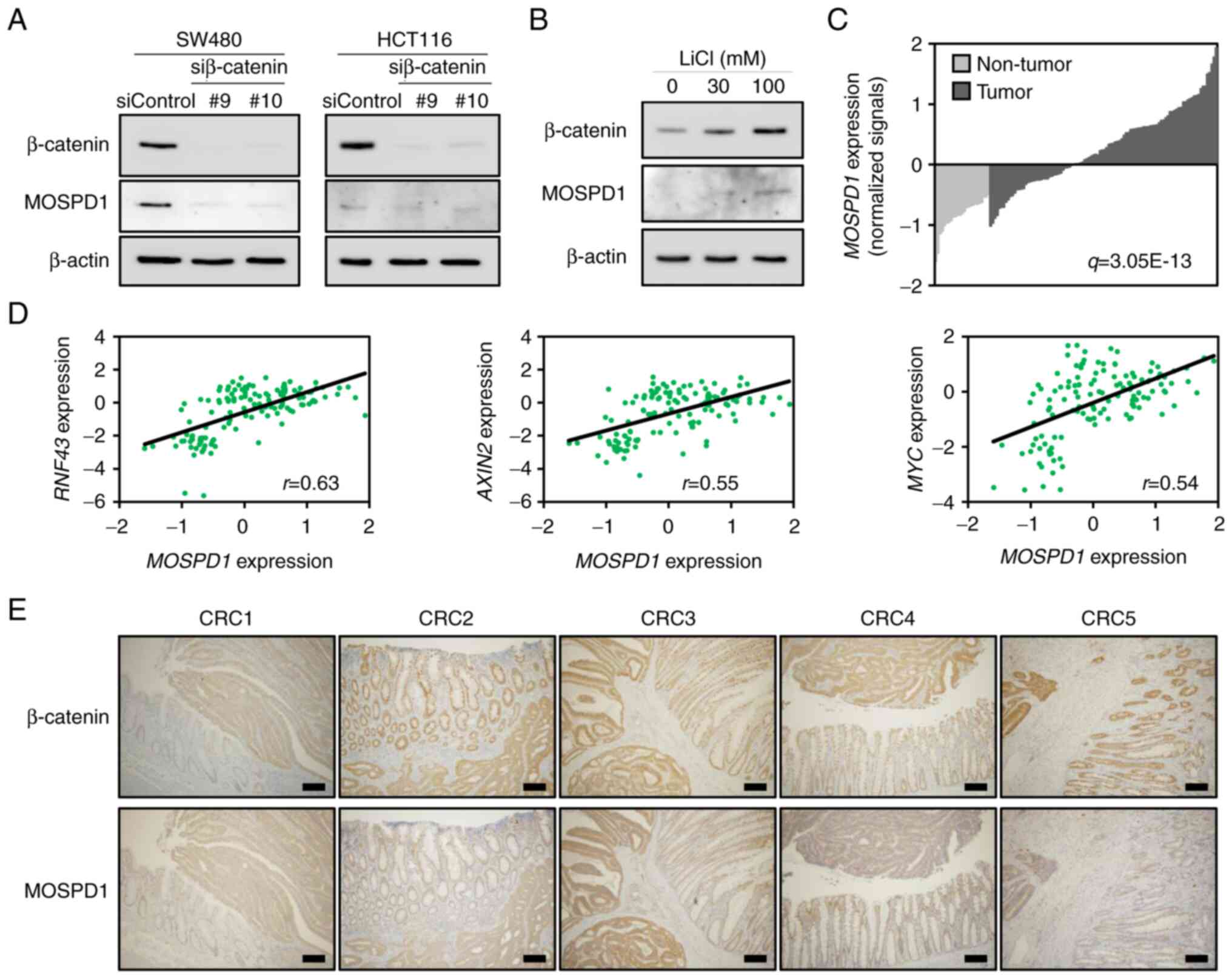
Wnt/β-catenin signaling, we performed western blot analysis using
lysates from SW480 and HCT116 cells treated with β-catenin or
control siRNA. In agreement with the qPCR data, treatment with two
independent β-catenin siRNAs decreased MOSPD1 expression in both
cells (Fig. 1A). In addition,
treatment of HeLa cells with lithium chloride (LiCl), a glycogen
synthase kinase 3 (GSK3) inhibitor that activates the Wnt/β-catenin
signaling, increased β-catenin and MOSPD1 expression (Fig. 1B). These data corroborated that
MOSPD1 is a downstream target of the Wnt/β-catenin signaling.
Since aberrant activation of the Wnt/β-catenin
signaling is involved in the majority of CRC (2,18),
we searched for gene expression data of colorectal tumors in NCBI
Gene Expression Omnibus. In a dataset (GSE21510) containing 104 CRC
tissues and 25 non-tumorous colonic tissues (17), the average MOSPD1 expression
was found to be 2.18-fold higher (q-value: 3.05E-13) in the tumor
tissues than in the non-tumorous tissues (Fig. 1C). In addition, the expression
levels showed a tendency of positive correlation with RNF43
(r=0.63), AXIN2 (r=0.55), and MYC
(r=0.54), three well-known Wnt targets (Fig. 1D). These data supported that
MOSPD1 expression is induced by the activation of Wnt
signaling.
We further carried out immunohistochemical staining
of β-catenin and MOSPD1 using 11 CRC tissues. As shown in Fig. 1E, β-catenin was stained in the
cytoplasm and/or nucleus of tumorous cells in all tumor tissues
tested. MOSPD1 was positively stained in tumor lesions of all CRC
cases tested. In addition, MOSPD1 was also positively stained in
the cytoplasm and/or nucleus of the tumorous cells (Fig. 1E, lower paned).
Identification of an enhancer in the
3′-flanking region of MOSPD1
In our previous study, ChIP-seq analysis showed a
region for the binding with TCF7L2 in the 3′-flaking region of
MOSPD1 (3′-putative enhancer,
GRCh38-chrX:134,885,306-134,886,672) (9). This region was overlapped with a peak
in ENCODE ChIP-seq data of TCF7L2 (ENCSR000EUV, Fig. 2A, upper panel). In addition to the
3′-region, the ENCODE data showed another peak in the 5′-flanking
region of MOSPD1 (GRCh38-chrX: 134,932,561-134,932,930).
These peaks were overlapped with peaks of histone modifications
(H3K4me1: ENCSR161MXP and H3K27Ac: ENCSR000EUT, Fig. 2A, middle and lower panels),
suggesting that these regions may have enhancer activity through
the interaction with TCF7L2. To investigate their enhancer
activity, these regions were cloned into reporter plasmids, and
reporter assays were performed using HCT116 cells. As a result,
both reporter plasmids, pGL4.23-MOSPD1-5′E and pGL4.23-MOSPD1-3′E,
showed increased reporter activity compared to the mock reporter
(empty vector control) by 1.29- and 5.62-fold, respectively
(Fig. 2B). Importantly,
co-transfection of the reporter plasmids with plasmids expressing
dnTCF7L2 significantly decreased the reporter activity of
pGL4.23-MOSPD1-3′E, but not the activity of pGL4.23-MOSPD1-5′E,
suggesting the enhancer activity of the 3′-flanking region through
the interaction with TCF7L2. In addition, knockdown of β-catenin by
two independent siRNAs markedly reduced the reporter activity of
pGL4.23-MOSPD1-3′E (Fig. 2C).
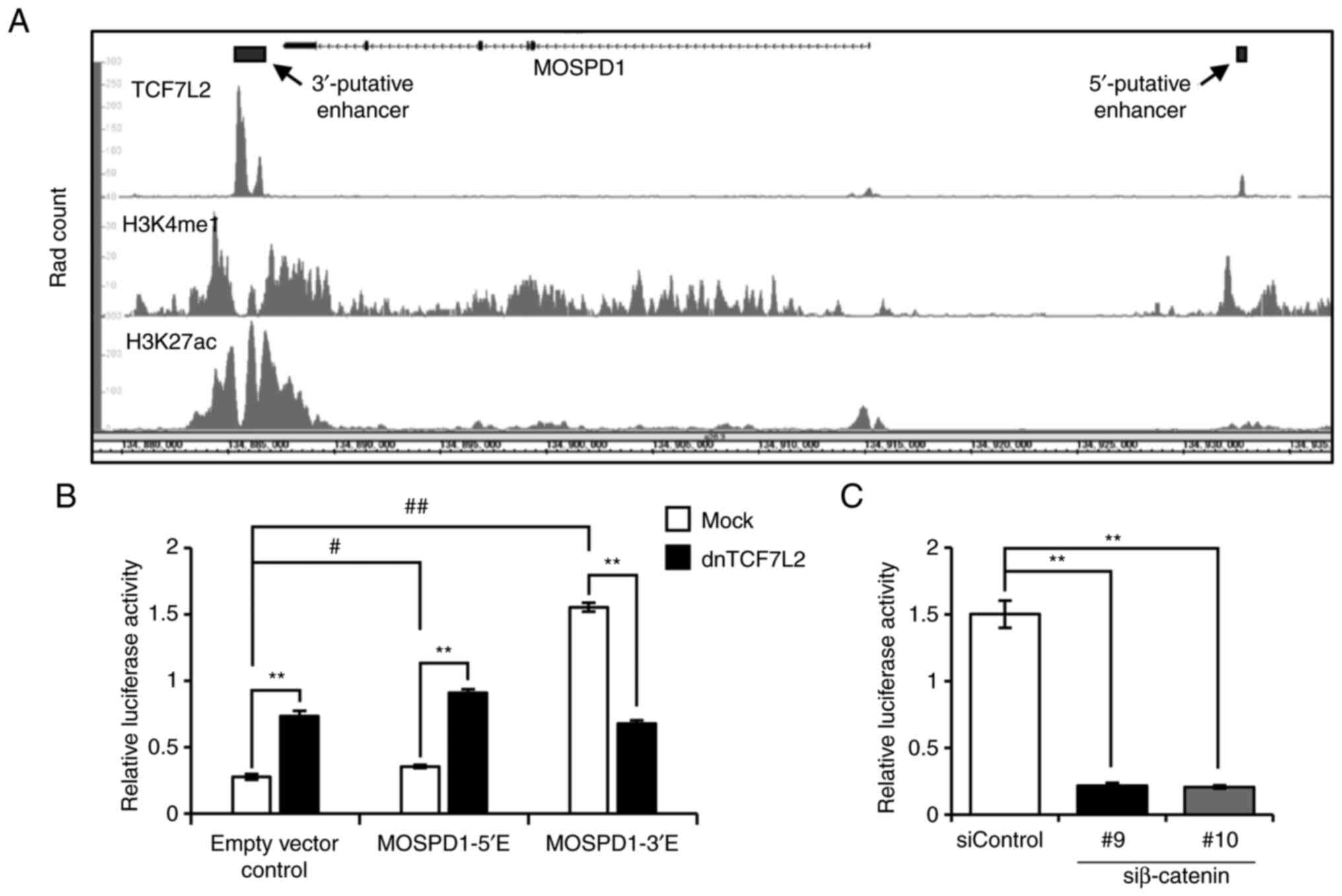 | Figure 2.TCF7L2-interacting region in the
3′-flanking region may play a role as an enhancer. (A) Schematic
representation of the ENCODE ChIP-seq data of TCF7L2 (ENCSR000EUV),
H3K4me1 (ENCSR161MXP), and H3K27ac (ENCSR000EUT) in HCT116 cells.
(B) Reporter activity of putative enhancer regions in the 5′- and
3′-flanking regions. HCT116 cells were transfected with pGL4.23
(Empty vector control), pGL4.23-MOSPD1-5′E (MOSPD1-5′E), or
pGL4.23-MOSPD1-3′E (MOSPD1-3′E) plasmids, in combination with
pRL-null reporter plasmids for the normalization of transfection.
The cells were co-transfected with pCAGGS-dnTCF7L2 plasmids
expressing a dominant-negative form of TCF7L2 or the empty plasmids
(Mock). Relative luciferase activities represent mean ± SD from
three independent cultures. Statistical significance was determined
by one-way ANOVA, followed by Turkey's test; **P<0.01, as
compared with Mock; #P<0.05, ##P<0.01,
as compared with Empty vector control. (C) Effect of β-catenin
siRNA on the reporter activity of MOSPD1-3′E in HCT116 cells.
Relative luciferase activities represent mean ± SD from three
independent cultures. Statistical significance was determined by
one-way ANOVA, followed by Dunnett's test; **P<0.01, as compared
with siControl. TCF7L2, transcription factor 7 like 2; ENCODE, The
Encyclopedia of DNA Elements; ChIP, chromatin immunoprecipitation;
MOSPD1, motile sperm domain containing 1; H3K4me1, histone H3
lysine 4 mono-methylation; H3K27ac, histone H3 lysine 27
acetylation. |
Involvement of three TCF-binding
motifs in the enhancer activity
We further searched for TCF-binding elements (TBE)
in the 3′-enhancer region using JASPAR, a database for
transcription factor binding profiles (14), and identified eight candidate TBEs
(Table SVI). Among the eight, we
focused on three TBEs with a similarity score greater than 10; TBE1
(GRCh38-chrX: 134,885,716-134,885,729), TBE2 (GRCh38-chrX:
134,885,543-134,885,556), and TBE3 (GRCh38-chrX:
134,885,482-134,885,495). To investigate the involvement of these
motifs in the enhancer activity, we prepared mutant reporter
plasmids containing two-nucleotide substitutions in each
TCF-binding motif (TBE1-mut, TBE2-mut, and TBE3-mut) of
pGL4.23-MOSPD1-3′E and reporter plasmids containing these
substitutions in the three motifs (TBEall-mut) (Fig. 3A). A reporter assay determined that
the reporter activity of mutant plasmids (TBE1-mut, TBE2-mut, and
TBE3-mut) was significantly reduced compared to the wild type
plasmids (pGL4.23-MOSPD1-3′E) by 9.87, 35.3, and 35.0%,
respectively. In addition, the activity of TBEall-mut plasmids was
markedly decreased compared to the wild type by 85.6% (Fig. 3B). Treatment of the cells
expressing TBE1-mut, TBE2-mut, or TBE3-mut with β-catenin siRNA
suppressed the activity by 50.2, 52.2, and 42.3%, respectively,
compared to the cells with control siRNA. These data indicated that
the three motifs are, at least in part, associated with the
enhancer activity of TCF7L2.
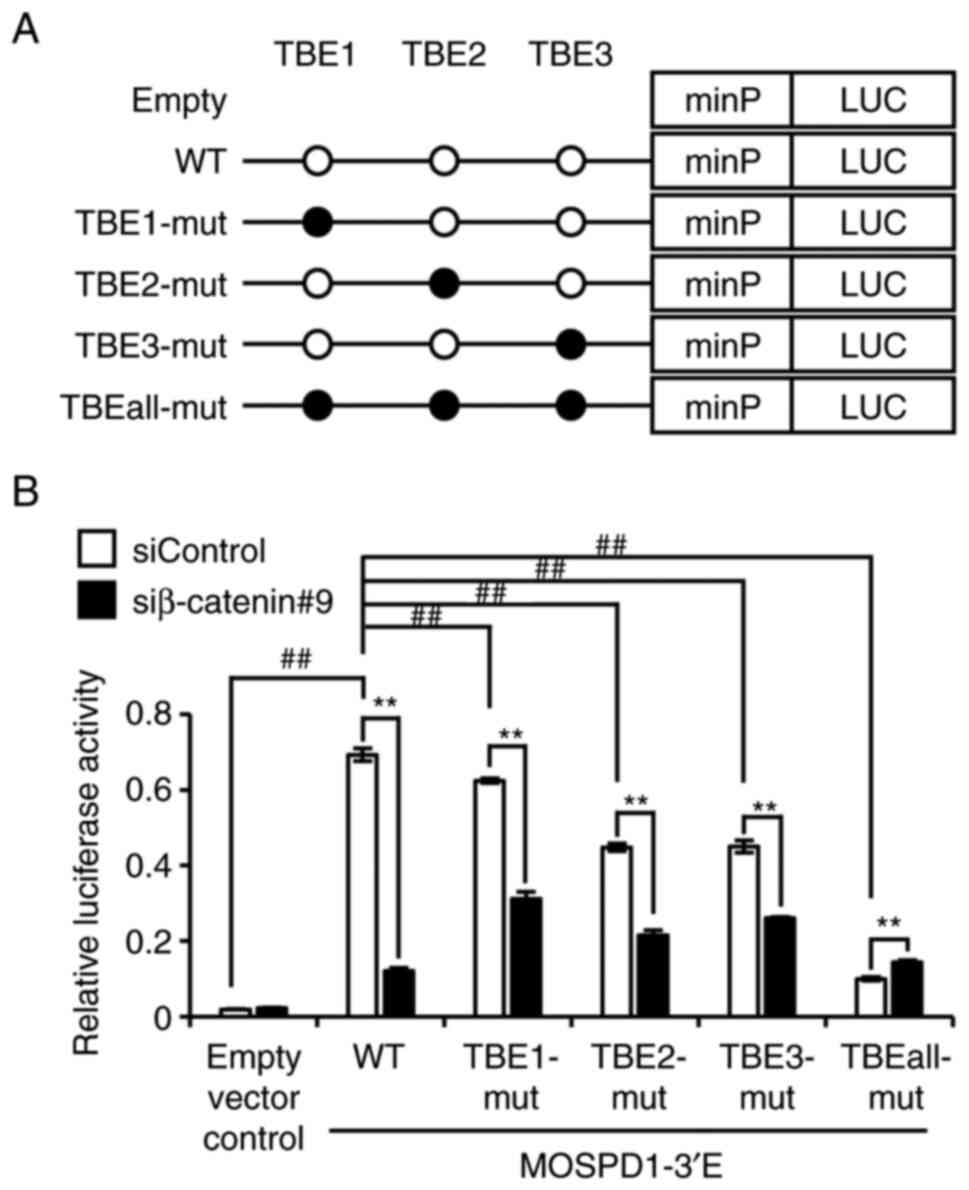 | Figure 3.Involvement of the three TBEs in the
reporter activity. (A) Schematic representation of wild type (open
circle: WWCAAAG, W: A/T) and mutant (closed circle: WWCAGCG)
TCF-binding motifs in pGL4.23-MOSPD1-3′E reporter plasmids. (B)
Relative reporter activity of empty, wild type and mutant reporter
plasmids in HCT116 cells (Empty vector control, WT, TBE1-mut,
TBE2-mut, TBE3-mut, or TBEall-mut). The data represent mean ± SD
from three independent cultures. Statistical significance was
determined by one-way ANOVA, followed by Turkey's test;
**P<0.01, as compared with siControl; ##P<0.01, as
compared with pGL4.23-MOSPD1-3′E WT. TBEs, TCF-binding elements;
WT, wild type; MOSPD1, motile sperm domain containing 1. |
Identification of the interaction
between 3′-putative enhancer and promoter region of MOSPD1
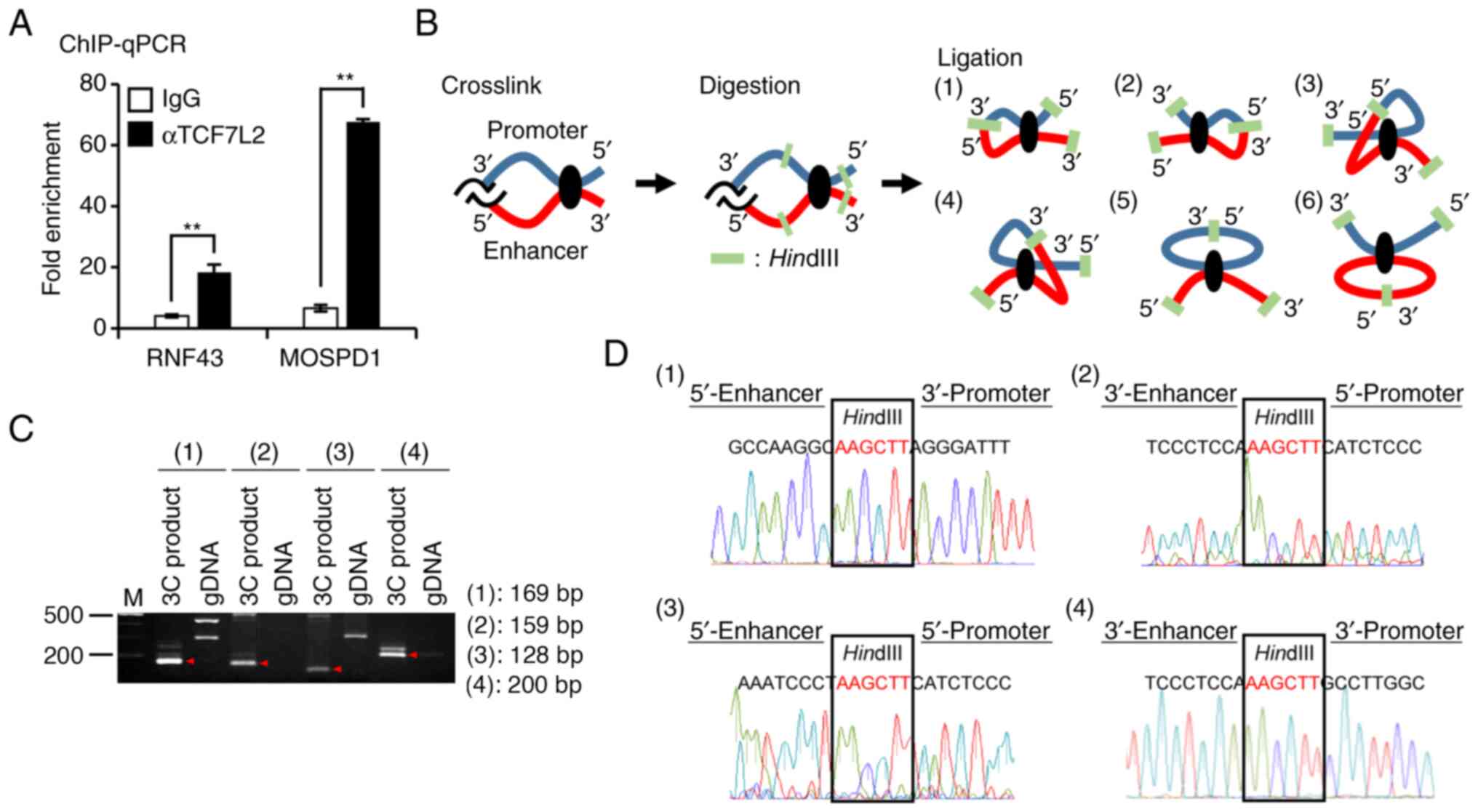
To confirm the interaction between the 3′-flanking
region of MOSPD1 and TCF7L2, we performed a ChIP-qPCR assay
using anti-TCF7L2 antibody and region-specific primer sets for the
3′-enhancer region of MOSPD1. An enhancer region in intron 2
of RNF43 was recruited as a positive control (19). This assay detected an enrichment of
the enhancer region in RNF43 by 4.53-fold in the
precipitants with the anti-TCF7L2 antibody compared to those with
normal IgG. DNA fragments containing the 3′-enhancer region of
MOSPD1 were enriched by 10.3-fold in the precipitants
(Fig. 4A).
To investigate whether the distal 3′-putative
enhancer region interacts with the promoter region of
MOSPD1, we performed a 3C assay. We used DNA from SW480
cells after the fixation with formaldehyde and subsequent digestion
with a restriction enzyme HindIII. Self-ligation of the DNA
was expected to produce six types of chromatin loops when the two
regions were closely associated (Fig.
4B). We designed four sets of 1st and nested primers that can
detect the associations between the promoter and enhancer regions
(Table SIV). As a result,
amplification of the DNA with the four primer sets detected PCR
products with the expected size, but it failed to amplify control
DNA extracted from SW480 cells (Fig.
4C). Additional sequence analysis of the PCR products confirmed
the interactions of enhancer-promoter regions connected with a
HindIII restriction enzyme site (Fig. 4D). These data suggested that the
distal enhancer located in the 3′-flanking region interacts with
the promoter region through the formation of a chromatin loop.
Discussion
In this study, we revealed that MOSPD1 is
transcriptionally regulated by Wnt signaling through the three TBEs
located in its 3′-flanking region.
MOSPD1 is a member of major sperm protein
(MSP) domain-containing family that is highly conserved in many
species. There are three MSP domain-containing proteins (MOSPD1, 2,
and 3) in humans, and four (Mospd1, 2, 3, and 4) in mice and rats
(20). The similarities between
human MOSPD1 and human MOSPD2, and that between human MOSPD1 and
human MOSPD3 are 8 and 32%, respectively, at protein levels
(CLUSTALW, http://www.genome.jp/tools-bin/clustalw). In our
previous expression profile analysis, knockdown of β-catenin did
not show significant decrease of MOSPD2 or MOSPD3 in
SW480 cells. These data may imply that MOSPD1 has a specific
function that is linked with the canonical Wnt signaling pathway in
development.
The function of MOSPD1 is still largely unclarified.
In the early 1980s, MSP was isolated as a protein 15K from sperm
cells of Caenorhabditis elegans (21), implying its role in
spermatogenesis. Later, MSP was shown to function as a motility
apparatus in sperm locomotion (22,23).
In GTEx Portal, a public database of gene expression in normal
tissues (https://gtexportal.org/home/),
MOSPD1 is expressed in a variety of tissues including
esophageal mucosa, adrenal gland, testis, skin, and uterus,
suggesting that MOSPD1 should play physiological role(s) in various
tissues. In mice, Mospd1 is abundantly expressed in
mesenchymal tissues, and its expression is elevated during
differentiation in osteoblastic, myoblastic, and adipocytic cell
lines (20). Another study
revealed that Mospd1-null embryonic stem cells were able to
proliferate and that they were unable to differentiate to
osteoblasts, adipocytes, and hematopoietic progenitors (24). These data indicated that Mospd1
should be involved in the differentiation and proliferation of
mesenchymal cells. In addition, knockdown of Mospd1 induced
the expression of epithelial cadherin Cdh1, and decreased
the expression of Snai1, Snai2, and mesenchymal cadherin
Cdh11 in MC3T3-E1 cells established from mouse osteoblasts
(20). These results suggested
that Mospd1 may be associated with epithelial-mesenchymal
transition (EMT). EMT plays an important role in the invasion or
metastasis of cancer. Ovarian cancer cells with high
invasion-phenotype expressed significantly increased levels of
MOSPD1 compared to the cells with low invasion-phenotype (25). Besides, the activation of Wnt
signaling pathway lead to induce EMT in cancer (26). Thus, further functional analysis of
MOSPD1 in EMT may give us better understanding of the EMT-induction
mechanism by Wnt signaling pathway. Interestingly, expression of
Runx2 and Osteocalcin was also down-regulated by the
knockdown of Mospd1 in MC3T3-E1 cells (20). RUNX2, one of the transcription
factors required for osteoblastic differentiation is abundantly
expressed in the nucleus of osteoid osteoma cells (27). It is noteworthy that osteomas
frequently develop in the mandible bone of patients with germline
variants in the APC gene (28). The induction of RUNX2 and/or
osteocalcin by the increased expression of MOSPD1 in osteoblasts
may be involved in the development of osteomas in patients with
familial polyposis of the colon.
We identified a distant enhancer region for the
Wnt/β-catenin signaling in the 3′-flanking region of MOSPD1.
Enhancer regions that associate with β-catenin-TCF/LEF1 complexes
have been identified in various regions of the target genes. For
instance, the enhancer regions of MYC (6), CCND1 (7), claudin-1 (CLDN1) (11), membrane-type matrix
metalloproteinase (MT1-MMP) (12), and SP5 (29) are localized in their 5′-flanking
regions, and those of RNF43 (19) and FRMD5 (9) in intron 2 and intron 1, respectively.
Regarding AXIN2, several enhancer regions have been
discovered in its 5′-flanking region and in intron 1 (30). It is of note that, in addition to
the 5′-flanking enhancer region, MYC has another enhancer
element in its 3′-flanking region (31). Therefore, MOSPD1 may have
additional enhancer region(s) in addition to the one identified
here.
In conclusion, we have discovered that MOSPD1
is a novel target gene of the Wnt signaling pathway in CRC. Further
analysis of MOSPD1 function will elucidate the precise molecular
mechanism underlying the development and progression of CRC, and
may contribute to the development of therapeutic strategies against
their invasion and metastasis.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Seira Hatakeyama
(The University of Tokyo) for their technical assistance.
Funding
This work was supported by the Japan Society for the Promotion
of Science, Grant-in-Aid for Scientific Research [grant no.
JP20K07563 (K.Y.)].
Availability of data and materials
Gene expression values of human colorectal tumors
analyzed during the current study are available in the Gene
Expression Omnibus repository under accession number GSE21510
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21510).
The other datasets used and /or analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
The experiments were designed by KY and YF. The
experiments were performed by CH, CZ, SN, YI and YT. Data analysis
was performed by CH, CZ, KT, TI, YO, SA, YI, GT, YA and DS. CH, CZ,
KY and YF confirm the authenticity of all the raw data. The
manuscript was written by CH, KY and YF. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
This study was approved by the ethical committee of
the Institute of Medical Science, The University of Tokyo (approval
nos. IMSUT-IRB, 21-14-0806 and 2020-78-0318). All colorectal tumor
tissues and corresponding non-cancerous tissues were obtained with
written informed consent from the resected specimens of patients
who underwent surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar
|
|
2
|
Voloshanenko O, Erdmann G, Dubash TD,
Augustin I, Metzig M, Moffa G, Hundsrucker C, Kerr G, Sandmann T,
Anchang B, et al: Wnt secretion is required to maintain high levels
of Wnt activity in colon cancer cells. Nat Commun. 4:26102013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang W, Xu L, Liu P, Jairam K, Yin Y, Chen
K, Sprengers D, Peppelenbosch MP, Pan Q and Smits R: Blocking wnt
secretion reduces growth of hepatocellular carcinoma cell lines
mostly independent of β-catenin signaling. Neoplasia. 18:711–723.
2016. View Article : Google Scholar
|
|
4
|
Cleary AS, Leonard TL, Gestl SA and
Gunther EJ: Tumour cell heterogeneity maintained by cooperating
subclones in Wnt-driven mammary cancers. Nature. 508:113–117. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Behrens J, Von Kries JP, Kühl M, Bruhn L,
Wedlich D, Grosschedl R and Birchmeier W: Functional interaction of
beta-catenin with the transcription factor LEF- 1. Nature.
382:638–642. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
He TC, Sparks AB, Rago C, Hermeking H,
Zawel L, da Costa LT, Morin PJ, Vogelstein B and Kinzler KW:
Identification of c-MYC as a target of the APC pathway. Science.
281:1509–1512. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Frietze S, Wang R, Yao L, Tak YG, Ye Z,
Gaddis M, Witt H, Farnham PJ and Jin VX: Cell type-specific binding
patterns reveal that TCF7L2 can be tethered to the genome by
association with GATA3. Genome Biol. 13:R522012. View Article : Google Scholar
|
|
9
|
Zhu C, Yamaguchi K, Ohsugi T, Terakado Y,
Noguchi R, Ikenoue T and Furukawa Y: Identification of FERM
domain-containing protein 5 as a novel target of β-catenin/TCF7L2
complex. Cancer Sci. 108:612–619. 2017. View Article : Google Scholar
|
|
10
|
Fujita M, Furukawa Y, Tsunoda T, Tanaka T,
Ogawa M and Nakamura Y: Up-regulation of the ectodermal-neural
cortex 1 (ENC1) gene, a downstream target of the β-catenin/T-cell
factor complex, in colorectal carcinomas. Cancer Res. 61:7722–7726.
2001.PubMed/NCBI
|
|
11
|
Miwa N, Furuse M, Tsukita S, Niikawa N,
Nakamura Y and Furukawa Y: Involvement of claudin-1 in the
beta-catenin/Tcf signaling pathway and its frequent upregulation in
human colorectal cancers. Oncol Res. 12:469–476. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takahashi M, Tsunoda T, Seiki M, Nakamura
Y and Furukawa Y: Identification of membrane-type matrix
metalloproteinase-1 as a target of the beta-catenin/Tcf4 complex in
human colorectal cancers. Oncogene. 21:5861–5867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamaguchi K, Zhu C, Ohsugi T, Yamaguchi Y,
Ikenoue T and Furukawa Y: Bidirectional reporter assay using HAL
promoter and TOPFLASH improves specificity in high-throughput
screening of Wnt inhibitors. Biotechnol Bioeng. 114:2868–2882.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sandelin A, Alkema W, Engström P,
Wasserman WW and Lenhard B: JASPAR: An open-access database for
eukaryotic transcription factor binding profiles. Nucleic Acids
Res. 32:D91–D94. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamaguchi K, Yamaguchi R, Takahashi N,
Ikenoue T, Fujii T, Shinozaki M, Tsurita G, Hata K, Niida A, Imoto
S, et al: Overexpression of cohesion establishment factor DSCC1
through E2F in colorectal cancer. PLoS One. 9:e857502014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hagege H, Klous P, Braem C, Splinter E,
Dekker J, Cathala G, de Laat W and Forné T: Quantitative analysis
of chromosome conformation capture assays (3C-qPCR). Nat Protoc.
2:1722–1733. 2007. View Article : Google Scholar
|
|
17
|
Tsukamoto S, Ishikawa T, Iida S, Ishiguro
M, Mogushi K, Mizushima H, Uetake H, Tanaka H and Sugihara K:
Clinical significance of osteoprotegerin expression in human
colorectal cancer. Clin Cancer Clin. 17:2444–2450. 2011. View Article : Google Scholar
|
|
18
|
Schatoff EM, Leach BI and Dow LE: Wnt
Signaling and colorectal cancer. Curr Colorectal Cancer Rep.
13:101–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takahashi N, Yamaguchi K, Ikenoue T, Fujii
T and Furukawa Y: Identification of two Wnt-responsive elements in
the intron of RING finger protein 43 (RNF43) Gene. PLoS One.
9:e865822014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Thaler R, Rumpler M, Spitzer S, Klaushofer
K and Varga F: Mospd1, a new player in mesenchymal versus epidermal
cell differentiation. J Cell Physiol. 226:2505–2515. 2011.
View Article : Google Scholar
|
|
21
|
Klass MR and Hirsh D: Sperm isolation and
biochemical analysis of the major sperm protein from caenorhabditis
elegans. Dev Biol. 84:299–312. 1981. View Article : Google Scholar
|
|
22
|
Sepsenwol S, Ris H and Roberts TM: A
unique cytoskeleton associated with crawling in the amoeboid sperm
of the nematode, Ascaris suum. J Cell Biol. 108:55–66. 1989.
View Article : Google Scholar
|
|
23
|
Nelson GA, Roberts TM and Ward S:
Caenorhabditis elegans spermatozoan locomotion: Amoeboid movement
with almost no actin. J Cell Biol. 92:121–131. 1982. View Article : Google Scholar
|
|
24
|
Kara M, Axton RA, Jackson M, Ghaffari S,
Buerger K, Watt AJ, Taylor AH, Orr B, Hardy WR, Peault B and
Forrester LM: A role for MOSPD1 in mesenchymal stem cell
proliferation and differentiation. Stem Cells. 33:3077–3086. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Puiffe ML, Le Page C, Filali-Mouhim A,
Zietarska M, Ouellet V, Tonin PN, Chevrette M, Provencher DM and
Mes-Masson AM: Characterization of ovarian cancer ascites on cell
invasion, proliferation, spheroid formation, and gene expression in
an in vitro model of epithelial ovarian cancer. Neoplasia.
9:820–829. 2007. View Article : Google Scholar
|
|
26
|
Puisieux A, Brabletz T and Caramel J:
Oncogenic roles of EMT–inducing transcription factors. Nat Cell
Biol. 16:488–494. 2014. View Article : Google Scholar
|
|
27
|
Dancer JY, Henry SP, Bondaruk J, Lee S,
Ayala AG, de Crombrugghe B and Czerniak B: Expression of master
regulatory genes controlling skeletal development in benign
cartilage and bone forming tumors. Hum Pathol. 41:1788–1793. 2010.
View Article : Google Scholar
|
|
28
|
Groen EJ, Roos A, Muntinghe FL, Enting RH,
de Vries J, Kleibeuker JH, Witjes MJH, Links TP and van Beek AP:
Extra-intestinal manifestations of familial adenomatous polyposis.
Ann Surg Oncol. 15:2439–2450. 2008. View Article : Google Scholar
|
|
29
|
Takahashi M, Nakamura Y, Obama K and
Furukawa Y: Identification of SP5 as a downstream gene of the
beta-catenin/Tcf pathway and its enhanced expression in human colon
cancer. Int J Oncol. 27:1483–1487. 2005.
|
|
30
|
Jho EH, Zhang T, Domon C, Joo CK, Freund
JN and Costantini F: Wnt/beta-catenin/Tcf signaling induces the
transcription of axin2, a negative regulator of the signaling
pathway. Mol Cell Biol. 22:1172–1183. 2002. View Article : Google Scholar
|
|
31
|
Yochum GS, Cleland R and Goodman RH: A
genome-wide screen for beta-catenin binding sites identifies a
downstream enhancer element that controls c -myc gene expression.
Mol Cell Biol. 28:7368–7379. 2008. View Article : Google Scholar
|