Introduction
Lung cancer is the leading cause of
cancer-associated mortality worldwide, with 40–50% of patients
already at advanced stages (IIIB or IV) when first diagnosed, which
precludes surgical resection (1).
Non-small cell lung cancer (NSCLC) is the most common histological
phenotype of lung cancer, accounting for 80–85% of all patients
with lung cancer. EGFR is one of the most frequently observed
drivers of NSCLC, with ~50% of Asian and 11.9-33.0% of non-Asian
patients harboring activating EGFR mutations (2). Over the past decade,
molecular-targeted therapy has greatly improved the prognosis of
patients with NSCLC carrying EGFR mutations (3–5). In
particular, tyrosine kinase inhibitors (TKIs) that can target
activated EGFR, particularly those cases caused by exon 19 deletion
and/or exon 21 L858R point mutations, are recommended as the
standard therapeutic option for the management of NSCLCs positive
for EGFR mutations according to the clinical guidelines of the
National Comprehensive Cancer Network (6).
Although EGFR-TKIs exert strong efficacy in NSCLCs
with EGFR mutations, 20–30% of EGFR-mutant NSCLCs eventually
develop resistance to this treatment, while highly variable
outcomes are observed in EGFR-TKI responders (7). In particular, certain responders may
benefit for years, whilst others suffer from disease progression
and recurrence within weeks. Therefore, the presence of
non-responders and the heterogeneous prognosis of responders
demonstrate that EGFR-TKI monotherapy is not always the optimal
treatment strategy for EGFR-mutant NSCLCs. Further studies focusing
on the genomic landscape of NSCLC are necessary to identify
additional mechanisms of TKI resistance. However, there is evidence
to suggest that multiple concurrent genetic alterations resulting
in inhibitory PTEN mutations, increased programed death-ligand 1
expression, MET alterations, Bcl-2-like protein gene polymorphisms
and/or PI3K/AKT pathway activation, are associated with primary
resistance to EGFR-TKI treatment (8,9).
The p53 protein is a tumor suppressor that is
encoded by the tumor protein p53 (TP53) gene and is a master
regulator of cellular processes, including DNA damage response, DNA
repair, cell-cycle arrest, cell senescence and apoptosis, which
suppresses tumorigenesis (10).
Under physiological conditions, wild-type p53 protein is a
stress-responsive transcription factor with a sequence-specific
DNA-binding domain, two N-terminal transactivation domains and an
oligomerization domain required for transcriptional activity
(11). When DNA damage occurs, the
DNA damage response is triggered by the activation of
ataxia-telangiectasia mutated (ATM) or Rad3-related protein (ATR)
kinases. The activated ATM and/or ATR kinases then phosphorylate
wild-type p53 protein via checkpoint kinase (CHK)1 and CHK2,
respectively. Phosphorylated p53 recognizes specific promoter sites
and halts the cell cycle at the G1 phase via the
transcriptional activation of cyclin-dependent kinase inhibitor 1A
(11). This inhibits the process
of cell division when DNA damage occurs, thereby preventing the
proliferation of genetically unstable cells and transformation to a
potentially cancerous phenotype (10). TP53 has been recognized as one of
most frequently mutated genes in various types of human cancer. In
particular, ~73% of the somatic TP53 alterations detected in all
types of malignancies are missense mutations (12). TP53 missense mutations can disrupt
the biological function of the p53 DNA-binding domain by blocking
its ability to transcriptionally activate downstream target genes
(12). In patients with NSCLC,
co-existing TP53 mutations have been detected in 55–65% of EGFR
mutation-positive cases; they are particularly prevalent in
individuals who smoke and highly associated with the histological
type of squamous cell carcinoma (13–16).
Deleterious mutant p53 proteins acquire oncogenic properties that
promote the proliferation, invasion, survival and metastasis of
cancer cells (17). Consequently,
alterations to the TP53 genetic structure are proposed to serve a
key role in the clinical and molecular heterogeneity of
oncogene-driven lung cancer subgroups, due to their effects on drug
resistance and genomic instability (18).
Although previous studies have reported that mutant
TP53 can be used to predict inferior clinical outcomes with trends
of lower objective response rates (ORRs) and shorter
progression-free survival (PFS) and overall survival (OS) after
initial treatment with TKIs, ambiguities remain in the
epidemiological data. Almost all previous studies on this subject
are cohort studies with a small number of included cases. Although
the cohort studies may have low heterogeneity with regard to the
included cases, the relatively small number of cases in these
studies limits their results and conclusions, which are therefore
inconsistent. For example, Yu et al (19) found that TP53 mutations were not
significantly associated with PFS, despite predicting a shorter OS
in patients treated with gefitinib. By contrast, Tsui et al
(20) reported that TP53 mutations
were significantly associated with a shorter OS but did not alter
the PFS following EGFR-TKI treatment.
Therefore, the present meta-analysis was performed
to investigate the potential association between the prognosis of
TKI treatment for patients with advanced EGFR mutation-positive
NSCLCs and the presence or absence of concurrent TP53
mutations.
Materials and methods
Search strategy
All relevant articles published on dates prior to
and including January 30, 2022 were searched for in the PubMed
(https://pubmed.ncbi.nlm.nih.gov), Embase
(www.embase.com) and Cochrane databases
(www.cochrane.org) using a combination of ‘lung
cancer’ and ‘EGFR’ and ‘TP53’ with associated terms (Table SI, Table SII, Table SIII). The present meta-analysis
was performed in accordance with the Meta-Analysis of Observational
Studies in Epidemiology checklist (21). The abstracts of the identified
articles were screened before assessment of the corresponding full
texts of the eligible articles using the inclusion/exclusion
criteria.
Inclusion and exclusion criteria
Potentially eligible studies were required to meet
the following criteria: i) All included patients were
pathologically diagnosed with advanced NSCLC and surgery was no
longer an option; ii) EGFR mutation was confirmed by gene
sequencing methods; iii) patients were treated with EGFR-TKIs
regardless of the line of treatment; iv) the status of the TP53
gene was analyzed by gene sequencing methods; and v) at least one
set of survival and associated prognostic data was presented in the
study. The mandatory types of survival or prognostic data were the
PFS or OS for the TP53 mutant group vs. the TP53 wild-type group
presented as hazard ratios (HRs) with associated 95% confidence
intervals (CIs) or Kaplan-Meier (KM) curves, or ORRs in the TP53
mutant and wild-type TP53 groups.
Studies were excluded if they met the following
criteria: i) Non-original research studies, such as reviews,
editorials or expert opinions; ii) insufficient data for the
extraction or calculation of ORRs or HRs with 95% CIs; iii) the
status of the TP53 gene was not analyzed using a gene sequencing
method; iv) EGFR-TKIs were used for postoperative adjuvant therapy;
v) duplicate publications; vi) not published in the English
language; and vii) studies with low quality.
Study selection and data
abstraction
Two independent investigators (BLa and NZ) reviewed
the titles, abstracts and full-texts of all potential studies. A
third investigator (BLe) was responsible for resolving any
disagreements between the first two investigators. Information was
extracted from each eligible paper, including the name of the first
author, publication year, country, type of study, EGFR-TKI used,
histological type, EGFR mutation profile, line of TKI treatment,
methods of TP53 detection, detected exons of TP53, samples
extracted for TP53 detection, clinical outcomes and the number of
patients with EGFR and/or TP53 mutations. Since exon 21 L858R
mutation and exon 19 deletion are the most frequent genotypes of
EGFR mutations, the number of patients concurrently harboring at
least one of these mutations along with mutant TP53 was also
recorded from each paper to investigate the incidence of TP53
mutations among these two genotypes with mutant EGFR genes. ORR
data was extracted from the number of patients exhibiting a
complete response (CR) and partial response (PR) in the TP53
mutation and wild-type TP53 groups, respectively. PFS and OS data
were measured as HRs with 95% CIs for the TP53 mutant group vs. the
wild-type TP53 groups. In addition, HRs estimated using
multivariate models were selected if PFS or OS were both analyzed
using univariate and multivariate models. If the HR with 95% CI was
not reported in the original article, the KM curves were digitized
using Engauge Digitizer 4.1 software (http://markummitchell.github.io/engauge–digitizer/)
prior to recalculation of the HR using the approach previously
described by Guyot et al (22). To ensure consistency in the
collected results, each recalculated HR was evaluated twice
independently (BLa and NZ).
Quality appraisal
Quality appraisal was conducted using the
Newcastle-Ottawa Scale (NOS) and performed by two independent
investigators (BLa and NZ). The NOS evaluates the quality of each
included study from the perspectives of ‘selection’,
‘comparability’ and ‘outcome’ to provide a maximum total score of 9
points (23). Based on the final
score, the quality of each included study was classified as
follows: High quality (score ≥7); medium quality (7> score ≥5)’;
and low quality (score <5). Low quality indicates potential bias
and confounding in the study. Since the inclusion of such studies
in the analysis may affect the accuracy of the results, studies
categorized as low quality were excluded.
Statistical analysis
The primary outcomes assessed for the present
meta-analysis were ORR, PFS and OS. ORR was defined as the
percentage of patients who exhibited CR or PR. PFS was defined as
the period from the initiation of EGFR-TKI treatment until disease
progression, recurrence or mortality. OS was defined as the period
from the initiation of EGFR-TKI treatment to mortality. Pooled HRs
with 95% CIs were used to assess the association between TP53
status and survival outcomes (PFS and OS). The pooled odds ratios
(ORs) with 95% CIs were used for comparing the ORR between TP53
mutation-positive and -negative groups, in addition to the
frequency of TP53 mutation between patients harboring EGFR exon 21
L858R mutation and/or EGFR exon 19 deletion.
Statistical heterogeneity among the included studies
was measured using the χ2-based Q-test, where a Q-test
yielding P≤0.10 and/or an I2-value of >50% was
considered to indicate the existence of significant heterogeneity
(24). A random-effects model was
used to calculate the pooled OR or HR if significant heterogeneity
was detected. Otherwise, a fixed-effects model was applied.
Subgroups were stratified according to the type of study,
histological type, genotype of mutant EGFR, line of TKI treatment,
method of TP53 detection, detected exons of TP53 and type of
samples used for TP53 detection. In the present study, all data
synthesis was two-sided, where P≥0.05 or 95% CI crossing 1.00 was
not considered to be statistically significant.
For the evaluation of publication bias, Begg's
funnel plots and Egger's tests were performed. Egger's tests
yielding P≤0.10 were considered to suggest potential publication
bias. All data synthesis and analysis was performed using STATA
14.0 software (StataCorp LP).
Results
Study selection
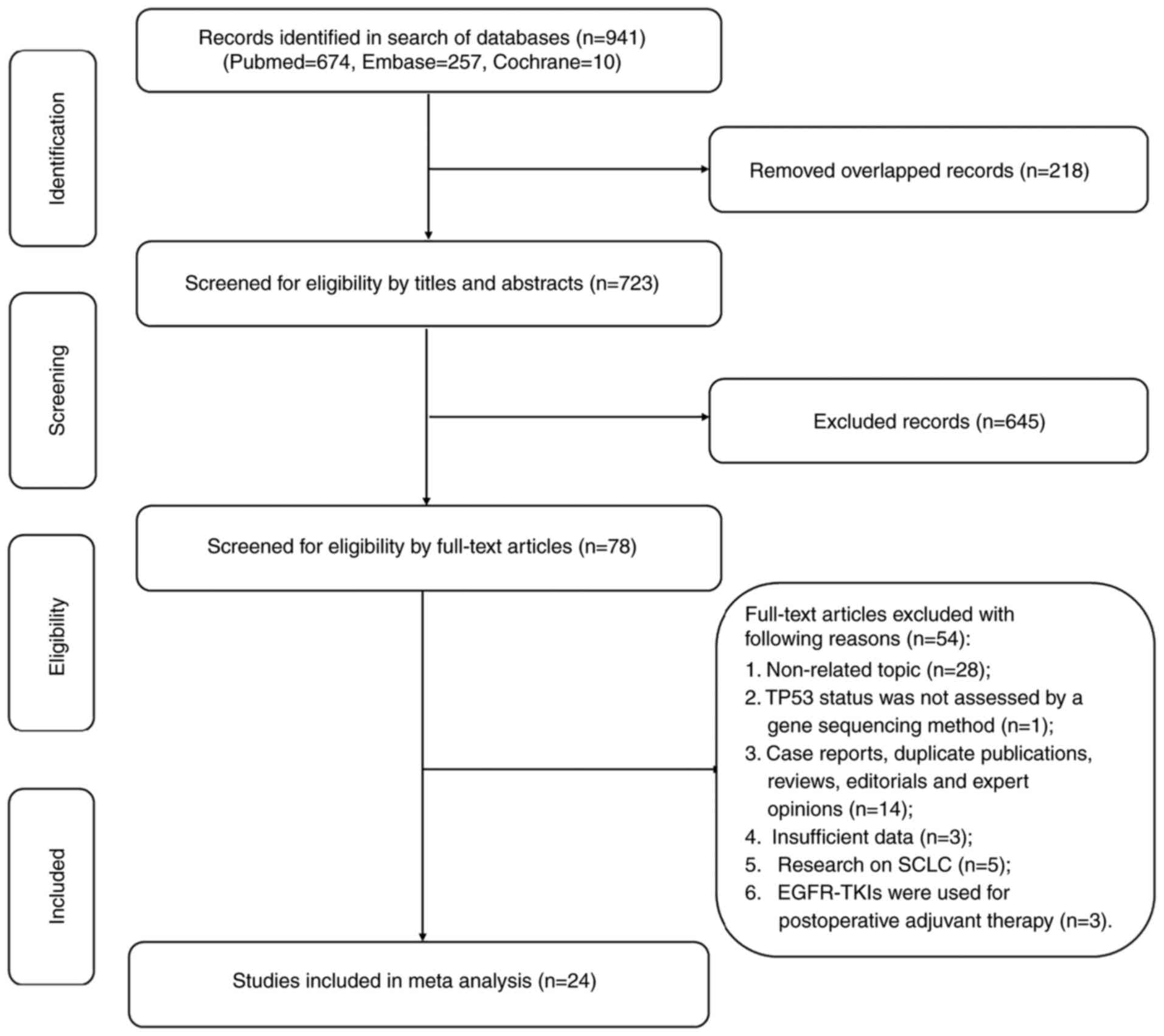
In total, 941 relevant records were identified in
the PubMed, Embase and the Cochrane library databases by January
30, 2022. Following screening of the titles and abstracts, 863
records were excluded and 78 full texts were reviewed. Finally, 24
studies involving a total of 2,227 patients with EGFR-mutant NSCLC
met the inclusion criteria and were included in the present
meta-analysis (Fig. 1).
Characteristics of the included
studies
The characteristics of each included study are
summarized in Table I. Of the
included studies, six were conducted as prospective studies and 18
were retrospective in design. In total, 14 cohorts included
patients who received EGFR-TKIs as the first-line treatment only,
two cohorts included patients with at least one prior treatment,
and nine cohorts analyzed patients receiving EGFR-TKIs as the
first, second or further line of treatment.
 | Table I.Basic characteristics of the included
studies. |
Table I.
Basic characteristics of the included
studies.
| First author,
year | Country | Type of study | Histological
type | Patients
included | Patients with EGFR
mutations | Patients with TP53
mutations | TKIs | Line of TKI
treatment | Outcomes | (Refs.) |
|---|
| CLCGP, 2013 | Europe,
Austria | Retrospective | NSCLC | 1,225 | 80 | 27 | Gefitinib,
erlotinib | 1st/≥2nd | OS | (43) |
| Molina-Vila,
2014 | Spain | Retrospective | Adenocarcinoma | 193 | 193 | 50 | Gefitinib,
erlotinib | 1st/≥2nd | ORR, OS | (44) |
| Bria, 2015 | Italy | Retrospective | Adenocarcinoma | 18 | 18 | 6 | Gefitinib | 1st | PFS | (45) |
| Canale, 2017 | Italy | Retrospective | Adenocarcinoma
(98%) | 123 | 123 | 37 | Gefitinib,
erlotinib, afatinib, dacomitinib | 1st | PFS, OS | (46) |
| VanderLaan,
2017 | USA | Retrospective | Adenocarcinoma | 171 | 16 | 7 | Gefitinib,
erlotinib, afatinib | 1st | PFS | (47) |
| Labbé, 2017 | Canada | Retrospective | Adenocarcinoma
(95%) | 105 | 60 | 24 | Gefitinib,
erlotinib, afatinib | 1st/≥2nd | ORR, PFS, OS | (32) |
| Tsui, 2018 | UK | Prospective | NSCLC | 50 | 30 | 12 | Gefitinib | 1st | ORR, PFS, OS | (20) |
| Aisner, 2018 | USA | Prospective | Adenocarcinoma | 904 | 60 | 34 | Not reported | 1st/≥2nd | OS | (48) |
| Aggarwal, 2018 | USA | Retrospective | Adenocarcinoma | 131 | 131 | 81 | Not reported | 1st | OS | (49) |
| Yu, 2018 | USA | Retrospective | Adenocarcinoma | 374 | 200 | 119 | Erlotinib,
dacomitinib, afatinib | 1st/≥2nd | PFS, OS | (50) |
| Kim, 2019 | Korea | Retrospective | Adenocarcinoma
(98%) | 307 | 157 | 101 | Cohort 1:
Gefitinib, erlotinib, afatinib; Cohort 2: osimertinib,
olmutinib | Cohort 1: 1st;
Cohort 2: ≥2nd | PFS, OS | (51) |
| Chang, 2019 | China | Retrospective | Adenocarcinoma | 33 | 33 | 10 | Gefitinib,
erlotinib, afatinib | 1st | PFS, OS | (52) |
| Hou, 2019 | China | Retrospective | Adenocarcinoma
(96%) | 163 | 71 | 43 | Gefitinib,
erlotinib, icotinib | 1st/≥2nd | PFS, OS | (53) |
| Rachiglio,
2019 | Italy | Retrospective | NSCLC | 133 | 133 | 23 | Gefitinib,
erlotinib, afatinib | 1st | PFS, OS | (54) |
| Canale, 2020 | Italy | Retrospective | NSCLC | 136 | 136 | 42 | Gefitinib,
erlotinib, afatinib | 1st | ORR, PFS, OS | (41) |
| Cheng, 2020 | China | Retrospective | Adenocarcinoma
(99%) | 179 | 76 | 53 | Gefitinib,
erlotinib, afatinib, dacomitinib, osimertinib, avitinib | 1st | PFS, OS | (55) |
| Li, 2021 | China | Retrospective | Adenocarcinoma
(96%) | 195 | 195 | 134 | Gefitinib,
erlotinib | 1st/≥2nd | ORR, PFS, OS | (56) |
| Steendam, 2020 | The
Netherlands | Prospective | NSCLC | 41 | 41 | 23 | Erlotinib,
osimertinib | 1st/≥2nd | PFS | (57) |
| Yang, 2021 | China | Retrospective | Adenocarcinoma
(97%) | 62 | 50 | 37 | Osimertinib | 1st/≥2nd | PFS | (58) |
| Yu, 2021 | China | Prospective | Adenocarcinoma | 180 | 180 | 115 | Gefitinib | 1st | ORR, PFS, OS | (19) |
| Wang, 2021 | China | Prospective | Adenocarcinoma | 135 | 54 | 25 | Gefitinib,
icotinib, afatinib | 1st | PFS | (42) |
| Tan, 2021 | China | Retrospective | Adenocarcinoma
(95%) | 180 | 51 | 30 | Gefitinib,
erlotinib, icotinib, afatinib | 1st | PFS | (40) |
| Wang, 2021 | China | Prospective | Adenocarcinoma | 106 | 62 | 26 | Mefatinib | 1st | ORR, PFS, OS | (59) |
| Roeper, 2022 | Germany | Retrospective | Adenocarcinoma
(99%) | 77 | 77 | 32 | Osimertinib | ≥2nd | ORR, PFS, OS | (60) |
Among the 2,227 patients who underwent TP53 gene
sequencing, alterations in the TP53 gene were detected in 1,091
cases. For efficacy evaluation, 8 included studies provided ORR
data, 20 studies reported PFS endpoints and 18 studies reported OS
endpoints. The majority of the studies (17/24) used next-generation
sequencing (NGS) as the sequencing method for detecting the status
of the TP53 gene. The samples used for TP53 gene sequencing were
tissue in 16 studies, both tissue and plasma in seven studies and
plasma in one study.
Study quality
The results of quality appraisal as assessed using
the NOS are shown in Table SIV.
According to the final scores, 20 studies were classified as high
quality, whilst the remaining four studies were classified as
medium quality. Low quality indicates the presence of potential
bias and confounding in the study. Therefore, no studies classified
as low quality were included in the present analysis.
Associations between TP53 mutations
and genotypes of mutant-EGFR
Exon 21 L858R mutation and exon 19 deletion are the
most frequently found genotypes of EGFR mutations. Although more
than eight studies included patients with the genotypes of EGFR
Exon 21 L858R mutation and exon 19 deletion (Table II), only eight studies with nine
cohorts included available data to calculate statistical
significance of the incidence of TP53 mutations in these two
genotypes. Among the 669 patients from the eight studies, the
incidence of TP53 mutations in the exon 21 L858R mutation group was
53.82% (148/275), whilst the incidence of TP53 mutations in the
exon 19 deletion group was 50.76% (200/394). No statistically
significant difference in the incidence of TP53 mutations between
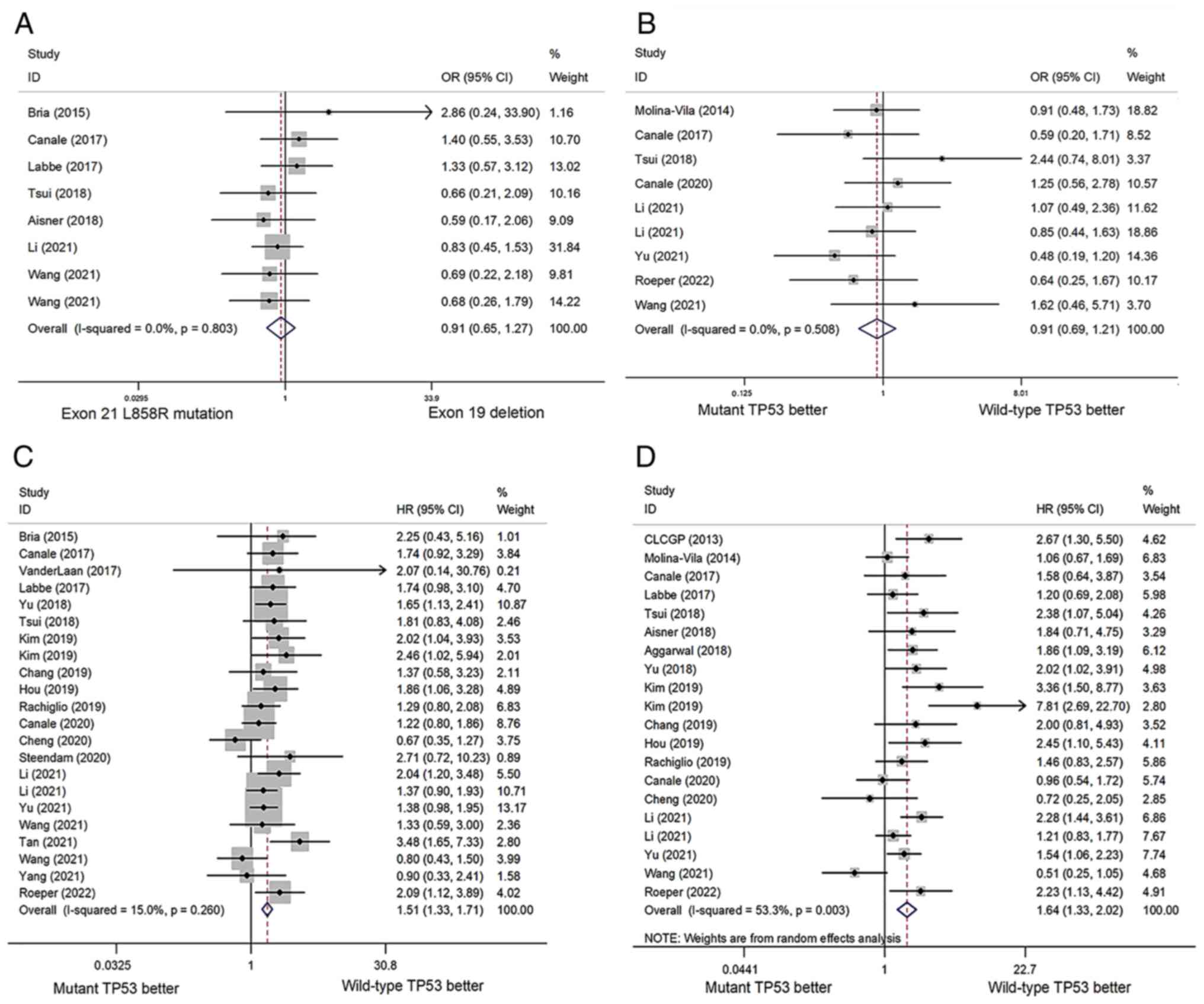
these two groups was identified (OR=0.91; 95% CI=0.65-1.27;
P=0.568; Fig. 2A). Q-test and
I2 analysis revealed no significant heterogeneity among
the eight included studies (I2=0%; P=0.803). Publication
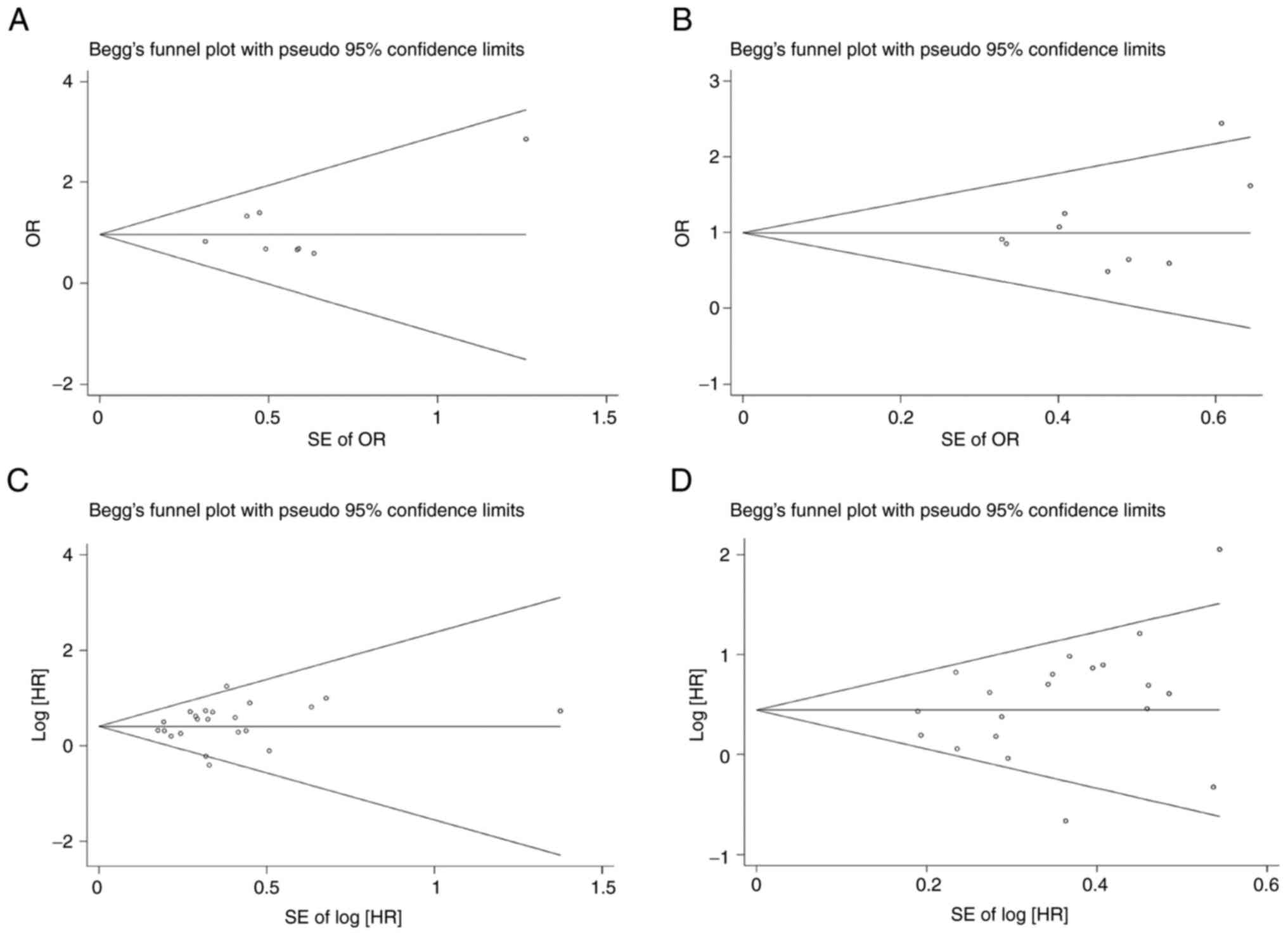
bias was not detected by Egger's test (P=0.386) or Begg's funnel
plot (Fig. 3A).
 | Table II.Details of gene alteration in the
included studies. |
Table II.
Details of gene alteration in the
included studies.
| First author,
year | EGFR mutation
profile | Methods of TP53
detection | Detected exons of
TP53 | Samples for TP53
detection | (Refs.) |
|---|
| CLCGP, 2013 | Not reported | PCR | Exons 5-9 | Tissue | (43) |
| Molina-Vila,
2014 | Not reported | High-resolution
melting | Exons 5-8 | Tissue | (44) |
| Bria, 2015 | Exon 19 deletion,
exon 21 L858R mutation | Sanger
sequencing | Exons 5-8 | Tissue | (45) |
| Canale, 2017 | Exon 18 point
mutation, exon 19 deletion, exon 21 point mutation, exon 21 L858R
mutation | Direct
sequencing | Exons 5-8 | Tissue | (46) |
| VanderLaan,
2017 | Exon 18-21
mutations | NGS/JAX-Cancer | Exons 5-8 | Tissue | (47) |
|
|
| Treatment
Profile |
|
|
|
| Labbé, 2017 | Exon 19 deletion,
exon 21 L858R mutation, exon 18 mutations, exon 19 insertion, exon
19 L747P mutation, multiple mutations | NGS or Sanger
sequencing | Exons 3-8 | Tissue | (32) |
| Tsui, 2018 | Exon 21 L858R
mutation, exon 19 deletion | Digital PCR | Exons 5-8 | Plasma | (20) |
| Aisner, 2018 | Exon 19 deletion,
exon 19 insertion, exon 18 G719X mutation, exon 21 L861Q
mutation | NGS | Exons 2-11 | Tissue | (48) |
| Aggarwal, 2018 | Exon 18-21
mutations | NGS | Exons 4-10 | Tissue, plasma | (49) |
| Yu, 2018 | Exon 19 deletion,
exon 20 insertion, exon 21 p.L858R, exon 18 deletion, exon 19
insertion, exon 21 L861Q mutation, exon 19 L747P mutation, exon 18
E709X + exon 18 G719X mutation, exon 18 G719X + exon 20 S768I
mutation, exon 18 G719X + exon 21 L861Q mutation | Illumina HiSeq400
platform | Exons 2-11 | Tissue | (50) |
| Kim, 2019 | Exon 21 L861Q
mutation, exon 19 deletion, exon 21 L858R mutation, exon 18 G719A
mutation, exon 21 L833V + exon 21 L858R mutation, exon 21 L833V +
exon 21 H835L mutation | NGS | Exons 2-11 | Tissue | (51) |
| Chang, 2019 | Exon 18 mutation,
exon 19 mutation, exon 21 mutation | NGS | Not reported | Tissue | (52) |
| Hou, 2019 | Exon 19 deletion,
exon 21 L858R mutation, uncommon mutations | NGS | Exons 1-10 | Tissue | (53) |
| Rachiglio,
2019 | Exon 19 deletion,
exon 21 L858R mutation, exon 20 T790M mutation | NGS | Exons 2-11 | Tissue | (54) |
| Canale, 2020 | Exon 19 deletion,
exon 21 L858R mutation, uncommon mutations | PCR/NGS | Exons 5-8 | Tissue | (41) |
| Cheng, 2020 | Exon 19 deletion,
exon 21 L858R mutation, exon 20 insertion, exon 18 G719X mutation,
exon 21 L861Q mutation | NGS | Not reported | Tissue, plasma | (55) |
| Li, 2021 | Exon 19 deletion,
exon 21 L858R mutation | NGS | Cohort 1: exons
4/7; Cohort 2: other exons | Tissue | (56) |
| Steendam, 2020 | Exon 19 deletion,
exon 19 deletion-insertion, exon 19 other mutation, exon 21 L858R
mutation, exon 21 other mutation, exon 20 T790M | NGS | Exons 5-8 | Tissue, plasma | (57) |
| Yang, 2021 | Exon 20
insertion | NGS | Exons 1-10 | Tissue, plasma | (58) |
| Yu, 2021 | Exon 19 deletion,
exon 21 L858R mutation | NGS | Not reported | Tissue, plasma | (19) |
| Wang, 2021 | Exon 19 deletion,
exon 21 L858R mutation, uncommon mutations |
ARMS-PCR/cSMART | Not reported | Tissue, plasma | (42) |
| Tan, 2021 | Exon 19 deletion,
exon 21 L858R mutation, uncommon mutations | NGS | Not reported | Tissue | (40) |
| Wang, 2021 | Exon 19 deletion,
exon 21 L858R mutation | NGS | Exons 5-8 | Tissue, plasma | (59) |
| Roeper, 2022 | Exon 19 deletion,
exon 21 L858R mutation, exon 20 T790M | NGS | Not reported | Tissue | (60) |
Associations between concurrent TP53
mutations and ORR
The analysis of ORR was obtained from eight eligible
studies with nine cohorts. The overall ORR to EGFR-TKI treatment
was 62% (95% CI=51–73%) and 63% (95% CI=51–75%) in the mutant TP53
and wild-type TP53 groups, respectively. However, this difference
in ORRs was not found to be statistically significant (OR=0.91; 95%
CI=0.69-1.21; P=0.529; Fig. 2B).
Statistically significant heterogeneity was not identified in this
analysis (I2=0%; P=0.508). Egger's test (P=0.265) and
Begg's funnel plot (Fig. 3B)
indicated the absence of publication bias. The results of specific
subgroup analyses are shown in Table
III, which reveal no significant associations between
concurrent TP53 mutations and ORR among the subgroups.
 | Table III.Subgroup analysis of ORR, PFS and
OS. |
Table III.
Subgroup analysis of ORR, PFS and
OS.
|
| ORR | PFS | OS |
|---|
|
|
|
|
|
|---|
| Subgroup | Cohorts (n) | Pooled OR
(95%CI) | I2
(%) | P-value | Cohorts (n) | Pooled HR
(95%CI) | I2
(%) | P-value | Cohorts (n) | Pooled HR
(95%CI) | I2
(%) | P-value |
|---|
| Line of TKI
treatment |
|
|
|
|
|
|
|
|
|
|
|
|
|
1st | 5 | 0.97
(0.63-1.50) | 37.7 | 0.895 | 13 | 1.37
(1.16-1.62) | 29.2 | <0.001 | 10 | 1.43
(1.06-1.94) | 50.8 | 0.020 |
|
≥2nd | - | - | - |
| 2 | 2.21
(1.33-3.67) | 0.0 | 0.002 | 2 | 3.89
(1.15-13.19) | 73.4 | 0.029 |
|
1st/≥2nd | 3 | 0.92
(0.62-1.37) | 0.0 | 0.695 | 7 | 1.63
(1.33-1.99) | 0 | <0.001 | 8 | 1.63
(1.25-2.12) | 41.4 | <0.001 |
| Samples for TP53
detection |
|
|
|
|
|
|
|
|
|
|
|
|
|
Tissue | 6 | 0.89
(0.65-1.23) | 0.0 | 0.490 | 15 | 1.66
(1.42-1.92) | 0 | <0.001 | 15 | 1.76
(1.39-2.23) | 49.0 | <0.001 |
|
Tissue/plasma | 3 | 0.99
(0.55-1.78) | 61.5 | 0.962 | 7 | 1.18
(0.93-1.49) | 26.9 | 0.164 | 5 | 1.28
(0.78-2.10) | 67.4 | 0.337 |
| Methods of TP53
sequencing |
|
|
|
|
|
|
|
|
|
|
|
|
|
NGS | 5 | 0.82
(0.56-1.19) | 0.0 | 0.070 | 14 | 1.49
(1.27-1.73) | 41.7 | <0.001 | 13 |
1.73(1.30-2.29) | 60.3 | <0.001 |
|
Other | 4 | 1.05
(0.69-1.60) | 12.6 | 0.806 | 8 | 1.55
(1.25-1.92) | 0 | <0.001 | 7 | 1.48
(1.09-2.00) | 36.3 | 0.012 |
| Detected exons of
TP53 |
|
|
|
|
|
|
|
|
|
|
|
|
| Exons
5-8 | 5 | 1.10
(074–1.64) | 0.0 | 0.638 | 7 | 1.35
(1.03–1.77) | 0 | 0.031 | 5 | 1.10
(0.7–1.72) | 55.6 | 0.665 |
| Exons
2–11 | - | - | - |
| 4 | 1.64
(1.26–2.12) | 0 | <0.001 | 5 | 2.44
(1.4–4.11) | 53.1 | 0.001 |
| Exons
1–10 | - | - | - |
| 2 | 1.56
(0.95–2.55) | 35.4 | 0.077 | - | - | - |
|
| Type of study |
|
|
|
|
|
|
|
|
|
|
|
|
|
Prospective | 3 | 0.99
(0.55–1.78) | 61.5 | 0.962 | 5 | 1.32
(1.02–1.72) | 5.7 | 0.036 | 4 | 1.35
(0.73–2.48) | 78.7 | 0.340 |
|
Retrospective | 6 | 0.89
(0.65–1.23) | 0.0 | 0.633 | 17 | 1.57
(1.36–1.81) | 16.6 | <0.001 | 16 |
1.71(1.36–2.15) | 49.8 | <0.001 |
| Histological
type |
|
|
|
|
|
|
|
|
|
|
|
|
|
NSCLC | 2 | 1.54
(0.79–2.98) | 0.0 | 0.205 | 4 | 1.36
(1.02–1.81) | 0.0 | 0.036 | 5 | 1.68
(1.18–2.38) | 36.5 | 0.004 |
|
Adenocarcinoma | 7 | 0.81
(0.60–1.11) | 0.0 | 0.286 | 18 | 1.54
(1.34–1.77) | 23.6 | <0.001 | 15 | 1.63
(1.25–2.11) | 59.0 | <0.001 |
Associations between concurrent TP53
mutations and PFS
The association between concurrent TP53 mutations
and PFS was analyzed using the data from 20 eligible studies with
22 cohorts. No statistically significant heterogeneity was observed
among these studies (I2=15.0%; P=0.260). Patients with
concurrent TP53 mutations showed a significantly shorter PFS
(HR=1.51; 95% CI=1.33-1.71; P<0.001) following EGFR-TKI
treatment (Fig. 2C). Among the
included studies, publication bias was not detected by Egger's test
(P=0.304) or Begg's funnel plot (Fig.
3C).
Subgroup analysis suggested that prospective
(HR=1.32; 95% CI=1.02-1.72; P=0.036) and retrospective (HR=1.57;
95% CI=1.36-1.81; P<0.001) studies demonstrated that TP53
mutations were significantly associated with a shorter PFS. In
terms of histological types, concurrent TP53 mutations predicted a
shorter PFS in patients with lung adenocarcinoma (HR=1.54; 95%
CI=1.34-1.77; P<0.001) and NSCLC (HR=1.36; 95% CI=1.02-1.81;
P=0.036). With the respect to the line of TKI treatment, TP53
mutations were significantly associated with a higher risk of
disease progression after first-line (HR=1.37; 95% CI=1.16-1.62;
P<0.001), all lines (HR=1.63; 95% CI=1.33-1.99; P<0.001) and
second line or further EGFR-TKI treatments (HR=2.21; 95%
CI=1.33-3.67; P=0.002). Whenever mutations were detected in exons
5-8 (HR=1.35; 95% CI=1.03-1.77; P=0.031) or exons 2-11 (HR=1.64;
95% CI=1.26-2.12; P<0.001), PFS was significantly shorter in the
TP53 mutant cohorts. However, the predictive value for PFS of TP53
mutations detected in both tissue and plasma specimens (HR=1.18;
95% CI=0.93-1.49; P=0.164) were not in concordance with TP53
mutations detected in tissue specimens alone (HR=1.66; 95%
CI=1.42-1.92; P<0.001).
Association between concurrent TP53
mutations and OS
In total, 18 studies with 20 cohorts were included
in the present analysis. Pooled results with a random-effects model
demonstrated a significantly shorter OS in patients harboring
concurrent TP53 mutations treated with EGFR-TKIs (HR=1.64; 95%
CI=1.33-2.02; P<0.001; Fig.
2D). Significant heterogeneity was observed among the included
studies (I2=53.3%; P=0.003). Begg's funnel plot
(Fig. 3D) and Egger's test
(P=0.183) indicated no publication bias.
Subgroup analysis was subsequently performed. In
retrospective studies, patients with TP53 mutants treated with
EGFR-TKIs exhibited poorer overall survival outcomes (HR=1.71; 95%
CI=1.36-2.15; P<0.001), as did those with adenocarcinoma
(HR=1.63; 95% CI=1.25-2.11; P<0.001) and NSCLC (HR=1.68; 95%
CI=1.18-2.38; P=0.004). TP53 mutations were also associated with a
shorter OS regardless of the TKI treatment line, namely first line
(HR=1.43; 95% CI=1.06-1.94; P=0.020), all lines (HR=1.63; 95%
CI=1.25-2.12; P<0.001) and second line or further (HR=3.89; 95%
CI, 1.15-13.19; P=0.029). Furthermore, OS was only shorter in the
TP53 mutant cohort if mutations were detected in exons 2-11
(HR=2.44; 95% CI=1.45-4.11; P<0.001).
Discussion
According to a previous study, the frequency of
concurrent TP53 and EGFR mutations in NSCLC is within the range of
55–65% (14). In the present
study, concurrent TP53 mutations were observed in 49% of cases
(1,091/2,227), which was similar to the previously reported
frequency (14). Elucidating the
role of concurrent TP53 mutations in EGFR-TKI resistance may be
beneficial for the precise identification of populations who are
most likely to benefit from TKI treatment. Evidence from previous
pre-clinical studies suggests that the TP53 gene status influences
the response to EGFR-TKIs. For example, wild-type TP53 was shown to
increase gefitinib sensitivity by facilitating apoptosis in
EGFR-mutant NSCLC cell lines, while the sensitivity to TKIs was
suppressed in TP53 mutant NSCLC cell lines (25,26).
In addition, numerous clinical studies have demonstrated that
EGFR-mutant NSCLCs with coexisting TP53 mutations treated with TKIs
exhibit a trend towards lower ORR, shorter PFS and OS compared with
those in NSCLCs with wild-type TP53. However, the results from
previous studies have exhibited inconsistencies.
According to a previous meta-analysis, which
included a fewer number of studies, patients with NSCLCs harboring
concurrent TP53 mutations have a significantly worse prognosis than
those without TP53 mutations when treated with EGFR-TKIs (27). The present study was performed with
the inclusion of updated data and more recent studies to provide a
more thorough analysis. In the present meta-analysis, the
association between concurrent TP53 mutations and the clinical
outcomes of patients with EGFR-mutant NSCLC treated with TKIs was
investigated. Concurrent TP53 mutations were found to be associated
with shorter PFS and OS but not with the ORR, suggesting that
concurrent TP53 mutations are associated with primary resistance to
TKI therapy. There are limited reports on the incidence of TP53
mutations in patients with the two classical genotypes of EGFR gene
alterations, namely exon 21 L858R mutation and exon 19 deletion.
The combined analysis of these reports in the present meta-analysis
identified no significant difference in the incidence of TP53
mutations between patients with exon 21 L858R mutation and exon 19
deletion.
It should be noted that there is no evidence
conclusively showing that TP53 mutations are directly involved in
the mechanism of resistance to EGFR-TKIs. Mutations in the TP53
gene can lead to p53 protein losing its function in the maintenance
of genomic stability, which has been previously reported to be
associated with a higher tumor mutational burden in cancers
(28). The development of drug
resistance in cancers is closely associated with genetic
alterations. Genomic instability and higher frequencies of gene
mutations facilitate the occurrence of resistance-associated
mutations at earlier stages of molecular-targeted therapy. Cancer
cells carrying resistance-associated mutations proliferate more
readily to form sub-clones, leading to clinical progression,
recurrence and metastasis (29).
This may explain the shorter PFS and OS following EGFR-TKI
treatment in the TP53 mutant group in the present meta-analysis,
although the ORRs for TP53 mutant and TP53 wild-type patient groups
were similar.
The DNA-binding domain encoded by exons 5-8 is the
main functional domain of the p53 protein. The binding of p53 to
specific DNA response elements promotes the expression of genes
that guard against malignant cell transformation and cancer
progression (30,31). Previous reports have revealed that
the frequency of mutations occurring in exons 5-8 is higher
compared with that in other coding regions of the TP53 gene
(19,32). Mutations occurring in exons 5-8 of
the TP53 gene may result in functional deficiency and
counterintuitive tumorigenic properties of the p53 protein
(33). In the present
meta-analysis, the association between TP53 mutations and the
response to TKI therapy varied among the subgroups stratified
according to mutant exons. According to the subgroup analysis,
mutations occurring in exons 5-8 predicted a poorer prognosis,
including shorter PFS and OS, following EGFR-TKI treatment, which
is consistent with the aforementioned studies. In terms of
influence on the function of p53 protein and the degree of
disturbance of the protein structure, TP53 gene alterations can be
classified into disruptive and non-disruptive mutations (34). However, due to the lack of studies
on this topic, the impact of disruptive/non-disruptive mutations on
the clinical outcomes could not be assessed in the present
study.
With the development of gene sequencing technology,
NGS has become the most frequently used method for detecting and
analyzing the tumor genotypes of NSCLC in clinical practice
(35). Although the consistency of
NGS and other methods for TP53 sequencing requires further
validation, the predictive value of NGS-detected TP53 mutations was
consistent with TP53 mutations detected using other methods
according to the current subgroup analysis. Likewise, the liquid
biopsy of circulating tumor DNA (ctDNA) has become widely used for
the identification of the real-time molecular characteristics of
advanced and metastatic NSCLC. TP53 mutations detected in ctDNA by
liquid biopsy appeared to have associations with PFS and OS, whilst
TP53 mutations in tissue specimens were predictors of shorter PFS
and OS in the present study. This discrepancy may be due to the
relatively small number of eligible studies focusing on liquid
biopsy. TKI treatment is recommended as the first line of treatment
for advanced and metastatic NSCLCs with sensitive EGFR mutations
according to various clinical guidelines (36). Based on the results of the subgroup
analysis in the present study, TP53 mutations predict a decreased
responsiveness of patients receiving EGFR-TKI therapy regardless of
the line of therapy.
It must be emphasized that the present study has a
number of limitations that must be addressed. The predictive values
of concurrent TP53 mutations on the efficacy of specific EGFR-TKIs,
including gefitinib, erlotinib, afatinib and osimertinib, remain
unclear. Similarly, there is insufficient evidence to verify
whether the effects of concurrent TP53 mutations on TKI efficacy
are consistent among populations with different genotypes of mutant
EGFR. However, it has been documented that various EGFR mutation
genotypes may lead to heterogeneous responsiveness to TKIs
(37–39). Yu et al (19) reported that patients harboring EGFR
exon 19 deletion and TP53 mutations treated with gefitinib had a
longer PFS and OS compared with those with EGFR L585R mutation and
TP53 mutations (19). In addition,
a retrospective study by Tan et al (40) demonstrated that the co-existence of
uncommon EGFR mutations and TP53 mutations is associated with a
shorter PFS. By contrast, Canale et al (41) observed significantly shorter PFS
and OS times in the subgroup of TKI-treated patients with EGFR exon
19 deletion compared with those without this deletion, whilst Wang
et al (42) identified no
significant difference in PFS between patients with EGFR exon 19
deletion and EGFR L585R mutations. Such evidence is not sufficient
to completely clarify the intrinsic association between TP53
mutations and EGFR-TKI efficacy. Finally, the present study
was not prospectively registered in an appropriate registry, such
as the National Institute for Health Research's PROSPERO
database.
Collectively, the present study suggests that
concurrent TP53 mutations are associated with the primary
resistance of NSCLC to EGFR-TKIs and predict poorer clinical
outcomes with shorter PFS and OS following EGFR-TKI treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BL and KD were responsible for the conception and
design of the study. BL, NZ and BLL collected data. BL, BLL and NZ
performed the data analysis and interpretation. All authors were
responsible for writing the manuscript. All authors read and
approved the final manuscript. BL and NZ confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TKI
|
tyrosine kinase inhibitor
|
|
NSCLC
|
non-small cell lung cancer
|
|
OR
|
odds ratio
|
|
CI
|
confidence interval
|
|
ORR
|
objective response rate
|
|
HR
|
hazard ratio
|
|
PFS
|
progression-free survival
|
|
OS
|
overall survival
|
|
CR
|
complete response
|
|
PR
|
partial response
|
|
NGS
|
next-generation sequencing
|
|
ctDNA
|
circulating tumor DNA
|
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Melosky B, Kambartel K, Häntschel M,
Bennetts M, Nickens DJ, Brinkmann J, Kayser A, Moran M and Cappuzzo
F: Worldwide prevalence of epidermal growth factor receptor
mutations in non-small cell lung cancer: A meta-analysis. Mol Diagn
Ther. 26:7–18. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et
al: Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): An open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rosell R, Carcereny E, Gervais R,
Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R,
Pallares C, Sanchez JM, et al: Erlotinib versus standard
chemotherapy as first-line treatment for European patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(EURTAC): A multicentre, open-label, randomised phase 3 trial.
Lancet Oncol. 13:239–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Soria JC, Ohe Y, Vansteenkiste J,
Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura
F, Nogami N, Kurata T, et al: Osimertinib in untreated EGFR-mutated
advanced non-small-cell lung cancer. N Engl J Med. 378:113–125.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
National Comprehensive Cancer Network
(NCCN), . non-small cell lung cancer, NCCN guidelines version 1.
2022.https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1450April
2–2022
|
|
7
|
Hirsch FR, Suda K, Wiens J and Bunn PA Jr:
New and emerging targeted treatments in advanced non-small-cell
lung cancer. Lancet. 388:1012–1024. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jin Y, Shi X, Zhao J, He Q, Chen M, Yan J,
Ou Q, Wu X, Shao YW and Yu X: Mechanisms of primary resistance to
EGFR targeted therapy in advanced lung adenocarcinomas. Lung
Cancer. 124:110–116. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo Y, Song J, Wang Y, Huang L, Sun L,
Zhao J, Zhang S, Jing W, Ma J and Han C: Concurrent genetic
alterations and other biomarkers predict treatment efficacy of
EGFR-TKIs in EGFR-mutant non-small cell lung cancer: A review.
Front Oncol. 10:6109232020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marei HE, Althani A, Afifi N, Hasan A,
Caceci T, Pozzoli G, Morrione A, Giordano A and Cenciarelli C: p53
signaling in cancer progression and therapy. Cancer Cell Int.
21:7032021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gomes AS, Ramos H, Inga A, Sousa E and
Saraiva L: Structural and drug targeting insights on mutant p53.
Cancers (Basel). 13:33442021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vaddavalli PL and Schumacher B: The p53
network: Cellular and systemic DNA damage responses in cancer and
aging. Trends Genet. 38:598–612. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kumari S, Sharma V, Tiwari R, Maurya JP,
Subudhi BB and Senapati D: Therapeutic potential of p53
reactivation in prostate cancer: Strategies and opportunities. Eur
J Pharmacol. 919:1748072022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hong S, Gao F, Fu S, Wang Y, Fang W, Huang
Y and Zhang L: Concomitant genetic alterations with response to
treatment and epidermal growth factor receptor tyrosine kinase
inhibitors in patients with EGFR-mutant advanced non-small cell
lung cancer. JAMA Oncol. 4:739–742. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Deben C, Deschoolmeester V, Lardon F,
Rolfo C and Pauwels P: TP53 and MDM2 genetic alterations in
non-small cell lung cancer: Evaluating their prognostic and
predictive value. Crit Rev Oncol Hematol. 99:63–73. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mogi A and Kuwano H: TP53 mutations in
nonsmall cell lung cancer. J Biomed Biotechnol. 2011:5839292011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Muller PAJ and Vousden KH: p53 mutations
in cancer. Nat Cell Biol. 15:2–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Skoulidis F and Heymach JV: Co-occurring
genomic alterations in non-small-cell lung cancer biology and
therapy. Nat Rev Cancer. 19:495–509. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu R, Bai H, Li T, Gao B, Han J, Chang G,
Zhang P, Fei K, He X and Wang J: TP53 mutations in circulating
tumor DNA in advanced epidermal growth factor receptor-mutant lung
adenocarcinoma patients treated with gefitinib. Transl Oncol.
14:1011632021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tsui DWY, Murtaza M, Wong ASC, Rueda OM,
Smith CG, Chandrananda D, Soo RA, Lim HL, Goh BC, Caldas C, et al:
Dynamics of multiple resistance mechanisms in plasma DNA during
EGFR-targeted therapies in non-small cell lung cancer. EMBO Mol
Med. 10:e79452018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stroup DF, Berlin JA, Morton SC, Olkin I,
Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA and Thacker
SB: Meta-analysis of observational studies in epidemiology: A
proposal for reporting. Meta-analysis of observational studies in
epidemiology (MOOSE) group. JAMA. 283:2008–2012. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guyot P, Ades AE, Ouwens MJ and Welton NJ:
Enhanced secondary analysis of survival data: Reconstructing the
data from published Kaplan-Meier survival curves. BMC Med Res
Methodol. 12:92012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wells G, Shea BJ and O'Connell J: The
Newcastle-Ottawa scale (NOS) for assessing the quality of
nonrandomised studies in meta-analyses. 2014.
|
|
24
|
Fletcher J: What is heterogeneity and is
it important? BMJ. 334:94–96. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chang GC, Hsu SL, Tsai JR, Liang FP, Lin
SY, Sheu GT and Chen CY: Molecular mechanisms of ZD1839-induced
G1-cell cycle arrest and apoptosis in human lung adenocarcinoma
A549 cells. Biochem Pharmacol. 68:1453–1464. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rho JK, Choi YJ, Ryoo BY, Na II, Yang SH,
Kim CH and Lee JC: p53 enhances gefitinib-induced growth inhibition
and apoptosis by regulation of Fas in non-small cell lung cancer.
Cancer Res. 67:1163–1169. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qin K, Hou H, Liang Y and Zhang X:
Prognostic value of TP53 concurrent mutations for EGFR-TKIs and
ALK-TKIs based targeted therapy in advanced non-small cell lung
cancer: A meta-analysis. BMC Cancer. 20:3282020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ulivi P, Urbini M, Petracci E, Canale M,
Dubini A, Bartolini D, Calistri D, Cravero P, Fonzi E, Martinelli
G, et al: Wide next-generation sequencing characterization of young
adults non-small-cell lung cancer patients. Cancers (Basel).
14:23522022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Park S, Shim JH, Lee B, Cho I, Park WY,
Kim Y, Lee SH, Choi Y, Han J, Ahn JS, et al: Paired genomic
analysis of squamous cell carcinoma transformed from EGFR-mutated
lung adenocarcinoma. Lung Cancer. 134:7–15. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Riley T, Sontag E, Chen P and Levine A:
Transcriptional control of human p53-regulated genes. Nat Rev Mol
Cell Biol. 9:402–412. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Levine AJ: The many faces of p53:
Something for everyone. J Mol Cell Biol. 11:524–530. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Labbé C, Cabanero M, Korpanty GJ, Tomasini
P, Doherty MK, Mascaux C, Jao K, Pitcher B, Wang R, Pintilie M, et
al: Prognostic and predictive effects of TP53 co-mutation in
patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung
Cancer. 111:23–29. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Petitjean A, Mathe E, Kato S, Ishioka C,
Tavtigian SV, Hainaut P and Olivier M: Impact of mutant p53
functional properties on TP53 mutation patterns and tumor
phenotype: Lessons from recent developments in the IARC TP53
database. Hum Mutat. 28:622–629. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Poeta ML, Manola J, Goldwasser MA,
Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D,
Saunders J, et al: TP53 mutations and survival in squamous-cell
carcinoma of the head and neck. N Engl J Med. 357:2552–2561. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
De Maglio G, Pasello G, Dono M, Fiorentino
M, Follador A, Sciortino M, Malapelle U and Tiseo M: The storm of
NGS in NSCLC diagnostic-therapeutic pathway: How to sun the real
clinical practice. Crit Rev Oncol Hematol. 169:1035612022.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hayashi H, Nadal E, Gray JE, Ardizzoni A,
Caria N, Puri T and Grohe C: Overall treatment strategy for
patients with metastatic NSCLC with activating EGFR mutations. Clin
Lung Cancer. 23:e69–e82. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Y, Sheng J, Kang S, Fang W, Yan Y,
Hu Z, Hong S, Wu X, Qin T, Liang W and Zhang L: Patients with exon
19 deletion were associated with longer progression-free survival
compared to those with L858R mutation after first-line EGFR-TKIs
for advanced non-small cell lung cancer: A meta-analysis. PLoS One.
9:e1071612014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hong W, Wu Q, Zhang J and Zhou Y:
Prognostic value of EGFR 19-del and 21-L858R mutations in patients
with non-small cell lung cancer. Oncol Lett. 18:3887–3895.
2019.PubMed/NCBI
|
|
39
|
Naidoo J, Sima CS, Rodriguez K, Busby N,
Nafa K, Ladanyi M, Riely GJ, Kris MG, Arcila ME and Yu HA:
Epidermal growth factor receptor exon 20 insertions in advanced
lung adenocarcinomas: Clinical outcomes and response to erlotinib.
Cancer. 121:3212–3220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tan J, Hu C, Deng P, Wan R, Cao L, Li M,
Yang H, Gu Q, An J and Jiang J: The predictive values of advanced
non-small cell lung cancer patients harboring uncommon EGFR
mutations-the mutation patterns, use of different generations of
EGFR-TKIs, and concurrent genetic alterations. Front Oncol.
11:6465772021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Canale M, Petracci E, Delmonte A, Bronte
G, Chiadini E, Ludovini V, Dubini A, Papi M, Baglivo S, De Luigi N,
et al: Concomitant TP53 mutation confers worse prognosis in
EGFR-mutated non-small cell lung cancer patients treated with TKIs.
J Clin Med. 9:10472020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang X, Liu Y, Meng Z, Wu Y, Wang S, Jin
G, Qin Y, Wang F, Wang J, Zhou H, et al: Plasma EGFR mutation
abundance affects clinical response to first-line EGFR-TKIs in
patients with advanced non-small cell lung cancer. Ann Transl Med.
9:6352021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Clinical Lung Cancer Genome Project
(CLCGP); Network Genomic, Medicine (NGM), . A genomics-based
classification of human lung tumors. Sci Transl Med.
5:209ra1532013.PubMed/NCBI
|
|
44
|
Molina-Vila MA, Bertran-Alamillo J, Gascó
A, Mayo-de-las-Casas C, Sánchez-Ronco M, Pujantell-Pastor L,
Bonanno L, Favaretto AG, Cardona AF, Vergnenègre A, et al:
Nondisruptive p53 mutations are associated with shorter survival in
patients with advanced non-small cell lung cancer. Clin Cancer Res.
20:4647–4659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bria E, Pilotto S, Amato E, Fassan M,
Novello S, Peretti U, Vavalà T, Kinspergher S, Righi L, Santo A, et
al: Molecular heterogeneity assessment by next-generation
sequencing and response to gefitinib of EGFR mutant advanced lung
adenocarcinoma. Oncotarget. 6:12783–12795. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Canale M, Petracci E, Delmonte A, Chiadini
E, Dazzi C, Papi M, Capelli L, Casanova C, De Luigi N, Mariotti M,
et al: Impact of TP53 mutations on outcome in EGFR-mutated patients
treated with first-line tyrosine kinase inhibitors. Clin Cancer
Res. 23:2195–2202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
VanderLaan PA, Rangachari D, Mockus SM,
Spotlow V, Reddi HV, Malcolm J, Huberman MS, Joseph LJ, Kobayashi
SS and Costa DB: Mutations in TP53, PIK3CA, PTEN and other genes in
EGFR mutated lung cancers: Correlation with clinical outcomes. Lung
Cancer. 106:17–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Aisner DL, Sholl LM, Berry LD, Rossi MR,
Chen H, Fujimoto J, Moreira AL, Ramalingam SS, Villaruz LC,
Otterson GA, et al: The impact of smoking and TP53 mutations in
lung adenocarcinoma patients with targetable mutations-the lung
cancer mutation consortium (LCMC2). Clin Cancer Res. 24:1038–1047.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Aggarwal C, Davis CW, Mick R, Thompson JC,
Ahmed S, Jeffries S, Bagley S, Gabriel P, Evans TL, Bauml JM, et
al: Influence of TP53 mutation on survival in patients with
advanced EGFR-mutant non-small-cell lung cancer. JCO Precis Oncol.
2018.PO.18.00107, 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yu HA, Suzawa K, Jordan E, Zehir A, Ni A,
Kim R, Kris MG, Hellmann MD, Li BT, Somwar R, et al: Concurrent
alterations in EGFR-mutant lung cancers associated with resistance
to EGFR kinase inhibitors and characterization of MTOR as a
mediator of resistance. Clin Cancer Res. 24:3108–3118. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kim Y, Lee B, Shim JH, Lee SH, Park WY,
Choi YL, Sun JM, Ahn JS, Ahn MJ and Park K: Concurrent genetic
alterations predict the progression to target therapy in
EGFR-mutated advanced NSCLC. J Thorac Oncol. 14:193–202. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chang SC, Lai YC, Chang CY, Huang LK, Chen
SJ, Tan KT, Yu PN and Lai JI: Concomitant genetic alterations are
associated with worse clinical outcome in EGFR mutant NSCLC
patients treated with tyrosine kinase inhibitors. Transl Oncol.
12:1425–1431. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hou H, Qin K, Liang Y, Zhang C, Liu D,
Jiang H, Liu K, Zhu J, Lv H, Li T and Zhang X: Concurrent TP53
mutations predict poor outcomes of EGFR-TKI treatments in Chinese
patients with advanced NSCLC. Cancer Manag Res. 11:5665–5675. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rachiglio AM, Fenizia F, Piccirillo MC,
Galetta D, Crinò L, Vincenzi B, Barletta E, Pinto C, Ferraù F,
Lambiase M, et al: The presence of concomitant mutations affects
the activity of EGFR tyrosine kinase inhibitors in EGFR-mutant
non-small cell lung cancer (NSCLC) patients. Cancers (Basel).
11:3412019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cheng Y, Ma L, Liu Y, Zhu J, Xin Y, Liu X,
Wang Y, Zhang T, Yang C, Wang S, et al: Comprehensive
characterization and clinical impact of concomitant genomic
alterations in EGFR-mutant NSCLCs treated with EGFR kinase
inhibitors. Lung Cancer. 145:63–70. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li XM, Li WF, Lin JT, Yan HH, Tu HY, Chen
HJ, Wang BC, Wang Z, Zhou Q, Zhang XC, et al: Predictive and
prognostic potential of TP53 in patients with advanced
non-small-cell lung cancer treated with EGFR-TKI: Analysis of a
phase III randomized clinical trial (CTONG 0901). Clin Lung Cancer.
22:100–109.e3. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Steendam CMJ, Veerman GDM, Pruis MA,
Atmodimedjo P, Paats MS, van der Leest C, von der Thüsen JH, Yick
DCY, Oomen-de Hoop E, Koolen SLW, et al: Plasma predictive features
in treating EGFR-mutated non-small cell lung cancer. Cancers
(Basel). 12:31792020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang GJ, Li J, Xu HY, Sun Y, Liu L, Li HS,
Yang L, Zhang Y, Li GH and Wang Y: Osimertinib for Chinese advanced
non-small cell lung cancer patients harboring diverse EGFR exon 20
insertion mutations. Lung Cancer. 152:39–48. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang P, Li Y, Lv D, Yang L, Ding L, Zhou
J, Hong W, Chen Y, Zhang D, He S, et al: Mefatinib as first-line
treatment of patients with advanced EGFR-mutant non-small-cell lung
cancer: A phase Ib/II efficacy and biomarker study. Signal
Transduct Target Ther. 6:3742021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Roeper J, Christopoulos P, Falk M, Heukamp
LC, Tiemann M, Stenzinger A, Thomas M and Griesinger F: TP53
co-mutations as an independent prognostic factor in 2nd and further
line therapy-EGFR mutated non-small cell lung cancer IV patients
treated with osimertinib. Transl Lung Cancer Res. 11:4–13. 2022.
View Article : Google Scholar : PubMed/NCBI
|