Introduction
The incidence of thyroid cancer is reported to have
increased at an annual rate of 3.6% between 1974 and 2013 in the
USA (1,2). Patients with differentiated thyroid
cancer (DTC), including papillary thyroid cancer (PTC) and
follicular thyroid cancer, usually have a favorable prognosis due
to the good efficacy of conventional treatments, particularly
postoperative radiotherapy (3),
which plays an important role in eradicating any residual tumor and
targeting distant metastases (4).
When patients with DTC begin to resist the uptake of radioiodine
during radiotherapy, radioiodine-refractory DTC (RR-DTC) develops,
which occurs in 15–20% cases of thyroid cancer (5). Patients with RR-DTC usually have a
poor prognosis due to their impaired radioiodine uptake; it has
been reported that patients with RR-DTC have a median survival time
of only 3–6 years once diagnosed (6). The pathology of radioiodine
resistance may be associated with tumor cell dedifferentiation,
which greatly increases malignancy (7,8). It
would be of great benefit to explore the exact underlying molecular
mechanism of tumor cell dedifferentiation and iodine resistance in
order to improve the survival rate of patients with RR-DTC.
Generally, patients with DTC have a favorable
prognosis. Traditional radiotherapy can effectively remove tumor
tissue remaining following thyroidectomy and any distant metastases
via the action of beta-rays emitted during radioiodine decay
(9). However, the typical
pathophysiological feature of RR-DTC is the reduced or deficient
capacity for iodine uptake, which results in the lack of efficacy
of iodine-dependent regimens in its treatment. However, it has been
reported that when artificially engineered to bind to tumor
targets, radiopharmaceuticals are able to eliminate tumor cells in
a similar manner to radioiodine (10,11).
Considering the advantages of radiotherapy in the clinical
treatment of DTC, the present study aimed to identify a potential
molecular target to which radiopharmaceuticals could be directed
for the radiotherapy of RR-DTC.
The process of iodine accumulation depends on the
sodium iodine symporter (NIS), a protein complex in the plasma
membrane of thyroid epithelial cells (12). Studies have shown that in RR-DTC,
low expression levels of NIS pose a great challenge to the
eradication of residual tumor and distant metastases, which can
lead to the spread of the tumor (13). Therefore, the expression level of
NIS is responsible for iodine uptake capacity to a certain extent
and its relationship with the pathogenesis of RR-DTC requires
further exploration.
Owing to the high-throughput and stability of
proteomic technology, it is widely used in cancer research to
identify differentially expressed proteins (DEPs) for diagnostic
and therapeutic purposes. Gene Ontology (GO) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) databases can be used to cluster these
identified DEPs in order to screen for potential biomarkers and
dynamically monitor the progression of diseases. On this basis,
previous studies have revealed that activation of the MAPK and PI3K
signaling pathways are closely associated with tumorigenesis since
they change the cellular microenvironment, promote tumor
neovascularization and stimulate tumor cell proliferation (14,15).
In addition, the MAPK/ERK signaling pathway has been found to have
regulatory effect on cell differentiation in studies of breast
cancer and rhabdomyosarcoma, which could potentially be the
mechanism underlying the dedifferentiation of tumor cells in RR-DTC
(16,17). However, investigations of the
specific DEPs and signaling pathways associated with RR-DTC are
lacking, and were therefore performed in the present study.
To the best of our knowledge, the present study is
the first to perform a proteomic analysis of the tumor tissue of
patients with RR-DTC. Tumor tissue from patients with PTC, the most
common type of DTC, was employed as a control. Isobaric tags for
relative and absolute quantification (iTRAQ) technology was used to
identify the DEPs. Further functional analysis of DEPs was
performed with the use of GO and KEGG databases. Proteins of
interest were verified by immunohistochemistry (IHC) and western
blotting.
Materials and methods
Sample collection
Inclusion criteria for sample collection of RR-DTC
were as follows: i) Patients who had undergone a total
thyroidectomy, with lesions confirmed as PTC pathologically; ii)
patients who had undergone >3 cycles of radioiodine therapy,
with metastasis confirmed as not avid for radioiodine on the last
whole-body scan (WBS); and iii) the concentration of thyroglobulin
increased every 3 months during the long-term follow-up. In
accordance with the inclusion criteria, 3 metastatic lymph nodes
from 3 patients with RR-DTC and 3 metastatic lymph nodes from 3
patients with PTC were collected. Samples were collected from the
First Affiliated Hospital of Chongqing Medical University
(Chongqing, China) between August 2019 and September 2020. The
exclusion criteria were as follows: i) Patients whose lesion was
avid for radioiodine at any rate despite receiving >600 mCi
radioiodine in total; ii) patients who had ever taken targeted
drugs such as sorafinib or lentatinib; iii) patients with multiple
distant metastases or who were in a poor condition; and iv)
patients whose lesions were not avid for radioiodine but were in a
stable condition. All lymph nodes were rinsed with PBS three times,
after which each lymph node was incised to provide two parts, one
of which was used to prepare formalin-fixed paraffin-embedded
tissue (FFPET) for IHC while the other was stored at −80°C for
proteomic analysis. In addition, to enlarge the sample size, a
further 6 metastatic lymph nodes from patients with PTC and RR-DTC
(3 of each) were included in addition to those from the
aforementioned patients. These samples were collected from the
Department of Pathology of the First Affiliated Hospital of
Chongqing Medical University between April 2019 and January 2020.
The study was approved by Chongqing Medical University Ethics
Committee and all patients involved provided written informed
consent to participate.
Proteomics technology and
bioinformatics analysis
All samples were subjected to proteomic analysis
using iTRAQ technology coupled with mass spectrometry (MS)
according to the manufacturer's protocol. For MS, polarity was set
in a positive ion mode and data-dependent manner, with full MS
scans from 350 to 2,000 m/z, full scan resolution set at 70,000,
ion source voltage set at 1.8 eV and MS/MS scan resolution set at
17,500. The raw MS data were loaded into Proteome Discoverer (PD)
1.4 (Thermo Fisher Scientific, Inc.), which selected mass spectra
using preset criteria as follows: Mass range of parent ion, 350 to
6,000 Da; S/N, 1.5. The selected mass spectra were searched using
Mascot (version 2.3.0; Matrix Science) to identify proteins. After
that, the PD software was used to perform a quantitative analysis
based on the search results and selected mass spectra. In the
proteomic analysis, a fold change >1.2 was regarded to indicate
a DEP between two groups according to previous studies (18–20).
According to the outcome of the quantitative analysis, all
identified DEPs were subjected to functional annotation using the
GO (http://geneontology.org/) and KEGG
databases (https://www.kegg.jp).
IHC
FFPET sections for the IHC assay were provided by
the Department of Pathology of the First Affiliated Hospital of
Chongqing Medical University. The thickness of each section was 5
µm. For antigen retrieval, all sections were heated by water bath
at 95°C with 0.01 mol/l sodium citrate buffer for 20 min, and then
cooled to room temperature and washed with PBS 3 times. The
expression of NIS and CHI3L1 was examined following quenching of
endogenous peroxidase activity and blocking the sections using an
immunohistochemical kit at room temperature for 15 min (cat. no.
SP9000; Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.).
Antibodies targeting NIS (bs-0048R; Bioss) and CHI3L1 (ab77528;
Abcam) were diluted to optimal concentrations (1:300 and 1:1,000,
respectively). The primary antibodies were incubated at 4°C
overnight. All sections were incubated for 30 min at 37°C after
addition of the secondary antibody, which was diluted by PBS at
1:100 (part of kit; cat. no. SP9000; Beijing Zhongshan Golden
Bridge Biotechnology Co., Ltd.). Each section was observed under a
microscope after being processed with DAB solution, counterstaining
with hematoxylin at room temperature for 30 sec, dehydration, and
transparentization using ethanol and xylene. ImageJ software
(version 1.53; National Institutes of Health) was used to evaluate
the immunostaining intensity.
Cell culture and lentiviral
transfection
PTC-K1 cells were purchased from Otwo Biotech. The
PTC-K1 cells were authenticated by STR profiling; they are
considered as a subpopulation of the Glag-66 cell line, and both
cell lines are papillary thyroid carcinoma cells. The cells were
cultured in complete DMEM, comprising 89% DMEM supplemented with
10% FBS and 1% penicillin-streptomycin by volume, and maintained in
an incubator with 5% carbon dioxide at 37.0°C and 95% humidity. A
CHI3L1 overexpression lentiviral vector (PSE3748; 2nd generation)
and control vector (PMT222; 2nd generation), obtained from SANGON
Biotech, were used to infect PTC-K1 cells following the
manufacturer's protocol. pLvx-AcGEP (3 µg) was used for
transfection and the ratio of pLvx-AcGFP, psPAX and pMD2.G was set
at 4:3:1. Cells were transfected at 37°C for 48 h. The multiplicity
of infection was set at 30. Cells were cultured in 2 µg/ml
puromycin for 2 weeks until the CHI3L1 overexpression and control
transfection were stable.
Western blotting
Total protein was extracted from the cells using
RIPA buffer (P0013B; Beyotime Institute of Biotechnology)
containing protease inhibitor (100:1 by volume). The concentration
of total protein was then measured using a BCA protein assay kit. A
sample containing 40 µg protein per lane was loaded onto a 10%
SDS-PAGE gel (P0012A; Beyotime Institute of Biotechnology).
Proteins were transferred to a PVDF membrane and blocked using 5%
BSA blocking buffer (cat. no. SW3015; Beijing Solarbio Science
& Technology Co., Ltd.) for 2 h at room temperature. Antibodies
targeting CHI3L1, phosphorylated MEK1 (p-MEK1; cat. no. ab96379;
Abcam), phosphorylated ERK1/2 (p-ERK1/2; cat. no. ab201015; Abcam)
and GAPDH (AB-P-R001; Hangzhou Goodhere Biotechnology Co., Ltd.)
were used at dilutions of 1:1,000, 1:500, 1:1,000, and 1:500,
respectively. The primary antibodies were incubated at 4°C
overnight, and the secondary antibody diluted at 1:2,000 (cat. no.
A0279; Beyotime Institute of Biotechnology) was incubated at 37°C
for 1 h. Visualization reagent (cat. no. P0018S; BeyoECL plus) was
obtained from Beyotime Institute of Biotechnology. ImageJ software
(version 1.53) was used to measure the band density for
quantitative analysis.
Statistical analysis
Statistical analysis was performed using Student's
unpaired t-test or one-way analysis of variance followed by Tukey's
multiple comparisons test for post hoc testing. SPSS 22.0 (IBM
Corp.) and GraphPad Prism 7.00 (GraphPad Software, Inc.) software
were used to perform the analyses. All data are presented as the
mean ± SD. P<0.05 was considered to indicate a statistically
significant difference.
Results
Clinical characteristics of patients
included for proteomic analysis
All patients with RR-DTC involved were eligible for
inclusion based on a radioiodine WBS and pathological sections.
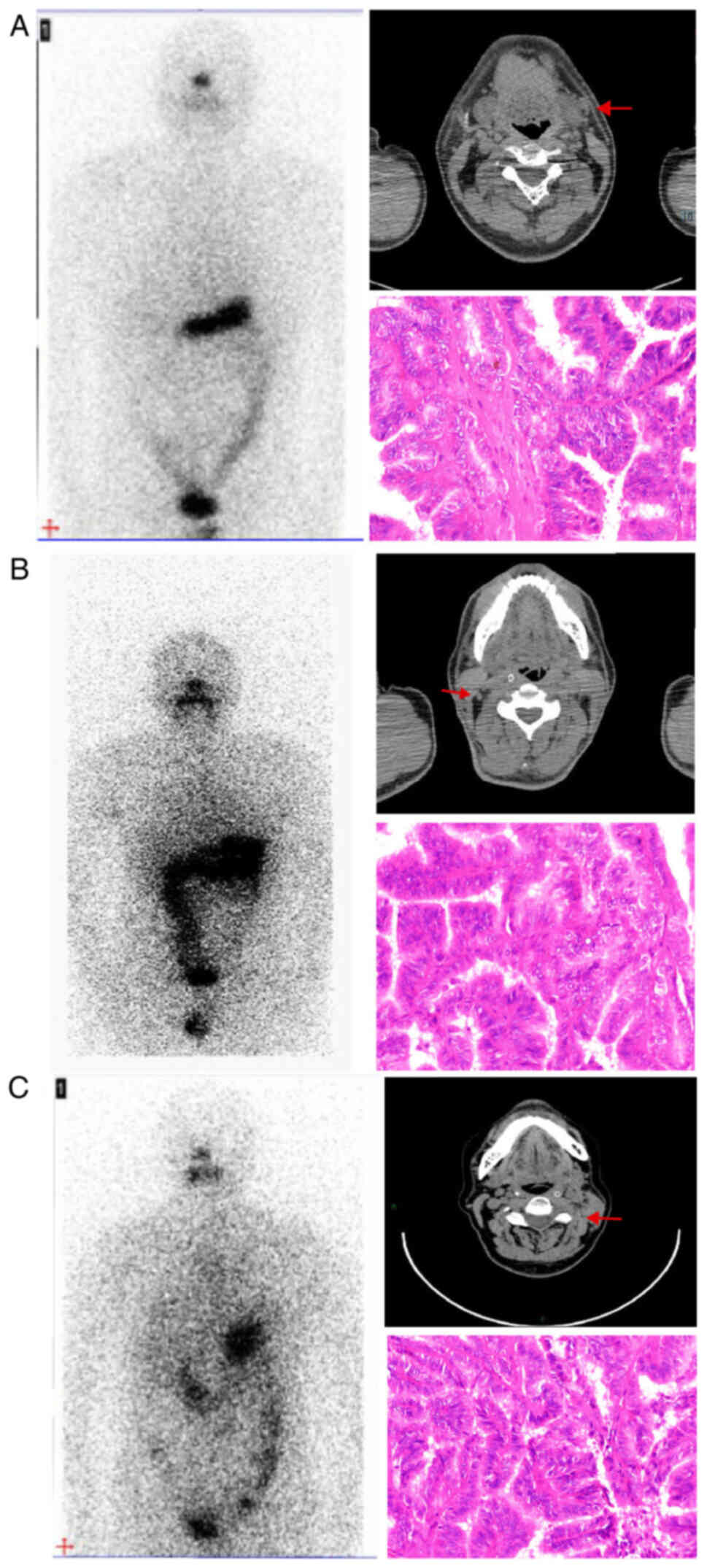
Computed tomography imaging demonstrated the presence of metastatic
lymph nodes in cervical locations, while no abnormal radioiodine
uptake was detected in the corresponding locations according to the
WBS. Following cervical lymph node dissection, the metastatic lymph
nodes were identified to be PTC by inspection of the pathological
sections (Fig. 1). The clinical
characteristics of the patients included in the proteomic analysis
are shown in Table I.
 | Table I.Clinical characteristics of the
patients for proteomic analysis. |
Table I.
Clinical characteristics of the
patients for proteomic analysis.
| Characteristics | RR-DTC | PTC |
|---|
| Sex | Male | Male | Female | Male | Male | Female |
| Age (years) | 31 | 49 | 52 | 38 | 29 | 35 |
| TNM stage |
T2N1bM0 |
T2N1bM0 |
T1N1bM0 |
T1N1bM0 |
T1N1bM0 |
T1N1bM0 |
| 131I
therapies (n) | 4 | 5 | 2 | 2 | 2 | 2 |
| Pathological
type | PTC | PTC | PTC | PTC | PTC | PTC |
| BRAF mutation | - | + | + | - | + | + |
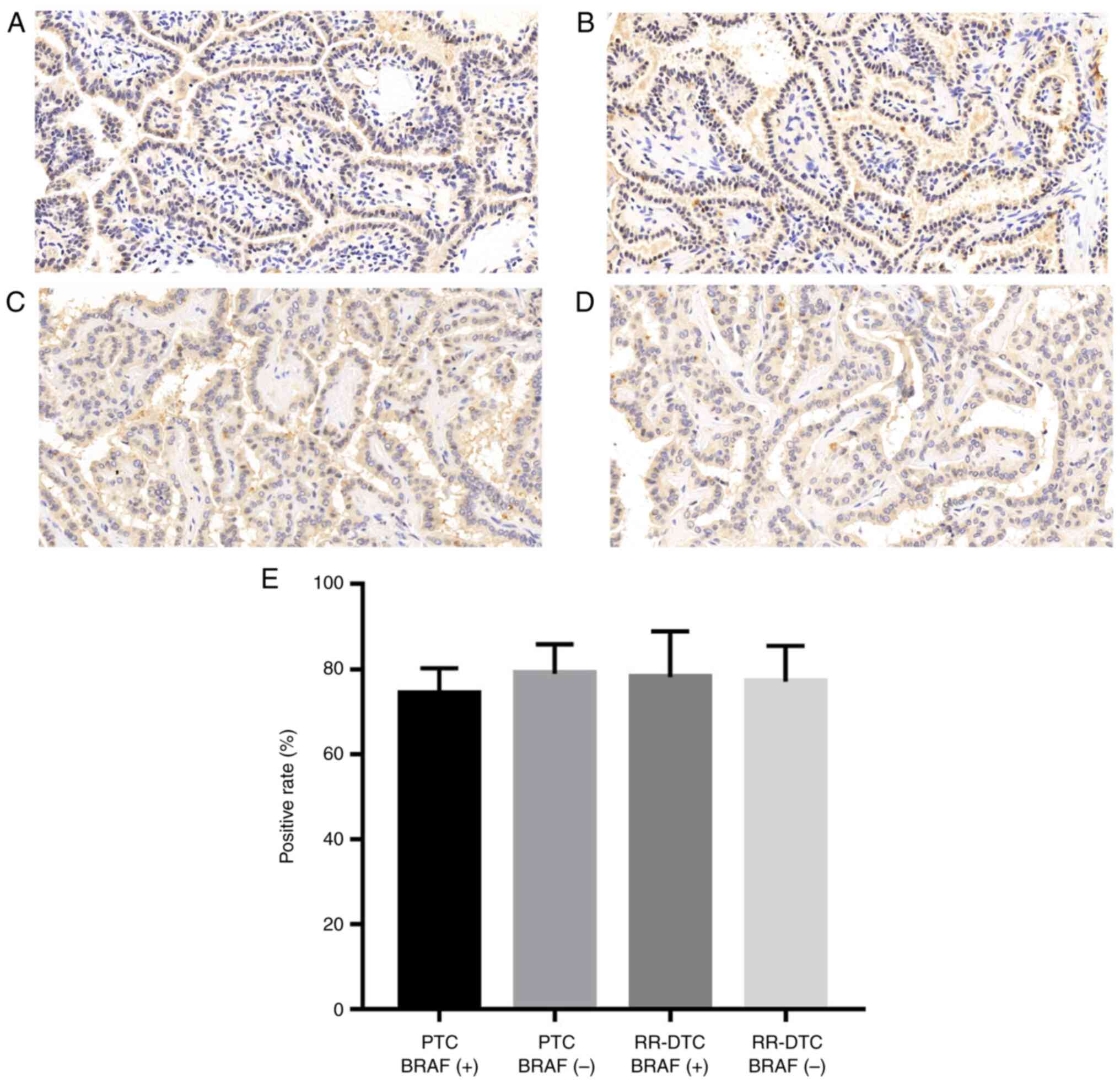
BRAF mutation is not closely
associated with NIS expression
To investigate whether BRAF mutation inhibits NIS
expression in RR-DTC, the immunohistochemical analysis of NIS was
performed in 24 FFPET sections collected from different patients,
including PTC BRAF (+) tissues, PTC BRAF (−) tissues, RR-DTC BRAF
(+) tissues, and RR-DTC BRAF (−) tissues (6 sections/group). As
shown in Fig. 2, no significant
difference was detected among the four groups with regard to the
positive rate of NIS expression.
DEPs between RR-DTC and PTC
The proteomic analysis detected 665 DEPs between the
RR-DTC and PTC groups. A total of 327 proteins were upregulated in
RR-DTC compared with PTC, including CHI3L1, extracellular matrix
protein 1 (ECM1), 14-3-3 ε and 14-3-3 σ, while 338 proteins were
downregulated, including myosin-9, cell division control protein 42
(CDC42) and Ras-related C3 botulinum toxin substrate (RAC2).
Notably, cancer stem cell markers including aldehyde dehydrogenase
(ALDH) and cluster of differentiation 44 (CD44) were found to be
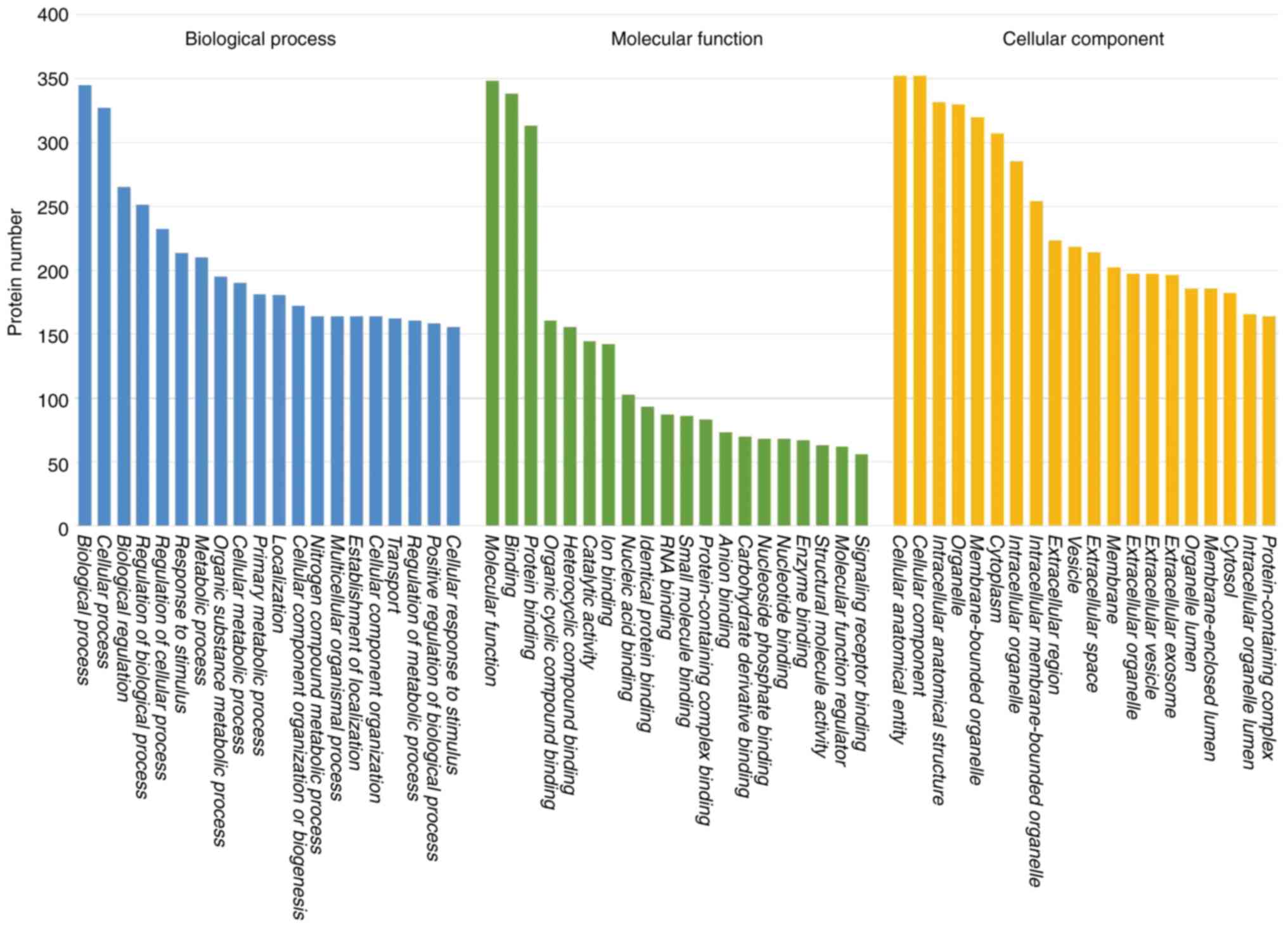
upregulated in RR-DTC compared with DTC. GO functional annotation
demonstrated that DEPs were enriched in ‘biological process’,
‘cellular process’, ‘biological regulation’, ‘regulation of
biological process’, ‘regulation of cellular process’ and ‘response
to stimulus’ (Fig. 3). KEGG
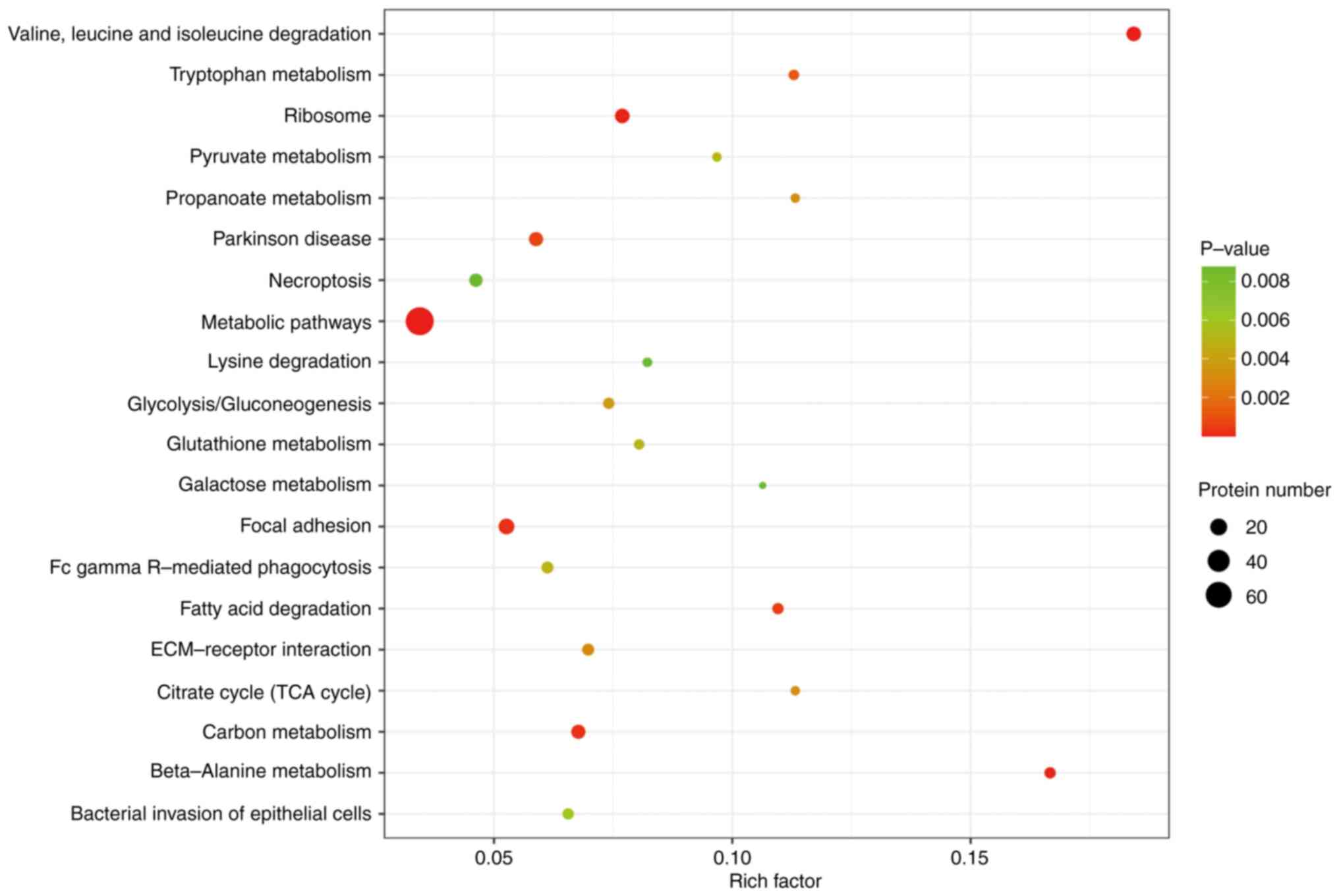
analysis revealed that the DEPs were enriched in 59 signaling
pathways, which included ‘valine, leucine and isoleucine
degradation’, ‘beta-alanine metabolism’, ‘focal adhesion’, ‘carbon
metabolism’ and ‘fatty acid degradation’ (Fig. 4).
CHI3L1 is significantly upregulated in
RR-DTC compared with PTC and activates the MEK/ERK1/2 signaling
pathway
CHI3L1 expression was approximately twice as high in
RR-DTC than in DTC according to the results of the proteomic
analysis. For the purpose of enlarging the sample size and
verifying the reliability of the proteomic results, IHC for CHI3L1
was performed on 12 FFPET sections collected from 6 patients with
RR-DTC and 6 patients with PTC. The clinical characteristics of the
patients included for IHC are shown in Table II. Consistent with the results of
the proteomic analysis, CHI3L1 expression was significantly
stronger in RR-DTC than in PTC. In addition, CHI3L1 was observed to
be located in the cytoplasm and membrane by IHC (Fig. 5).
 | Table II.Clinical characteristics of patients
for IHC of CHI3L1. |
Table II.
Clinical characteristics of patients
for IHC of CHI3L1.
|
Characteristics | RR-DTC | PTC |
|---|
| Sex | Male | Male | Male | Male | Female | Female |
| Age (years) | 45 | 56 | 34 | 27 | 38 | 42 |
| TNM stage |
T1N1bM0 |
T2N1bM0 |
T1N1bM0 |
T1N1aM0 |
T1N1bM0 |
T1N1bM0 |
| 131I
therapies (n) | 4 | 3 | 5 | 2 | 2 | 2 |
| Pathological
type | PTC | PTC | PTC | PTC | PTC | PTC |
| BRAF mutation | - | + | + | + | - | - |
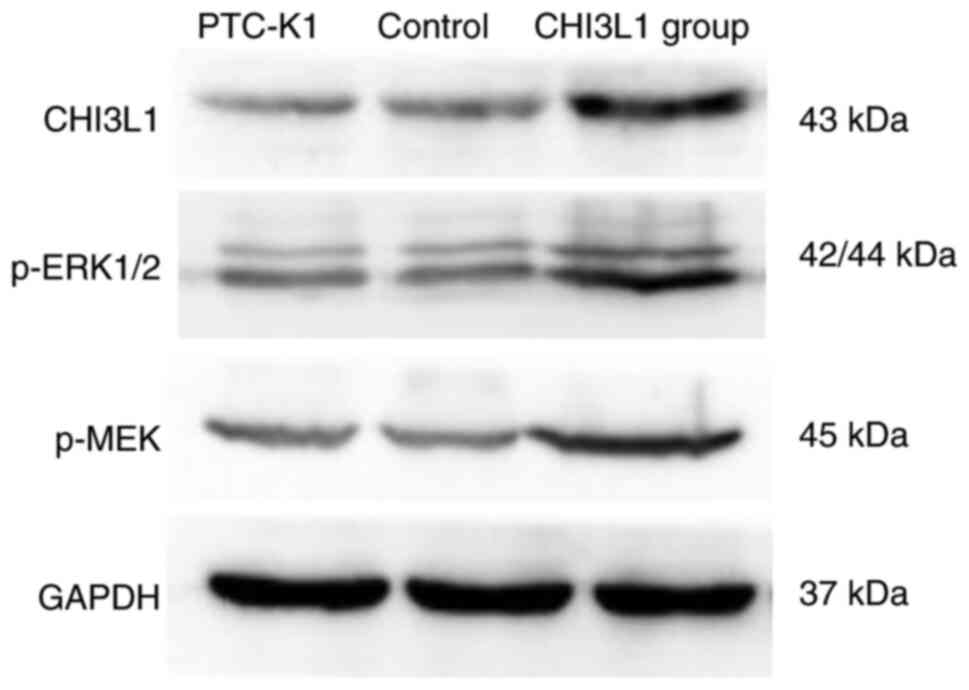
Finally, as shown in Fig. 6, PTC-K1 cells were successfully
transfected with CHI3L1 overexpression vector. The levels of p-MEK1
and p-ERK1/2 were higher in the cells stably transfected with
CHI3L1 overexpression vector than in those stably transfected with
the empty vector and the untransfected PTC-K1 control. No marked
difference in the levels of p-MEK1 and p-ERK1/2 were detected
between the empty vector and untransfected control.
Discussion
In the present study, proteomic analysis was
performed in patients with RR-DTC to investigate the DEPs
associated with the pathogenesis of RR-DTC and screen them for a
potential therapeutic target. The results revealed 327 upregulated
and 338 downregulated DEPs that were mainly enriched in 59
signaling pathways in RR-DTC. Among these DEPs, CHI3L1 was verified
to be significantly upregulated and its transmembrane structure may
indicate that this protein is a potential therapeutic target for
radiotherapy in patients with RR-DTC.
Notably, the tumor tissue collected for the RR-DTC
group in this study was all from patients with PTC. Therefore, PTC
tissue was selected as the control group for the proteomic analysis
of RR-DTC, while most tumor studies select normal tissues adjacent
to tumors as the control (21,22).
The DEPs obtained in the present study may provide an improved
reflection of the discrepancy between tumor cells of the same
pathological type and with different degrees of differentiation.
Markers for cell dedifferentiation, including ALDH and CD44 were
found to be overexpressed in RR-DTC, which is consistent with
previous literature (23–25).
NIS is considered to be strongly associated with
radioiodine uptake in thyroid cancer (26). However, no significant difference
was detected in the NIS expression level between the RR-DTC and PTC
groups in the present study. Notably, numerous DEPs were found to
be enriched in biological processes associated with NIS, including
‘ion transport’ (GO:0006811), ‘sodium ion transport’ (GO:0006814)
and ‘transmembrane transport’ (GO:0007571), suggesting that even
though the total amount of NIS was not reduced, the NIS may be in a
non-functional state. The low expression of some trans-membrane
transport-associated proteins detected in RR-DTC, including 14-3-3
η, mitochondrial ATP synthase subunit-E and receptor tyrosine
protein phosphatase, may also be associated with NIS dysfunction.
However, the specific mechanism requires further investigation.
Numerous DEPs were identified in the proteomic
analysis, including ECM1, glutathione S-transferase, RAC2, CDC42,
myosin regulatory light polypeptide 9 and actinin. These proteins
are able to regulate tumor angiogenesis, invasion, metastasis and
cell dedifferentiation (27,28),
whereby they may potentially modulate the progression of RR-DTC.
For example, RAC2 and CDC42 were found to be significantly
downregulated in RR-DTC. We hypothesize that these proteins are
positively associated with cell differentiation via the promotion
of the expression of c-Jun N-terminal kinase 2. This may be one of
the molecular mechanisms underlying the poor differentiation of
RR-DTC.
Previous studies have demonstrated that CHI3L1 is
highly expressed in PTC compared with adjacent normal tissues
(29,30), and it was further identified that
CHI3L1 is significantly upregulated in RR-DTC compared with PTC in
the present study. These findings suggest that CHI3L1 may serve an
important role in thyroid cancer and could be associated with the
dedifferentiation of tumor cells in RR-DTC. Furthermore, p-MEK1 and
p-ERK1/2 levels were markedly higher in stably CHI3L1
overexpressing PTC-K1 cells compared with empty vector transfected
controls, indicating activation of the MEK/ERK1/2 signaling pathway
in RR-DTC. KEGG analysis suggests that activation of the MAPK/ERK
signaling pathway leads to poor cell differentiation in tumor
tissues, which is basically consistent with the results of the
present study. Therefore, we hypothesize that CHI3L1 leads to poor
cell differentiation via activation of the MEK/ERK1/2 signaling
pathway in RR-DTC, and the upregulation of CHI3L1 in RR-DTC might
explain the poor prognosis of RR-DTC compared with PTC.
Ideally, all the identified DEPs should be screened
to identify whether they meet the criteria of a radiotherapeutic
target. An eligible target would be upregulated in RR-DTC to bind
with its specific probe. Also, probe-receptor combination is
favored when the target protein is located on the membrane surface
and has a transmembrane structure. Using the protein structure
homology-modeling server SWISS-MODEL (https://swissmodel.expasy.org/), the structure of
CHI3L1 (accession no. P36222) makes it a suitable radiotherapy
target. With seven transmembrane domains, CHI3L1 has the same
properties as the NIS. The transmembrane domains ensure that CHI3L1
adheres well to the membrane and its membrane location readily
allows the binding of a specific polypeptide or antibody. The IHC
results in the present study revealed that CHI3L1 is located in the
cytoplasm and membrane, which is consistent with the predicted
structure. Furthermore, the upregulation of CHI3L1 in RR-DTC may
allow engagement with a specific polypeptide or antibody around the
lesion, thereby reducing radiation damage to normal tissue. In a
previous study on osteosarcoma, a specific ligand for CHI3L1 was
successfully designed, prepared and verified to inhibit cell
migration and invasion (31).
Therefore, we hypothesize that a novel agent combining a
radionuclide-labeled specific polypeptide or antibody targeting
CHI3L1 has the potential to achieve targeted radiotherapy for
RR-DTC, which may improve the poor prognosis of patients and
alleviate their anxiety.
The present study has certain limitations. Most
cases of RR-DTC lose the opportunity to undergo surgery because of
distant metastases, which creates a major barrier to sample
collection. Therefore, the identified DEPs were not extensive
enough to fully elucidate the pathogenesis of RR-DTC due to the
small sample size and individual heterogeneity. In addition, the
number of screened DEPs was so large that it was not possible to
process and analyze them all, and so other potential therapeutic
targets for RR-DTC may have been missed. Also, it should be noted
that only the PTC-K1 cell line was included in the in vitro
assays; the inclusion of more PTC cell lines might improve the
reliability of the study. Furthermore, the total amounts of MEK and
ERK1/2 were not detected by western blotting, so the ratio of
phosphorylated protein/total protein could not be determined and
the activation level of this signaling pathway could not be
evaluated properly.
In conclusion, the present study is the first to
obtain the DEPs between PTC and RR-DTC. These DEPs may provide an
improved perspective for analysis of the pathogenesis of RR-DTC. In
addition, it revealed that significantly upregulated CHI3L1 may be
responsible for the poor cell differentiation in RR-DTC and could
be an appropriate target for radiotherapy in the future.
Acknowledgements
Not applicable.
Funding
The study was financially supported by the General Program of
Natural Science Foundation of Chongqing
(cstc2019jcyj-msxmX0327).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The proteomics datasets generated and/or analyzed during
the present study are available in the ProteomeXchange (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD037968).
Authors' contributions
YL performed the proteomic analysis and was a major
contributor to writing the manuscript. FH analyzed the proteomic
data and chose the target for further analysis. JD and XH collected
samples for proteomic analysis and performed the western blotting.
CZ and MW performed IHC and statistical analysis. DD conceived and
designed the study. All authors read and approved the final version
of the manuscript. DD and YL confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
The study was approved by Chongqing Medical
University Ethics Committee and all patients provided written
informed consent to participate.
Patient consent for publication
All patients involved in the study provided written
informed consent for the publication of any data and accompanying
images.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RR-DTC
|
radioiodine-refractory differentiated
thyroid cancer
|
|
CHI3L1
|
chitinase-3-like 1
|
|
NIS
|
sodium-iodine symporter
|
|
IHC
|
immunohistochemistry
|
|
DEPs
|
differentially expressed proteins
|
|
DTC
|
differentiated thyroid cancer
|
|
PTC
|
papillary thyroid cancer
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
MS
|
mass spectrometry
|
|
FFPET
|
formalin-fixed paraffin-embedded
tissue
|
|
ECM1
|
extracellular matrix protein 1
|
|
CDC42
|
cell division control protein 42
|
|
RAC2
|
Ras-related C3 botulinum toxin
substrate
|
|
ALDH
|
aldehyde dehydrogenase
|
|
CD44
|
cluster of differentiation 44
|
|
p-
|
phosphorylated
|
References
|
1
|
Lim H, Devesa SS, Sosa JA, Check D and
Kitahara CM: Trends in thyroid cancer incidence and mortality in
the United States, 1974–2013. JAMA. 317:1338–1348. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dal Maso L, Tavilla A, Pacini F, Serraino
D, van Dijk BAC, Chirlaque MD, Capocaccia R, Larrañaga N, Colonna
M, Agius D, et al: Survival of 86,690 patients with thyroid cancer:
A population-based study in 29 European countries from EUROCARE-5.
Eur J Cancer. 77:140–152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sparano C, Moog S, Hadoux J, Dupuy C, Al
Ghuzlan A, Breuskin I, Guerlain J, Hartl D, Baudin E and Lamartina
L: Strategies for radioiodine treatment: What's new. Cancers
(Basel). 14:38002022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jin Y, Van Nostrand D, Cheng L, Liu M and
Chen L: Radioiodine refractory differentiated thyroid cancer. Crit
Rev Oncol Hematol. 125:111–120. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mu ZZ, Zhang X and Lin YS: Identification
of radioactive iodine refractory differentiated thyroid cancer.
Chonnam Med J. 55:1272019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu Q, Zhang X, Li L, Zhang C, Huang J and
Huang W: Molecular basis and targeted therapies for radioiodine
refractory thyroid cancer. Asia Pac J Clin Oncol. 10:ajco.13836.
2022.
|
|
8
|
Jögi A, Vaapil M, Johansson M and Påhlman
S: Cancer cell differentiation heterogeneity and aggressive
behavior in solid tumors. Ups J Med Sci. 117:217–224. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Haugen BR, Alexander EK, Bible KC, Doherty
GM, Mandel SJ, Nikiforov YE, Pacini F, Randolph GW, Sawka AM,
Schlumberger M, et al: 2015 American thyroid association management
guidelines for adult patients with thyroid nodules and
differentiated thyroid cancer: The american thyroid association
guidelines task force on thyroid nodules and differentiated thyroid
cancer. Thyroid. 26:1–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pirooznia N, Abdi K, Beiki D, Emami F,
Arab SS, Sabzevari O and Soltani-Gooshkhaneh S: 177Lu-labeled
cyclic RGD peptide as an imaging and targeted radionuclide
therapeutic agent in non-small cell lung cancer: Biological
evaluation and preclinical study. Bioorg Chem. 102:1041002020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schuchardt C, Zhang J, Kulkarni HR, Chen
X, Müller D and Baum RP: Prostate-specific membrane antigen
radioligand therapy using 177 Lu-PSMA I&T and
177 Lu-PSMA-617 in patients with metastatic
castration-resistant prostate cancer: Comparison of safety,
biodistribution, and dosimetry. J Nucl Med. 63:1199–1207. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dai G, Levy O and Carrasco N: Cloning and
characterization of the thyroid iodide transporter. Nature.
379:458–460. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paladino S and Melillo RM: Editorial:
Novel mechanism of radioactive iodine refractivity in thyroid
cancer. J Natl Cancer Inst. 109:10.1093/jnci/djx106. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo G, Zhou J, Li G, Hu N, Xia X and Zhou
H: Retracted: Ferruginol diterpenoid selectively inhibits human
thyroid cancer growth by inducing mitochondrial dependent
apoptosis, endogenous reactive oxygen species (ROS) production,
mitochondrial membrane potential loss and suppression of
mitogen-activated protein kinase (MAPK) and PI3K/AKT signaling
pathways. Med Sci Monit. 27:e9323412021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ban Z, He J, Tang Z, Zhang L and Xu Z:
LRG-1 enhances the migration of thyroid carcinoma cells through
promotion of the epithelial-mesenchymal transition by activating
MAPK/p38 signaling. Oncol Rep. 41:3270–3280. 2019.PubMed/NCBI
|
|
16
|
Li W, Qian C, Ma F, Liu M, Sun X, Liu X,
Liu C, Chen Z, Ma W, Liu J, et al: MAPK/ERK-CBP-RFPL-3 mediates
adipose-derived stem cell-induced tumor growth in breast cancer
cells by activating telomerase reverse transcriptase expression.
Stem Cells Int. 2022:85405352022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ahmed AA, Farooqi MS, Habeebu SS, Gonzalez
E, Flatt TG, Wilson AL and Barr FG: NanoString digital molecular
profiling of protein and microrna in rhabdomyosarcoma. Cancers
(Basel). 14:5222022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Zhang R, Liu F, Jiang P, Xu J, Cao
H, Du X, Ma L, Lin F, Cheng L, et al: TMT-based quantitative
proteomic analysis reveals proteomic changes involved in longevity.
Prot Clin Appl. 13:18000242019. View Article : Google Scholar
|
|
19
|
Kroksveen AC, Aasebø E, Vethe H, Van Pesch
V, Franciotta D, Teunissen CE, Ulvik RJ, Vedeler C, Myhr KM,
Barsnes H and Berven FS: Discovery and initial verification of
differentially abundant proteins between multiple sclerosis
patients and controls using iTRAQ and SID-SRM. J Proteomics.
78:312–325. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li XJ, Pan HT, Chen JJ, Fu YB, Fang M, He
GH, Zhang T, Ding HG, Yu B, Cheng Y, et al: Proteomics of
uterosacral ligament connective tissue from women with and without
pelvic organ prolapse. Prot Clin Appl. 13:18000862019. View Article : Google Scholar
|
|
21
|
Zhou Y, Lih TM, Pan J, Höti N, Dong M, Cao
L, Hu Y, Cho KC, Chen SY, Eguez RV, et al: Proteomic signatures of
16 major types of human cancer reveal universal and
cancer-type-specific proteins for the identification of potential
therapeutic targets. J Hematol Oncol. 13:1702020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang JH, Lin Y, Ouyang T, Tang W, Huang
Y, Ye W, Zhao JY, Wang ZN and Ma CC: Nuclear magnetic
resonance-based metabolomics and metabolic pathway networks from
patient-matched esophageal carcinoma, adjacent noncancerous tissues
and urine. World J Gastroenterol. 25:3218–3230. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fukui R, Saga R, Matsuya Y, Tomita K,
Kuwahara Y, Ohuchi K, Sato T, Okumura K, Date H and Fukumoto M:
Tumor radioresistance caused by radiation-induced changes of
stem-like cell content and sub-lethal damage repair capability. Sci
Rep. 12:10562022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Suwiwat S, Tungsinmunlong K and
Siriaungkul S: Expression of CD44v6 and RCAS1 in uterine cervical
carcinoma infected with human papillomavirus and its effect on cell
proliferation and differentiation. Asian Pac J Cancer Prev.
23:2431–2439. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cui Y, Liu Y, Mu L, Li Y and Wu G:
Transcriptional expressions of ALDH1A1/B1 as independent indicators
for the survival of thyroid cancer patients. Front Oncol.
12:8219582022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ravera S, Reyna-Neyra A, Ferrandino G,
Amzel LM and Carrasco N: The Sodium/Iodide Symporter (NIS):
Molecular physiology and preclinical and clinical applications.
Annu Rev Physiol. 79:261–289. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Z, Zhou Q, Li A, Huang W, Cai Z and
Chen W: Extracellular matrix protein 1 (ECM1) is associated with
carcinogenesis potential of human bladder cancer. Onco Targets
Ther. 12:1423–1432. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feng M, Dong N, Zhou X, Ma L and Xiang R:
Myosin light chain 9 promotes the proliferation, invasion,
migration and angiogenesis of colorectal cancer cells by binding to
Yes-associated protein 1 and regulating Hippo signaling.
Bioengineered. 13:96–106. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luo D, Chen H, Lu P, Li X, Long M, Peng X,
Huang M, Huang K, Lin S, Tan L, et al: CHI3L1 overexpression is
associated with metastasis and is an indicator of poor prognosis in
papillary thyroid carcinoma. Cancer Biomark. 18:273–284. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dimitrova I, Shinkov A, Dodova R, Ivanova
R, Kirilov G, Kyurkchiyan S, Kaneva R and Kovatcheva R: Increased
gene expression of TIMP1 and CHI3L1 in fine-needle aspiration
biopsy samples from papillary thyroid cancer as compared to benign
nodules. Diagn Cytopathol. 49:1045–1051. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moore RG, Blackman A, Miller MC, Robison
K, DiSilvestro PA, Eklund EK, Strongin R and Messerlian G: Multiple
biomarker algorithms to predict epithelial ovarian cancer in women
with a pelvic mass: Can additional makers improve performance?
Gynecol Oncol. 154:150–155. 2019. View Article : Google Scholar : PubMed/NCBI
|