Introduction
Maspin, a serine protease inhibitor that is encoded
by the human SERPINB5 gene, is a tumor suppressor that was
discovered in 1994 (1). As a tumor
inhibitor, it can impair the growth of tumor cells, increase
adhesion between tumor cells and inhibit tumor angiogenesis
(2). The SERPINB5 gene is mainly
distributed on human chromosome 18q21.3f9, with a cDNA sequence of
2,548 bp and a coding protein containing 375 amino acids. This
protein belongs to the serine protease inhibitor
superfamily-ovalbumin subfamily, which includes an amino-bound
terminal methionine and a carboxyl-bound terminal valine. The eight
cysteine residues present in it can form two or more disulfide
bonds to form a stable tertiary structure. It has a stable and
specific three-dimensional structure that can inhibit the activity
of serine protease (3). Maspin
inhibits tumor metastasis by reducing the ability of tumor cells to
move and invade, increasing intercellular adhesion (4).
Silencing of the maspin gene leads to a decrease in
Bax-mediated cell apoptosis and tumor suppression, resulting in a
weakened inhibitory effect on cancer cells (5). Maspin can act on fibroblast growth
factor and vascular endothelial growth factor, blocking cell
mitosis and pipeline formation, inhibiting the transfer of cultured
endothelial cells to the matrix and thus inhibiting the growth of
tumors in blood vessels (6). Yin
et al (7) found that maspin
delayed Ca2+ reduction-induced disengagement through a
new interaction with urokinase type plasmin activator/urokinase
type plasmin. Endsley et al (8) discovered that maspin, as a bridge
between the plasmin activator system and β1 integrin, promotes cell
adhesion, also in breast epithelial cells. Studies have found that
maspin can increase cell adhesion and reduce cell mobility. Maspin
can inhibit cell mobility by regulating the activities of G protein
Racl and Pakl (p21 active kinase), and regulate cell adhesion
through phosphoinositol 3 kinase and extracellular signal regulated
kinase pathways (9). It can also
inhibit the regulatory effect of urokinase-type plasminogen
activator around cells on protein hydrolysis and cell movement,
thereby blocking cell infiltration and movement (10). Maspin expression is directly
regulated by wild-type p53 and there is a p53 binding site upstream
of the maspin promoter. p53 can directly bind to this site to
regulate the expression of its mRNA. When wild-type p53 binds to
the p53 binding site of maspin, it activates the maspin promoter
and enhances transcription (11).
In order to examine the expression of maspin in
breast cancer, a meta-analysis was conducted in the present study,
and the odds ratio (OR) of risk factors affecting abnormal
expression of maspin, including TNM stage and lymph node
metastasis, was analyzed. Furthermore, a bioinformatics analysis
was used to estimate the relationship between maspin expression and
prognosis.
Materials and methods
Identification of eligible studies and
data extraction
A publication search was performed using PubMed
(https://www.ncbi.nlm.nih.gov/) and the
China National Knowledge Infrastructure (CNKI; http://www.cnki.net/) database updated on March 20,
2023. The following search terms were used: (‘maspin’) AND
(‘breast’) AND (‘cancer’ OR ‘carcinoma’). The inclusion criteria
were as follows: i) Patients with breast cancer; ii)
immunohistochemical staining was used to detect the expression of
maspin; and iii) none of the patients received chemotherapy or
radiotherapy before surgery. The exclusion criteria for articles
were as follows: i) Abstract, case report, meeting abstract or
review; ii) protein blotting and reverse transcription PCR (RT-PCR)
were used to detect the expression of maspin; and iii) duplicate
publications. Meta-analysis was performed following the Preferred
Reporting Items for Systematic reviews and Meta-Analyses checklist
(12). The results of Egger's test
indicated no significant publication bias in the present
meta-analysis.
Data extraction and quality score
assessment
The main information in the article was extracted by
two reviewers (SS and HYM). As presented in Table I, the main information included in
the article was as follows: Name of first author, year of
publication, country, antibody supplier, number of cases and
controls, expression change and quality assessment score. The
quality of the research articles included was independently
evaluated by two reviewers based on the Newcastle-Ottawa scale
(NOS) (13).
 | Table I.Main characteristics of eligible
studies. |
Table I.
Main characteristics of eligible
studies.
| First author | Year | Country | Antibody
supplier | Cases, n | Controls, n | Risk of cancer | Quality (NOS
standard) | (Refs.) |
|---|
| Liu | 2005 | China | Santa Cruz | 137 | - | NS | 8 | (14) |
|
|
|
| Biotechnology, |
|
|
|
|
|
|
|
|
| Inc. |
|
|
|
|
|
| Liu | 2004 | China | Santa Cruz | 104 | 10 | Down | 8 | (15) |
|
|
|
| Biotechnology, |
|
|
|
|
|
|
|
|
| Inc. |
|
|
|
|
|
| Hu | 2006 | China | MXB | 34 | - | NS | 8 | (16) |
|
|
|
|
Biotechnologies |
|
|
|
|
|
| Zhang | 2014 | China | Lab Vision | 60 | 20 | Down | 8 | (17) |
| Liu | 2013 | China | Dako | 96 | 25 | Down | 8 | (18) |
| Sun | 2014 | China | Nevomarkers | 98 | 96 | Down | 8 | (19) |
| Yang | 2009 | China | Mindray | 65 | 12 | Down | 7 | (20) |
| Ding | 2007 | China | Lab Vision | 82 | 15 | Down | 8 | (21) |
| Cao | 2006 | China | MXB | 40 | - | NS | 7 | (22) |
|
|
|
|
Biotechnologies |
|
|
|
|
|
| Zhu | 2007 | China | Novoprotein | 57 | - | NS | 8 | (23) |
| Chen | 2015 | China | Nevomarkers | 60 | 35 | Down | 8 | (24) |
| Pei | 2011 | China | Nevomarkers | 53 | 34 | Down | 8 | (25) |
| Fang | 2009 | China | MXB | 30 | 30 | Down | 8 | (26) |
|
|
|
|
Biotechnologies |
|
|
|
|
|
| Zhang | 2012 | China | Nevomarkers | 90 | 90 | Down | 8 | (27) |
| Zhang | 2022 | China | MXB | 80 | 30 | Down | 8 | (28) |
|
|
|
|
Biotechnologies |
|
|
|
|
|
| Liu | 2008 | China | Nevomarkers | 60 | - | NS | 8 | (29) |
| Wang | 2005 | China | BD Pharmingen | 21 | - | NS | 8 | (30) |
| Wakahara | 2017 | Japan | Leica | 164 | - | NS | 7 | (31) |
|
|
|
| Biosystems |
|
|
|
|
|
| Tuncel | 2020 | Turkey | Abcam | 200 | - | NS | 7 | (32) |
| Feng | 2008 | China | Lab Vision | 80 | - | NS | 7 | (33) |
| Helal | 2017 | Egypt | Santa Cruz | 45 | - | NS | 7 | (34 |
|
|
|
| Biotechnology, |
|
|
|
|
|
|
|
|
| Inc. |
|
|
|
|
|
| Lee | 2006 | China | Nevomarkers | 80 | - | NS | 8 | (35) |
| Kim | 2002 | Korea | BD Pharmingen | 162 | - | NS | 7 | (36) |
| Umekita | 2003 | Japan | BD Pharmingen | 92 | - | NS | 7 | (37) |
| Joensuu | 2009 | Finland | Dako | 73 | - | NS | 7 | (38) |
| Prasad | 2009 | India | BD Pharmingen | 59 | - | NS | 7 | (39) |
Bioinformatics analysis
The prognostic value of SERPINB5 mRNA expression in
breast cancer was evaluated using the Kaplan-Meier plotter database
(http://www.kmplot.com). The expression of
SERPINB5 mRNA in breast cancer and normal breast tissue and its
relationship with clinical characteristics were obtained from the
GEPIA (gepia.cancer-pku.cn/) and UALCAN (UALCAN.path.uab.edu/)
databases.
Statistical analysis
Revman (version 5.3; the Cochrane Collaboration) was
used for meta-analysis. According to the clinicopathological
parameters of patients with breast cancer, the ratio and 95%
confidence interval were used to estimate the expression of maspin.
If heterogeneity was not significant, a fixed-effects model
(Mantel-Haenszel method) is used. Otherwise, a random-effects model
was used (DerSimonian and Laird methods). The I2 test
was used to quantify heterogeneity effects. Based on three cutoff
values (25, 50 and 75%), heterogeneity was classified as low,
moderate or high. Funnel plots were used to evaluate publication
bias, and Begg's and Egger's tests were used to evaluate compliance
with funnel plots. A two-sided P<0.05 was considered to indicate
a statistically significant difference.
Results
Characteristics of eligible
studies
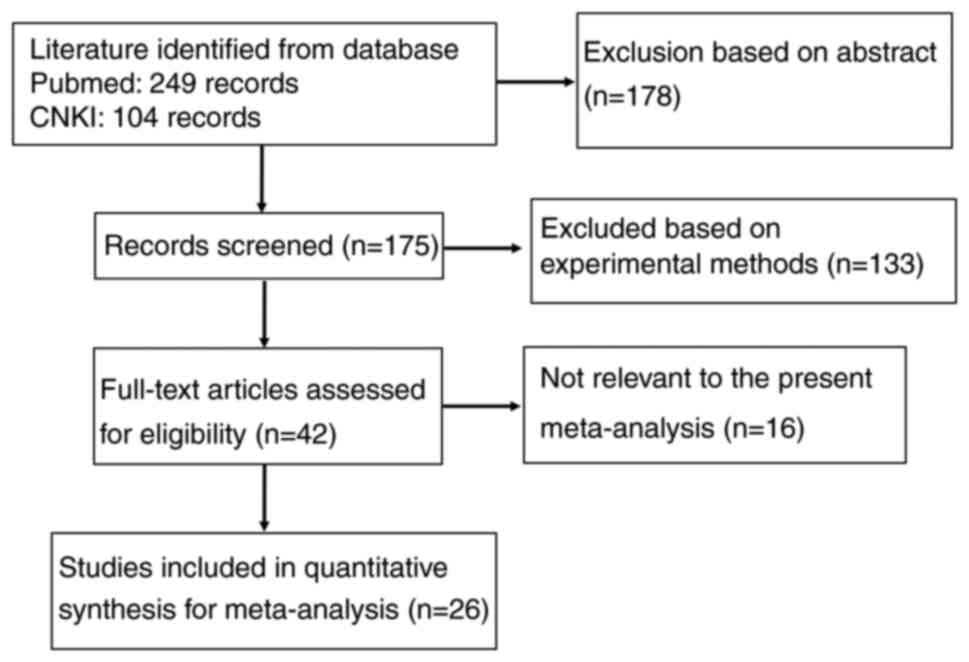
As presented in Fig.
1, 26 articles on the relationship between maspin expression
and clinicopathological characteristics of breast cancer evaluated
by immunohistochemical methods were retrieved from PubMed and CNKI
(14–39). Only 11 of these articles contained
an analysis of normal breast tissues (15,17–21,24–28).
The clinicopathological parameters included in the articles were
lymph node metastasis, TNM staging, estrogen receptor (ER)
expression, progesterone receptor (PR) expression and human EGFR2
(HER2) expression.
Forest plot of OR for the association
between maspin expression and the clinicopathological parameters of
patients with breast cancer
The association between maspin expression and cancer
susceptibility of normal breast tissue reported in 11 studies with
818 cancers and 397 controls was analyzed. It was found that maspin
expression was downregulated in breast cancer compared with normal
breast tissue (P<0.00001, Fig.
2). Maspin expression was not significantly associated with the
TNM stage (P>0.05, Table II).
Lower maspin expression was found in lymph node metastasis of
breast cancer (P<0.05, Table
II). Maspin expression was not significantly associated with
ER-positive (P>0.05, Table II),
PR-positive (P>0.05, Table II)
or HER2-positive (P>0.05, Table
II) patients with breast cancer.
 | Table II.Meta-analysis of the association
between maspin expression and clinicopathological parameters of
patients with breast cancer. |
Table II.
Meta-analysis of the association
between maspin expression and clinicopathological parameters of
patients with breast cancer.
|
| Heterogeneity | Test for overall
effect |
|---|
|
|
|
|
|---|
| Clinicopathological
features | I2
(%) | P-value | Odds ratio (95%
CI) | P-value |
|---|
| TNM staging
(I–II/III–IV) | 67 | <0.001 | 1.07
(0.69–1.65) | 0.770 |
| Lymph node
metastasis (LN+/LN-) | 60 | <0.001 | 0.36
(0.24–0.54) | <0.001 |
| ER (+/-) | 61 | 0.002 | 1.26
(0.77–2.07) | 0.360 |
| PR (+/-) | 70 | <0.001 | 1.08
(0.55–2.09) | 0.830 |
| HER2 (+/-) | 77 | <0.001 | 1.50
(0.48–4.71) | 0.490 |
Publication bias
As indicated in Fig.
3, heterogeneity testing was conducted on the included
articles. In the sensitivity analysis, one study was removed from
the summary analysis each time and the impact of each single study
on the summary results was thereby evaluated. The results of
Egger's test indicated no significant publication bias in the
present meta-analysis.
Clinicopathological and prognostic
significance of SERPINB5 expression in breast cancers
As indicated in Fig.
4, the analysis with the Kaplan-Meier plotter database
indicated that lower SERPINB5 expression was positively associated
with the overall survival rate of patients with ER-positive
(P<0.05, Fig. 4A and B), luminal
A (P<0.05, Fig. 4C) and grade 2
(P<0.05, Fig. 4D) breast cancer.
Patients with luminal A breast cancer with high SERPINB5 mRNA
expression had a longer relapse-free survival time than those with
low SERPINB5 expression (P<0.05, Fig. 4E) and until the end of the follow-up
time (100–200 months), there was an advantage. The post-progression
survival rate of patients with basal-like carcinoma or
HER2-positive carcinoma in the SERPINB5 mRNA high expression group
was lower than that in the low expression group (P<0.05,
Fig. 4F and H), but the opposite
was found in patients with PR-negative breast cancer (P<0.05,
Fig. 4G). There appeared to be a
negative relationship between low SERPINB5 mRNA expression and the
distant metastasis-free survival rate of patients with ER-positive
(P<0.05, Fig. 4I and J), luminal
A (P<0.05, Fig. 4K) and
PR-negative (P<0.05, Fig. 4L)
breast cancer.
 | Figure 4.Clinicopathological and prognostic
significance of SERPINB5 expression in breast cancers according to
the Kaplan-Meier plotter database. Overall survival rate: (A) Array
ER-positive, (B) IHC ER-positive, (C) luminal A, (D) grade 2.
Relapse-free survival rate: (E) Luminal A. Post-progression
survival rate: (F) Basal-like, (G) PR-negative, (H) HER2-positive.
Distant metastasis-free survival rate: (I) Array ER-positive, (J)
IHC ER-positive, (K) luminal A, (L) PR-negative. ER, estrogen
receptor; PR, progesterone receptor; HER2, human EGFR2; IHC,
immunohistochemistry. |
According to the GEPIA database, SERPINB5 mRNA
expression was higher in normal tissues than breast cancer
(P<0.05, Fig. 5A) and negatively
correlated with TNM staging (P<0.05, Fig. 5B). High expression of maspin was
also positively associated with the overall survival rate
(P<0.05, Fig. 5C). In addition,
SERPINB5 mRNA expression was also higher in normal tissues than
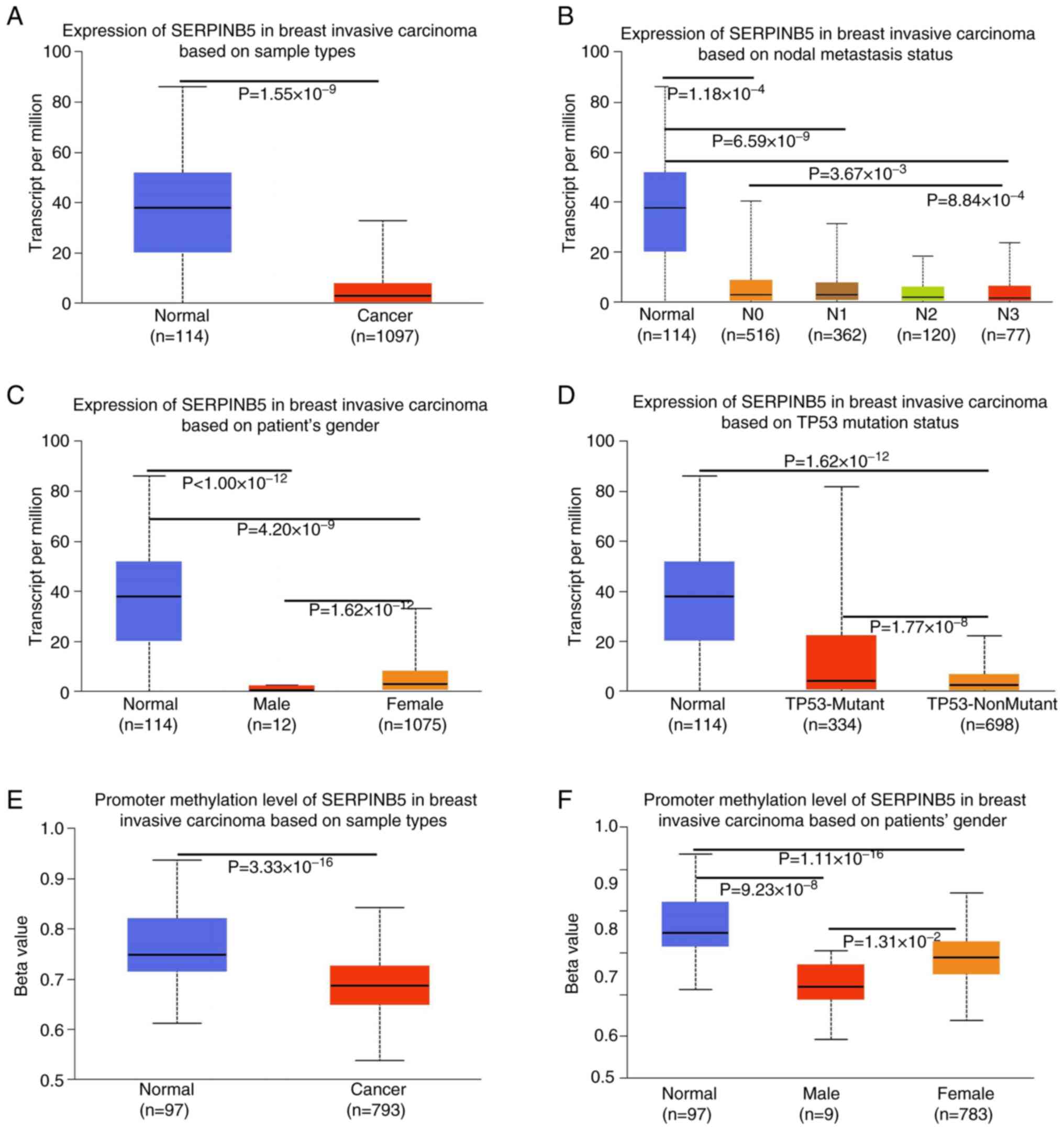
breast cancer (P<0.05, Fig. 6A)
in the UALCAN database. There appeared to be a positive
relationship between low SERPINB5 mRNA expression and lymph node
metastasis (P<0.05, Fig. 6B),
gender (P<0.05, Fig. 6C) and
tumor protein 53 mutation (P<0.05, Fig. 6D) in patients with breast cancer.
The promoter methylation level of SERPINB5 was higher in normal
tissue than breast cancer tissue (P<0.05, Fig. 6E). The promoter methylation level of
SERPINB5 was also associated with patients' gender (P<0.05,
Fig. 6F).
Discussion
The occurrence and development of breast cancer is a
process involving multiple genes and steps. The biological
characteristics of malignant tumors are invasive growth and
metastasis, and its mechanism comprises abnormal changes of gene
structure and function in cells caused by genetic defects and
epigenetic changes (40). The
expression of maspin mRNA in the human breast cancer cell line
MDA-M-B-435S is low, but after the treatment with
5-aza-2′-deoxycytidine, the expression of maspin mRNA was
significantly increased, which suggests that the decreased
expression of maspin in breast cancer cells may, at least in part,
be caused by abnormal methylation or deacetylation of the promoter
region (41). Maspin can
significantly inhibit the proliferation of glioma cells, causing
them to stagnate in the S phase, indicating that maspin may have
tumor suppressor gene characteristics in glioma. In glioma, the
inhibition of maspin expression is related to the methylation of
its promoter CpG island; 5-Aza-2′-deoxycitydine (5-aza-dC) was able
to restore the transcription of maspin in glioma cell lines
(42). When the recombinant maspin
gene was used to treat breast cancer cell lines, it was found that
their ability to transfer through laminin, type IV collagen and
gelatin matrix was significantly lower than that of control cells,
and this inhibitory effect could be blocked by maspin antibodies,
which showed that maspin is able to increase cell adhesion and
inhibit tumor metastasis and growth (9). In order to study the
clinicopathological and prognostic significance of maspin in breast
cancer, 26 articles that met specific inclusion criteria were
analyzed in the present study and the quality of these articles was
scored according to the NOS.
Previous studies have found that the synthesis of
nitric oxide (NO) in endothelial cells is related to the maspin
gene. NO can induce a decrease in maspin expression, thereby
reducing the motility and invasiveness of tumor cells and
increasing cell apoptosis (43).
Khalkhali-Ellis et al (44)
and Lim et al (45) also
found that the synthesis of NO is related to maspin gene in breast
cancer cells and human neuroepithelial tumor cells. Jiang et
al (46) proved that γ citric
acid can upregulate the expression of maspin and inhibit the
motility of colon cancer, breast cancer and melanoma cell lines.
Yin et al (47) found that
glutathione S-transferase interacts most with maspin. After maspin
transfection or recombinant maspin protein intervention, breast
cancer and prostate cancer cell lines showed higher glutathione
S-transferase activity and lower reactive oxygen species products.
This effect was reduced when the maspin gene is knocked out or
glutathione S-transferase is inhibited, which indicates that the
interaction between maspin and glutathione S-transferase may have a
role in reducing oxidative stress of cells (47).
The expression of maspin protein in oral squamous
cell carcinoma is lower than that in normal oral tissue and the low
expression of maspin is related to lymph node metastasis in oral
squamous cell carcinoma (48). The
present meta-analysis found that the low expression of maspin in
breast cancer tissue was associated with lymph node metastasis.
Maspin gene expression in gastric cancer tissue was significantly
lower than that in adenoma tissue and its low expression was
significantly associated with histological type, tumor stage and
invasion depth, indicating that loss of the maspin gene is
associated with the occurrence and progression of gastric cancer.
At the same time, it was found that when the methylation inhibitor
5-aza-2′-deoxycytidine was used to treat eight gastric cancer cell
lines with the loss of maspin gene expression, re-expression of the
maspin gene was found in five cell lines, indicating that DNA
methylation has a role in the loss of maspin gene expression, which
led to tumor progression and invasion (49). Zheng et al (50) found through bioinformatics databases
that the expression of maspin in gastric cancer tissue was lower
than that in normal gastric tissue. Through the UALCAN database, it
was found that the methylation level of SERPINB5 promoter of maspin
in normal tissues was higher than that in breast cancer tissues.
The results of this study are thus consistent with those of other
studies.
Maspin expression levels gradually decrease in
normal endometrium, atypical endometrial hyperplasia and
endometrial cancer. As the level of malignant biological behavior
in endometrial cancer increases, the expression of maspin
decreases. At the same time, maspin protein is lowly expressed in
endometrial cancer with high pathological staging and poor
differentiation, and vice versa (51). According to research, overexpression
of maspin is associated with better overall survival in esophageal
and oral squamous cell carcinoma (52,53).
Zheng et al (50) found that
SERPINB5 mRNA expression is positively associated with overall
survival and progression-free survival in patients with gastric
cancer, and even after stratification based on clinical and
pathological characteristics. The results of the present
bioinformatics analysis showed that the expression of maspin mRNA
was positively associated with the overall survival rate of
patients with breast cancer. This is contrary to the report by Lu
et al (54) on lung
adenocarcinoma. This abnormal phenomenon may be due to different
methods: Bioinformatics analysis is based on cDNA arrays, while
Lu's experiment is based on RT-PCR. As for the prognostic
significance of maspin expression in breast cancer, more cases of
breast cancer are crucial for future research.
In conclusion, at the protein and mRNA levels,
maspin is lowly expressed in breast cancer tissue and is negatively
associated with lymph node metastasis of breast cancer. High
expression of maspin mRNA is positively associated with the overall
survival rate of patients with breast cancer. Low expression of
maspin may be a good potential marker for poor prognosis of
patients with breast cancer.
Acknowledgements
Not applicable.
Funding
Cangzhou Science and Technology Bureau (NO:222001007).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SS and ZGZ designed the study. SS and HYM prepared
figures and tables, interpreted the data and wrote the main
manuscript. JS and YZS participated in the research of the study
and performed the statistical analysis. ZGZ and SS confirm the
authenticity of the raw data. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zou Z, Anisowicz A, Hendrix MJ, Thor A,
Neveu M, Sheng S, Rafidi K, Seftor E and Sager R: Maspin, a serpin
with tumor-suppressing activity in human mammary epithelial cells.
Science. 263:526–529. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bailey CM, Khalkhali-Ellis Z, Seftor EA
and Hendrix MJC: Biological functions of maspin. J Cell Physiol.
209:617–624. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sheng S, Carey J, Seftor EA, Dias L,
Hendrix MJ and Sager R: Maspin acts at the cell membrane to inhibit
invasion and motility of mammary and prostatic cancer cells. Proc
Natl Acad Sci USA. 93:11669–11674. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang W and Zhang M: Tissue microarray
analysis of maspin expression and its reverse correlation with
mutant p53 in various tumors. Int J Oncol. 20:1145–1150.
2002.PubMed/NCBI
|
|
5
|
Loo JA, Yan W, Ramachandran P and Wong DT:
Comparative human salivary and plasma proteomes. J Dent Res.
89:1016–1023. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sopel M, Surowiak P and Berdowska I:
Nuclear maspin expression as a good prognostic factor in human
epithelial ovarian carcinoma. Folia Morphol (Warsz). 69:204–212.
2010.PubMed/NCBI
|
|
7
|
Yin S, Lockett J, Meng Y, Biliran H Jr,
Blouse GE, Li X, Reddy N, Zhao Z, Lin X, Anagli J, et al: Maspin
retards cell detachment via a novel interaction with the
urokinase-type plasminogen activator/urokinase-type plasminogen
activator receptor system. Cancer Res. 66:4173–4181. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Endsley MP, Hu Y, Deng Y, He X, Warejcka
DJ, Twining SS, Gonias SL and Zhang M: Maspin, the molecular bridge
between the plasminogen activator system and beta1 integrin that
facilitates cell adhesion. J Biol Chem. 286:24599–24607. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Odero-Marah VA, Khalkhali-Ellis Z,
Chunthapong J, Amir S, Seftor RE, Seftor EA and Hendrix MJ: Maspin
regulates different signaling pathways for motility and adhesion in
aggressive breast cancer cells. Cancer Biol Ther. 2:398–403. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Biliran H Jr and Sheng S: Pleiotrophic
inhibition of pericellular urokinase-type plasminogen activator
system by endogenous tumor suppressive maspin. Cancer Res.
61:8676–8682. 2001.PubMed/NCBI
|
|
11
|
Alvarez Secord A, Darcy KM, Hutson A,
Huang Z, Lee PS, Jewell EL, Havrilesky LJ, Markman M, Muggia F and
Murphy SK: The regulation of MASPIN expression in epithelial
ovarian cancer: Association with p53 status, and MASPIN promoter
methylation: A gynecologic oncology group study. Gynecol Oncol.
123:314–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moher D, Liberati A, Tetzlaff J, Altman
DG, Antes G, Atkins D, Barbour V, Barrowman N, Berlin JA, Clark J,
et al: Preferred reporting items for systematic reviews and
meta-analyses: The PRISMA statement. Rev Esp Nutr Hum Diet.
18:172–181. 2014. View Article : Google Scholar
|
|
13
|
Luchini C, Stubbs B, Solmi M and Veronese
N: Assessing the quality of studies in meta-analyses: Advantages
and limitations of the Newcastle Ottawa Scale. World J Meta Anal.
5:80–84. 2017. View Article : Google Scholar
|
|
14
|
Liu W, Zhang XH, Zhang ZG, Wang XL, et al:
The relationship between the maspin, BCSG1 and c-erbB-2 expression
and the prognosis of breast cancer. Chin J Clin Oncol. 32:431–434.
2005.
|
|
15
|
Liu W, Zhang XH, Zhang ZG, Wang XL, et al:
The significance of expression of maspin in breast cancer and
lesions in the neighborhood of cancer. Chin J Clin Oncol.
31:1272–1276. 2004.
|
|
16
|
Hu W and Xing LQ: Significance of maspin
protein expression in breast cancer. Chin J Misdiagnostics.
6:4526–4528. 2006.
|
|
17
|
Zhang W, Wang RY, Li J and Zhang XY:
Expression and clinical significance of Maspin and MMP-2 in breast
infiltrative ductal carcinoma. Chin J Surg Oncol. 6:225–229.
2014.
|
|
18
|
Liu JC, Jiang ZM, Li Xin and Liu XZ:
Expressions of maspin and p53 in breast cancer and its clinical
signifi cance. Chin J Bases Clin Gen Surg. 20:547–550. 2013.
|
|
19
|
Sun G, Shi CQ, Zhang M and Qi Y:
Expression and clinical significance of maspin and p53 in breast
cancer. J Clin Exp Med. 13:328–330. 2014.
|
|
20
|
Yang WP, Wei CY, Zhou HJ, Ding ZL and Li
H: Expression of maspin in 65 cases with breast cancer and its
significance. J Oncol. 16:163–167. 2010.
|
|
21
|
Ding YF, Feng YZ, Ping JL, Wang XH and Li
F: The clinical significance of maspin expression in breast cancer.
Suzhou Univ J Med Sci. 27:396–398. 2006.
|
|
22
|
Cao GF, Zhang CH and You QH: Different
expressions of maspin in human mammary cancer of different ER
expressions. Med J Commun. 20:23–24. 2006.
|
|
23
|
Zhu XQ and Gong DS: Maspin expression in
breast cancer and its relationship with microvessel density. J Surg
Concepts Pract. 12:353–356. 2007.
|
|
24
|
Chen SP, Che AW, Tan XD, Li JL and Kong
JG: Expression and clinicopathological significance of Maspin and
Bmi-1 in breast cancer. Hebei Med J. 37:2774–2776. 2015.
|
|
25
|
Pei XH, Wang F and Yu Z: Expression and
significance of Maspin and uPA in breast cancer. 51:71–72.
2011.
|
|
26
|
Fang F, Li TC and Wu P: Analysis of maspin
protein expression and maspin gene promoter methylation in breast
cancer tissue. Carcinog Teratog Mutagen. 21:42009.
|
|
27
|
Zhang QY, Chen XD, Li JW, Yu HF, Liang QL,
Huang SC and Zhang Z: Expression of vascular endothelial growth
factor and Maspin in breast carcinoma and its clinical
significance. Prog Mod Biomed. 12:493–496. 2012.
|
|
28
|
Zhang W, Qiao SP, Shang PZ, Liu B, Li W
and Nan RL: Clinicopathologic study on expression sleX and maspin
in invasive ductal carcinoma tissue of breast. J Hebei North Univ.
38:1–8. 2022.
|
|
29
|
Liu XZ, Zhou SF, Cai FL, Wu YY and Jin LF:
Expression and clinical significance of maspin and uPA in breast
cancer. J Mod Oncol. 17:2139–2142. 2009.
|
|
30
|
Wang LX, Zhao PR, Wang B, Fan QX, Wang RL
and Zhang GM: The different expression of maspin in human mammary
cancer of different CerbB-2 expression. J Basic Clin Oncol.
18:85–86. 2005.
|
|
31
|
Wakahara M, Sakabe T, Kubouchi Y, Hosoya
K, Hirooka Y, Yurugi Y, Nosaka K, Shiomi T, Nakamura H and Umekita
Y: Subcellular localization of maspin correlates with histone
deacetylase 1 expression in human breast cancer. Anticancer Res.
37:50712017.PubMed/NCBI
|
|
32
|
Tuncel F, Bozkurt F and Berkesoglu M: The
value of maspin and PD-L1 expression and peritumoral lymphocytic
infiltration in breast tumors. Bratisl Lek Listy. 121:894–900.
2020.PubMed/NCBI
|
|
33
|
Feng Y, Zhu J, Shi JP, Wang HL and Zhang
Y: Expression and significances of inhibitors of DNA binding-1,
maspin in invasive ductal cancer with breast infiltrations. J
Lanzhou Univ. 34:42008.
|
|
34
|
Helal DS and El-Guindy DM: Maspin
expression and subcellular localization in invasive ductal
carcinoma of the breast: Prognostic significance and relation to
microvessel density. J Egypt Natl Canc Inst. 29:177–183. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee MJ, Suh CH and Li ZH:
Clinicopathological significance of maspin expression in breast
cancer. J Korean Med Sci. 21:309–14. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim DH, Yoon DS, Dooley WC, Nam ES, Ryu
JW, Jung KC, Park HR, Sohn JH, Shin HS and Park YE: Association of
maspin expression with the high histological grade and
lymphocyte-rich stroma in early-stage breast cancer.
Histopathology. 42:37–42. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Umekita Y and Yoshida H: Expression of
maspin is up-regulated during the progression of mammary ductal
carcinoma. Histopathology. 42:541–545. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Joensuu KM, Leidenius M, Andersson LC and
Heikkilä PS: High expression of maspin is associated with early
tumor relapse in breast cancer. Hum Pathol. 40:1143–1151. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Prasad CP, Rath G, Mathur S, Bhatnagar D
and Ralhan R: Expression analysis of maspin in invasive ductal
carcinoma of breast and modulation of its expression by curcumin in
breast cancer cell lines. Chem Biol Interact. 183:455–461. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Oliveira AM, Ross JS and Fletcher JA:
Tumor suppressor genes in breast cancer: The gatekeepers and the
caretakers. Am J Clin Pathol. 124 (Suppl):S16–S28. 2005.PubMed/NCBI
|
|
41
|
Zhang B, Liu K and Chen JY: Combination of
5-Aza-CdR and trichostatin A on cell proliferation and maspin gene
expression in breast cancer cell line MDA-MB-435S. Chin J Cancer
Prev Treat. 15:725–728. 2008.
|
|
42
|
Liu HY, Cheng G and Li ZP: The epigenetics
mechanism of maspin silencing in glioma. Yiayao Qianyan.
27:103–105. 2020.
|
|
43
|
Man XB, Tang L, Qiu XH, Yang LQ, Cao HF,
Wu MC and Wang HY: Expression of cytochrome P4502E1 gene in
hepatocellular carcinoma. World J Gastroenterol. 10:1565–1568.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Khalkhali-Ellis Z, Zhang M and Hendrix
MJC: Maspin and cathepsin d partnership in regulating mammary gland
development and breast cancer. Breast Cancer: Causes. Diagnosis and
Treatment. Romero MR: Nova Science Publishers, Inc.; pp. 161–176.
2011
|
|
45
|
Lim S, Hung AC and Porter AG: Focused PCR
screen reveals p53 dependence of nitric oxide-induced apoptosis and
up-regulation of maspin and plasminogen activator inhibitor-1 in
tumor cells. Mol Cancer Res. 7:55–66. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jiang WG, Hiscox S, Horrobin DF, Bryce RP
and Mansel RE: Gamma linolenic acid regulates expression of maspin
and the motility of cancer cells. Biochem Biophys Res Commun.
237:639–644. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yin S, Li X, Meng Y, Finley RL Jr, Sakr W,
Yang H, Reddy N and Sheng S: Tumor-suppressive maspin regulates
cell response to oxidative stress by direct interaction with
glutathione S-transferase. J Biol Chem. 280:34985–3496. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shui HH, Luo L, Liang SZ, ZHuang L and Li
W: The expression of Maspin protein in oral squamous cell carcinoma
and its significance. Hua Xi Kou Qiang Yi Xue Za Zhi. 26:604–606.
6102008.(In Chinese). PubMed/NCBI
|
|
49
|
Caro AA and Cederbaum AI: Role of
phospholipase A2 activation and calcium in CYP2E1-dependent
toxicity in HepG2 cells. J Biol Chem. 278:33866–33877. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zheng HC and Gong BC: The roles of maspin
expression in gastric cancer: A meta- and bioinformatics analysis.
Oncotarget. 8:66476–66490. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liang XF, Cui ZY, Peng BY, et al: Maspin
and p73 in endometrial cancer tissue Expression level and its
correlation analysis. World's Latest Med Inf Dig. 21:390–391.
2021.
|
|
52
|
Wang Y, Sheng S, Zhang J, Dzinic S, Li S,
Fang F, Wu N, Zheng Q and Yang Y: Elevated maspin expression is
associated with better overall survival in esophageal squamous cell
carcinoma (ESCC). PLoS One. 8:e635812013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yoshizawa K, Nozaki S, Okamune A, Kitahara
H, Ohara T, Kato K, Kawashiri S and Yamamoto E: Loss of maspin is a
negative prognostic factor for invasion and metastasis in oral
squamous cell carcinoma. J Oral Pathol Med. 38:535–539. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lu M, Li J, Huang Z, Du Y, Jin S and Wang
J: Aberrant maspin mRNA expression is associated with clinical
outcome in patients with pulmonary adenocarcinoma. Med Sci Monit.
22:134–139. 2016. View Article : Google Scholar : PubMed/NCBI
|