Introduction
According to the World Health Organization (WHO),
colorectal cancer (CRC) is a malignant neoplasm that develops when
normal epithelial cells lining the large intestine undergo
malignant transformation and give rise to adenocarcinoma (1). CRC is the third leading cause of
cancer-related mortality (1) and
accounted for 576,858 deaths and 1.15 million new cases in 2020,
based on the report of GLOBOCAN 2020 database for 185 countries and
36 cancer types (2). The number of
new cases of CRC is projected to rise to 1.92 million by 2040
(3).
According to previous statistics, CRC has a
mortality rate of 10.5% in men and 9% in women, making it the
second leading cause of cancer-related death in both sexes in
Jordan (4), and the 5-year survival
rate is 22.6% (5). Thus, to improve
survival in patients with advanced-stage disease, a further
understanding of the molecular mechanisms underlying CRC
pathogenesis and the application of targeted therapies in clinical
practice is required.
The pathophysiology of CRC is complex and
multifaceted, leading to the widespread acceptance that the causes
are heterogeneous (6). Changes at
the genomic, transcriptomic, epigenomic and metabolomic levels all
serve key roles in the etiology and progression of CRC, and our
understanding of this disease remains limited (7). Different molecular subtypes have
distinct clinical and pathological characteristics, and varying
responses to cytotoxic and targeted therapies. In metastatic CRC
(mCRC), it is currently advised that thorough RAS and v-Raf Murine
Sarcoma Viral Oncogene Homolog B (BRAF) mutation testing should be
performed before treatment (8). RAS
proteins are GTPases and there are three subtypes: Kirsten rat
sarcoma viral oncogene homolog (KRAS) and Harvey rat sarcoma viral
oncogene homolog were discovered in 1982 (9), whereas neuroblastoma RAS viral
oncogene homolog (NRAS) was not discovered until 1983 (10). NRAS mutations are infrequent,
occurring in ~4% of CRC cases (11). NRAS mutant tumors are predominantly
situated in the proximal colon and are more prevalent among older
patients (11). KRAS mutations are
linked to right-sided primary CRC, whereas NRAS mutations are more
prevalent in left-sided CRC, particularly in women (12). The prognosis for patients with RAS
mutations is worse compared with that of patients with a wild-type
RAS genotype. KRAS and NRAS mutations exert differing effects on
survival in mCRC; the prognosis for the aggressive NRAS mutant
molecular subgroup is unfavorable (13).
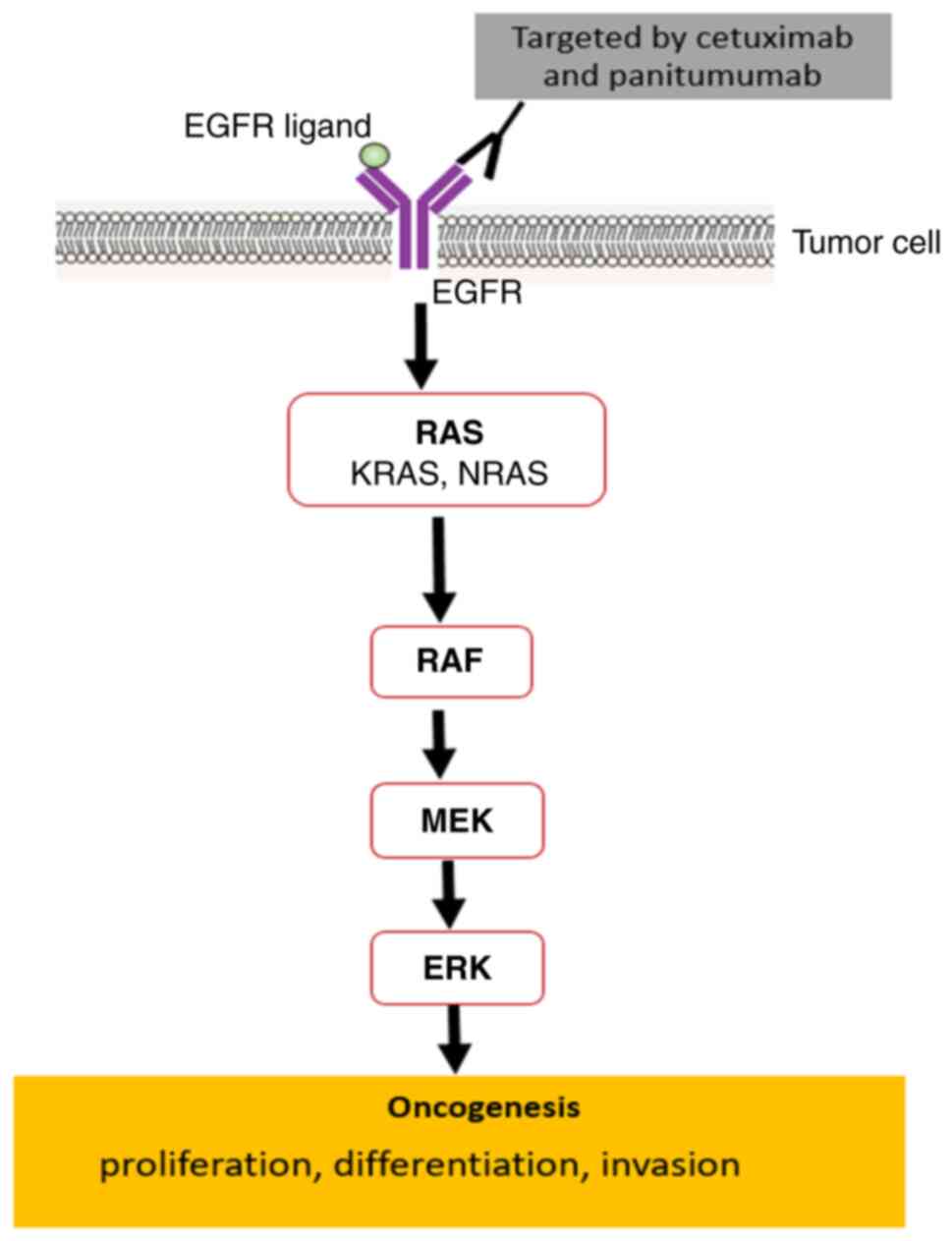
KRAS is a component located downstream of the
epidermal growth factor receptor (EGFR) signaling pathway. KRAS
operates as an intracellular signal transducer by linking signals
from cell surface receptors to intracellular targets that control
important functions for tumor progression, including proliferation,
differentiation and apoptosis (Fig.
1) (14).
The development of CRC is associated with the
presence of activating mutations in oncogenes; mutant RAS proteins
elicit downstream oncogenic signaling, resulting in tumor cells
acquiring aggressive characteristics such as increased cell mitosis
(15). Activating mutations in the
KRAS gene are associated with ~45% of colorectal malignancies, and
a large proportion of mutations are found in codons 12 (30%) and 13
(8%) of exon 2 (16). The presence
of these somatic mutations leads to continuous activation of the
EGFR pathway, resulting in the acquisition of resistance to
anti-EGFR therapies, including panitumumab or cetuximab (17). Previous research has demonstrated
that in mCRC, KRAS mutations in exon 2 were not associated with any
therapeutic benefit when using anti-EGFR monoclonal antibodies
(18). Consequently, the use of
cetuximab and panitumumab as anti-EGFR monoclonal antibodies is
restricted to patients with wild-type RAS mCRC and therefore, this
mandates that RAS mutation screening should be conducted to
determine whether anti-EGFR monoclonal antibodies should be
recommended. It has been demonstrated that KRAS mutations predict a
worse prognosis for patients with CRC (19). The FOCUS trial reported that the
presence of activating mutations in the KRAS and BRAF oncogenes is
associated with shorter overall survival. Patients with a KRAS
mutation had significantly worse overall survival compared with
patients with KRAS wild-type tumors [hazard ratio (HR), 1.24; 95%
confidence interval (CI), 1.06–1.46; P=0.008]. However, these KRAS
mutations did not significantly affect progression-free survival
(HR, 1.14; 95% CI, 0.98–1.33; P=0.09) (20). The Raf protein has also been
extensively studied and shown to be involved in signal
transduction, cellular proliferation and carcinogenesis (Fig. 1); ~8% of CRC tumors exhibit
activating mutations in BRAF, which predominantly impacts codon 600
(21). The presence of BRAF
mutations in mCRC is widely recognized to have a notable negative
prognostic effect (22). These
mutations require an alternative therapeutic strategy since
conventional anti-EGFR treatments are frequently ineffective
(23). However, numerous targeted
methods are now being investigated for various types of cancer with
BRAF mutations, owing to the potential significance of BRAF
mutations as a driving signal in tumor growth (24). Previous studies have shown that KRAS
mutations, as well as BRAF mutations, have distinguished
pathological and clinical characteristics (25,26).
For example, CRC with KRAS exon 2 mutations are more common in
elderly individuals, particularly males, and are commonly seen in
the proximal colon when compared with the wild-type exon (27). Mutations in the BRAF gene are
strongly associated with females, the elderly, mucinous
differentiation, low histological grade and tumors located in the
proximal region of the colon (28).
Thus, the molecular variations emphasize the significance of
tailored treatment strategies in mCRC.
Although routine testing for RAS and BRAF mutations
is conducted on patients with mCRC in Jordan before recommending
anti-EGFR therapy, to the best of our knowledge, no previous
studies have reported on the possible association between
clinicopathological features and complete RAS and BRAF status,
which could help researchers identify other factors that influence
therapeutic response to anti-EGFR antibodies. Therefore, the aim of
the present study was to identify mutations in the RAS and BRAF
genes in patients with sporadic CRC and investigate their
associations with clinicopathological characteristics.
Materials and methods
Study design
The present study retrospectively analyzed patients'
electronic medical records from a single center, the Jordanian
Military Cancer Center-Royal Medical Services (Amman, Jordan). The
present study included 262 patients diagnosed with mCRC between
January 2020 and January 2022. Data regarding age, sex, RAS and
BRAF status, primary tumor site, histological subtypes and grade
were acquired. The inclusion criteria for patients were as follows:
Age ≥18 years old and diagnosis of adenocarcinoma of the colon or
rectum shown histologically. The exclusion criteria were as
follows: Patients with tumor types other than CRC, patients in
which RAS and BRAF mutations could not be determined (wild-type or
mutant), and cases where data was missing from the patients'
electronic medical records. The primary tumor sites were
categorized as follows: Tumors on the left side of the body, which
are more likely to develop in the distal one-third transverse
colon, splenic flexure, descending colon, sigmoid colon and rectum,
and tumors on the right side of the body, which are more likely to
develop in the cecum, ascending colon, hepatic flexure and proximal
two-thirds of the transverse colon (29). Transverse colon cancer is
characterized by tumors situated between the hepatic and splenic
flexures, and is extremely uncommon, comprising 10% of all colon
malignancies (30).
Histological subtypes were classified as classical
adenocarcinoma, mucinous adenocarcinoma, tubuvillous adenoma,
invasive adenocarcinoma, medullary adenocarcinoma, adenosquamous
carcinoma or undifferentiated carcinoma. Additionally, the
histological grade was categorized as well-differentiated,
moderately differentiated or poorly differentiated.
Tumor molecular analysis
The KRAS, NRAS and BRAF mutation status of patients
with CRC were extracted retrospectively from the electronic medical
records of patients at the Jordanian Military Cancer Center-Royal
Medical Services. However, all patients underwent tumor tissue
biopsies to ascertain RAS and BRAF mutation status using
quantitative polymerase chain reaction and reverse hybridization in
an accredited diagnostic laboratory at Jordan University Hospital
(Amman, Jordan) and only the final results were sent to the Royal
Medical Services.
Statistical analysis
Statistical analysis was performed using SPSS
(version 23.0; IBM Corp.). Categorical variables are presented as
the frequency (n) and percentage. A χ2 test or Fisher's
exact test was used to determine the associations between
variables. The φ factor was used to examine the strength of
association (as a measure of effect size) as follows: 0, no
association; 0.1, small association; 0.3, medium association; 0.5,
strong association; and 1, complete association. Univariate
analysis was used to assess these associations, utilizing
statistical tests such as the χ2 or Fisher's exact test
when appropriate. In addition, a multivariate hierarchical
regression analysis was performed to examine the impact of
covariates on gene mutations. The regression analysis consisted of
three models, the first model included mutant or wild-type as
predictors, the second model included mutant or wild-type status,
age and sex as predictors, and the third model included
histological subtypes and tumor grade as predictors alongside
mutant or wild-type status, age and sex. P<0.05 was considered
to indicate a statistically significant difference.
Results
Sample characteristics
A total of 262 patient records of mCRC were analyzed
(Table I). The cohort's mean age
was 55.91±12.81 years, with an age range of 19–85 years, and
patients >50 years old accounted for 64.9% of the cases (n=170).
Of the 262 patients, 153 (58.4%) were male and 109 (41.6%) were
female. Regarding the tumor site, 224 (85.5%) samples were
left-site tumors, which was more frequent compared with right-site
tumors (n=30; 11.5%) or transverse site tumors (n=6; 2.3%). More
than one-half of tumor cases (n=139; 53%) had a mutation in the RAS
gene and BRAF, whereas 46.9% (n=123) of the cases had wild-type RAS
gene and BRAF. In terms of histological subtypes, a large
proportion of cases were adenocarcinoma (not specified; n=246;
93.9%), followed by mucinous adenocarcinoma (n=11; 4.2%), then
tubular adenocarcinoma (n=3; 1.1%) and signet adenocarcinoma (n=2;
0.8%).
 | Table I.Characteristics of patients with
metastatic CRC (n=262). |
Table I.
Characteristics of patients with
metastatic CRC (n=262).
|
Characteristics | Patients, n
(%) |
|---|
| Sex |
|
|
Female | 109 (41.6) |
|
Male | 153 (58.4) |
| Agea, years |
|
|
≤50 | 92 (35.1) |
|
>50 | 170 (64.9) |
| CRC site |
|
|
Right | 30 (11.5) |
|
Left | 224 (85.5) |
|
Transverse | 6 (2.3) |
|
Unknown | 2 (0.8) |
| Histological
subtype |
|
|
Mucinous adenocarcinoma | 11 (4.2) |
| Tubular
adenocarcinoma | 3 (1.1) |
| Signet
adenocarcinoma | 2 (0.8) |
|
Adenocarcinoma (not
specified) | 246 (93.9) |
| Tumor grade |
|
| Poorly
differentiated | 23 (8.8) |
|
Moderately differentiated | 193 (73.7) |
|
Well-differentiated | 11 (4.2) |
| Not
determined | 35 (13.4) |
| Mutation
status |
|
|
Wild-type | 123 (46.9) |
|
Mutated | 139 (53.1) |
The characteristics of patients with mCRC are shown
in Table I. According to the WHO
criteria (31), tumor histology was
graded as poorly differentiated, moderately differentiated and
well-differentiated at 8.8 (n=23), 73.7 (n=193) and 4.2% (n=11),
respectively; the histological grade was not determined in 13.4%
(n=35) of patients.
Mutational status and detailed
mutation
Out of 139 patients with mCRC carrying a mutation,
127 patients (48.5%) had KRAS mutations and 120 mutations (45.8%)
were detected in exon 2, with 10 mutations occurring outside of
exon 2 (exon 3 or exon 4). The most prevalent KRAS exon 2 mutations
were G12V (n=40; 15.3%), followed by G12D (n=32; 12.2%), while KRAS
K117× was the least common KRAS mutation (n=2; 0.8%). Notably, 4
patients with KRAS mutation exhibited 2 mutations each: 2 with G12C
and G12V mutations, 1 with G12V and G12A mutations, and 1 with G13D
and KRAS G12S mutations. Moreover, there is missing data regarding
detailed mutation for one patient thus, the total KRAS mutation
count was 130 (49.6%).
Regarding NRAS, 10 patients possessed a mutation of
this gene. Half of the cases demonstrated a substitution of glycine
to any amino acid mutation (G12×-G13×) accounting for 1.9% (n=5) of
mutated cases. Moreover, Q61K and Q61L mutations were present at
the same frequency and percentage (both n=2; 0.8%), and the A164×
mutation was the least common among all RAS mutations accounting
for 0.4% (n=1). For BRAF mutations, 2 cases of V600E were observed
(0.8%). Table II illustrates all
the mutations and the detailed classes. Moreover, none of the
patients carrying a mutation with the KRAS or NRAS genotype had a
simultaneous BRAF mutation (Fig.
2).
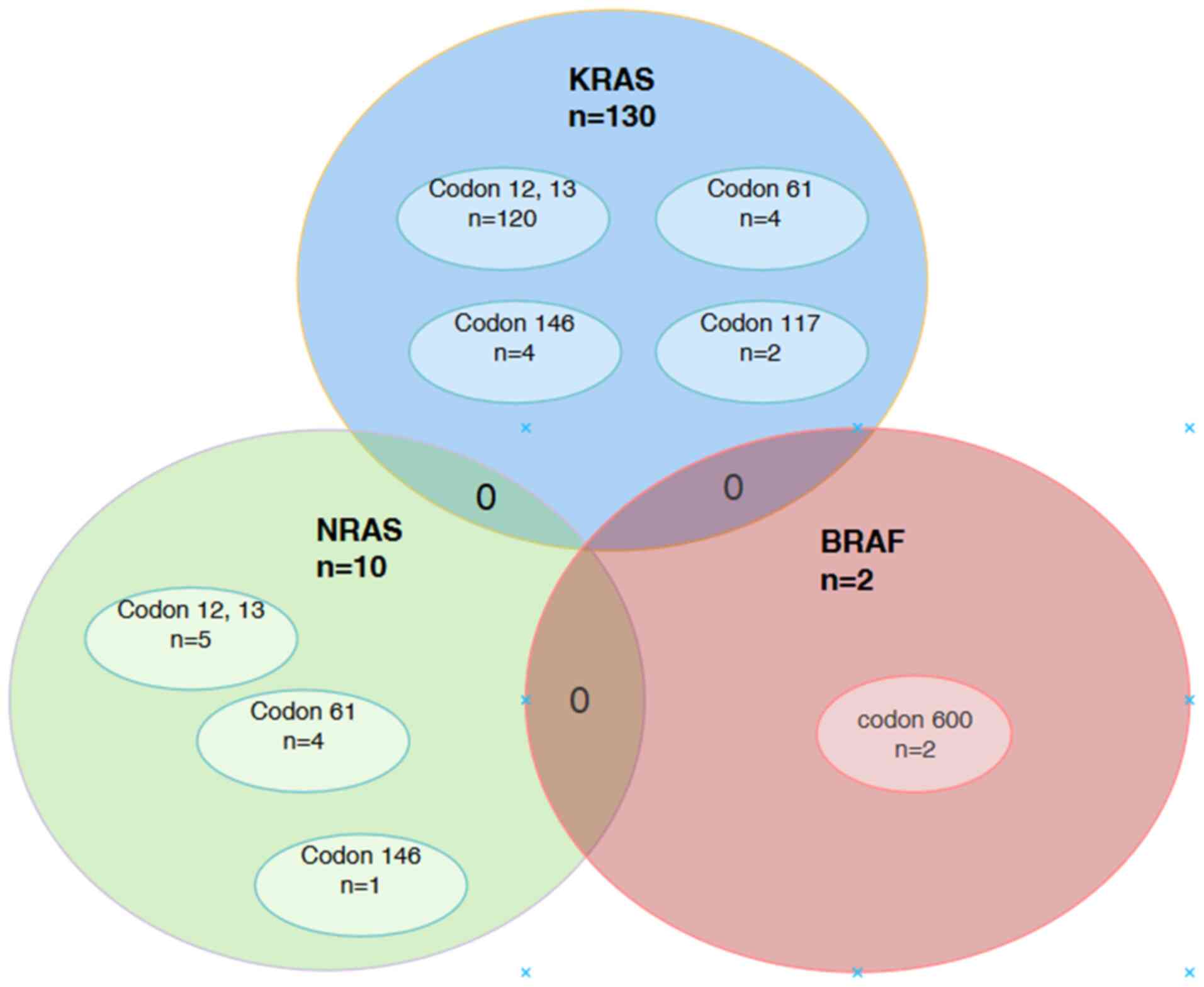 | Figure 2.Distribution of single and
concomitant mutations in the KRAS, NRAS and BRAF genes. In the
present study, 120 KRAS mutations were distributed across codons 12
and 13, codons 61 and 146, and codon 117. There were 10 NRAS
mutations distributed across codons 12 and 13, codon 61 and codon
146. There were 2 BRAF mutations in codon 600. Although overlapping
mutations between KRAS, NRAS and BRAF were not detected, 4 patients
did possess two KRAS mutations each, which made the total number of
KRAS mutations detected 130. KRAS, Kirsten rat sarcoma viral
oncogene homolog; NRAS, neuroblastoma RAS viral oncogene homolog;
BRAF, v-Raf murine sarcoma viral oncogene homolog B. |
 | Table II.Mutational status and detailed
mutation classes found in patients (n=262). |
Table II.
Mutational status and detailed
mutation classes found in patients (n=262).
| Mutational
status | Mutations, n
(%) | Exon | Codon |
|---|
| A, KRAS | 130a (49.6) |
|
|
| KRAS
G12A | 20 (7.6) | 2 | 12,13 |
| KRAS
G12D | 32 (12.2) | 2 | 12,13 |
| KRAS
G12V | 40 (15.3) | 2 | 12,13 |
| KRAS
G12S | 5 (1.9) | 2 | 12,13 |
| KRAS
G12C | 4 (1.5) | 2 | 12,13 |
| KRAS
G13A | 8 (3.1) | 2 | 12,13 |
| KRAS
G13D | 11 (4.2) | 2 | 12,13 |
| KRAS
Q61× | 4 (1.5) | 3 | 61 |
| KRAS
K117× | 2 (0.8) | 4 | 117 |
| KRAS
A146× | 4 (1.5) | 4 | 146 |
| B, NRAS | 10 (3.8) |
|
|
| NRAS
G12×-G13× | 5 (1.9) | 2 | 12,13 |
| NRAS
Q61K | 2 (0.8) | 3 | 61 |
| NRAS
Q61L | 2 (0.8) | 3 | 61 |
| NRAS
A146× | 1 (0.4) | 4 | 146 |
| C, BRAF | 2 (0.76) |
|
|
| BRAF
V600E | 2 (0.76) | 15 | 600 |
| Total
number of KRAS/NRAS/BRAF mutations | 142 (54.2) | - | - |
Association between RAS mutations,
wild-type status and the patients' clinicopathological
features
The associations between the mutational status of
the RAS gene and various clinicopathological characteristics were
assessed using univariate analyses, such as the χ2 or
Fisher's exact test, as appropriate. RAS mutations were more
prevalent in males (n=78; 56.9%), in patients >50 years old
(n=94; 68.6%) and in tumors located in the left side of the colon
(n=115; 83.9%). Additionally, RAS mutations were more frequently
observed, albeit not significantly, with the adenocarcinoma (not
specified) histological subtype (n=129; 94.2%) and moderately
differentiated tumor grade (n=102; 74.5%). No other
clinicopathological features had notable associations with RAS
mutations (Table III).
 | Table III.Univariate analysis of the
association between RAS mutation, wild-type status and
clinicopathological features. |
Table III.
Univariate analysis of the
association between RAS mutation, wild-type status and
clinicopathological features.
|
| RAS status |
|---|
|
|
|
|---|
|
Characteristics | Mutated, n (%) | Wild-type, n
(%) | P-value | Association
strength (φ) |
|---|
| Sex |
|
|
|
|
|
Female | 59 (43.1) | 50 (40) | 0.615 | −0.031 |
|
Male | 78 (56.9) | 75 (60) |
|
|
| Age, years |
|
|
|
|
|
≤50 | 43 (31.4) | 49 (39.2) | 0.186 | 0.082 |
|
>50 | 94 (68.6) | 76 (60.8) |
|
|
| CRC site |
|
|
|
|
|
Right | 17 (12.4) | 13 (10.4) | 0.687a | 0.091 |
|
Left | 115 (83.9) | 109 (87.2) |
|
|
|
Transverse | 3 (2.2) | 3 (2.4) |
|
|
|
Unknown | 2 (1.5) | - |
|
|
| Histological
subtype |
|
|
|
|
|
Mucinous adenocarcinoma | 5 (3.6) | 6 (4.8) | 0.953a | 0.042 |
| Tubular
adenocarcinoma | 2 (1.5) | 1 (0.8) |
|
|
| Signet
adenocarcinoma | 1 (0.7) | 1 (0.8) |
|
|
|
Adenocarcinoma (not
specified) | 129 (94.2) | 117 (93.6) |
|
|
| Tumor grade |
|
|
|
|
| Poorly
differentiated | 17 (12.4) | 6 (4.8) | 0.056 | 0.170 |
|
Moderately differentiated | 102 (74.5) | 91 (72.8) |
|
|
|
Well-differentiated | 4 (2.9) | 7 (5.6) |
|
|
| Not
determined | 14 (10.2) | 21 (16.8) |
|
|
Association between KRAS, NRAS, BRAF
mutations and wild-type status with the patients'
clinicopathological features
Univariate analysis was performed to investigate the
association between different subtypes of RAS mutations (KRAS and
NRAS) and BRAF mutations with various clinicopathological
characteristics (Table IV). Both
KRAS (n=86; 67.7%) and NRAS n=8; 80%) mutations were more frequent
in patients >50 years old and in tumors located in the left side
of the colon (KRAS n=106; 83.5%), (NRAS n=9; 90%) frequently, as
well as both mutations (KRAS n=120; 94.5%, and NRAS n=9; 90%) were
more prevalent in patients with adenocarcinoma (not specified);
however, these findings were not statistically significant. By
contrast, NRAS mutations were significantly associated with
moderately differentiated tumors (n=6; 60%, P<0.05). Similar to
KRAS mutations, BRAF mutations were more common in patients >50
years old (n=2; 100%) and in tumors located in the left side of the
colon (n=2; 0.9%), although the association was not significant
(Table IV).
 | Table IV.Univariate analysis of the
association between KRAS, NRAS, BRAF mutations, wild-type status
and clinicopathological features. |
Table IV.
Univariate analysis of the
association between KRAS, NRAS, BRAF mutations, wild-type status
and clinicopathological features.
| A, KRAS status |
|---|
|
|---|
|
Characteristics | Mutated, n (%) | Wild-type, n
(%) | P-value | Association
strength (φ) |
|---|
| Sex |
|
|
|
|
|
Female | 54 (42.5) | 55 (40.7) | 0.868a | −0.018 |
|
Male | 73 (57.5) | 80 (59.3) |
|
|
| Age, years |
|
|
|
|
|
≤50 | 41 (32.3) | 51 (37.8) | 0.352 | 0.058 |
|
>50 | 86 (67.7) | 84 (62.2) |
|
|
| CRC site |
|
|
|
|
|
Right | 17 (13.4) | 13 (9.6) | 0.354b | 0.117 |
|
Left | 106 (83.5) | 118 (87.4) |
|
|
|
Transverse | 2 (1.6) | 4 (3.0) |
|
|
|
Unknown | 2 (1.6) | - |
|
|
| Histological
subtype |
|
|
|
|
|
Mucinous adenocarcinoma | 5 (3.9) | 6 (4.4) | 0.682b | 0.094 |
| Tubular
adenocarcinoma | 2 (1.6) | 1 (0.7) |
|
|
| Signet
adenocarcinoma | - | 2 (1.5) |
|
|
|
Adenocarcinoma (not
specified) | 120 (94.5) | 126 (93.3) |
|
|
| Tumor grade |
|
|
|
|
| Poorly
differentiated | 13 (10.2) | 10 (7.4) | 0.499 | 0.095 |
|
Moderately differentiated | 96 (75.6) | 97 (71.9) |
|
|
|
Well-differentiated | 4 (3.1) | 7 (5.2) |
|
|
| Not
determined | 14 (11.0) | 21 (15.6) |
|
|
|
| B, NRAS
status |
|
|
Characteristics | Mutated, n
(%) | Wild-type, n
(%) | P-value | Association
strength (φ) |
|
| Sex |
|
|
|
|
|
Female | 5 (50) | 104 (41.3) | 0.746b | −0.034 |
|
Male | 5 (50.0) | 148 (58.7) |
|
|
| Age, years |
|
|
|
|
|
≤50 | 2 (20.0) | 90 (35.7) | 0.307 | 0.063 |
|
>50 | 8 (80.0) | 162 (64.3) |
|
|
| CRC site |
|
|
|
|
|
Right | - | 30 (11.9) | 0.215b | 0.123 |
|
Left | 9 (90.0) | 215 (85.3) |
|
|
|
Transverse | 1 (10.0) | 5 (2.0) |
|
|
|
Unknown | - | 2 (0.8) |
|
|
| Histological
subtype |
|
|
|
|
|
Mucinous adenocarcinoma | - | 11 (4.4) | 0.116b | 0.216 |
| Tubular
adenocarcinoma | - | 3 (1.2) |
|
|
| Signet
adenocarcinoma | 1 (10.0) | 1 (0.4) |
|
|
|
Adenocarcinoma (not
specified) | 9 (90.0) | 237 (94.0) |
|
|
| Tumor grade |
|
|
|
|
| Poorly
differentiated | 4 (40.0) | 19 (7.5) | 0.02b,c | 0.228 |
|
Moderately differentiated | 6 (60.0) | 187 (74.2) |
|
|
|
Well-differentiated | - | 11 (4.4) |
|
|
| Not
determined | - | 35 (13.9) |
|
|
|
| C, BRAF
status |
|
|
Characteristics | Mutated, n
(%) | Wild-type, n
(%) | P-value | Association
strength (φ) |
|
| Sex |
|
|
|
|
|
Female | 1 (50.0) | 108 (41.5) |
>0.999b | −0.015 |
|
Male | 1 (50.0) | 152 (58.5) |
|
|
| Age, years |
|
|
|
|
|
≤50 | - | 92 (35.4) | 0.543b | 0.065 |
|
>50 | 2 (100.0) | 168 (64.6) |
|
|
| CRC site |
|
|
|
|
|
Right | - | 30 (11.5) |
>0.999b | 0.036 |
|
Left | 2 (0.9) | 222 (85.4) |
|
|
|
Transverse | - | 6 (2.3) |
|
|
|
Unknown | - | 2 (0.8) |
|
|
| Histological
subtype |
|
|
|
|
|
Mucinous adenocarcinoma | 1 (50) | 10 (3.8) | 0.119b | 0.20 |
| Tubular
adenocarcinoma | - | 3 (1.2) |
|
|
| Signet
adenocarcinoma | - | 2 (0.8) |
|
|
|
Adenocarcinoma (not
specified) | 1 (50) | 245 (94.2) |
|
|
| Tumor grade |
|
|
|
|
| Poorly
differentiated | - | 23 (8.8) | 0.458b | 0.097 |
|
Moderately differentiated | 1 (50) | 192 (73.8) |
|
|
|
Well-differentiated | - | 11 (4.2) |
|
|
| Not
determined | 1 (50) | 34 (13.1) |
|
|
A multivariate hierarchical regression analysis was
performed to examine the impact of covariates on gene mutations.
For the KRAS gene, in the first model the predictor was the mutant
or wild-type. The result showed that the mutant or wild-type is a
significant predictor (B=0.914; β=0.912; P<0.001). The model
accounted for 83.2% of the variance in KRAS (R2=0.832;
adjusted R2=0.832), with a model significance
(F=1,291.813; P<0.001) (Table
V).
 | Table V.A multivariate hierarchical
regression analysis of the KRAS gene. |
Table V.
A multivariate hierarchical
regression analysis of the KRAS gene.
| A, Model 1 |
|---|
|
|---|
| Variable | B | SE.B | β |
|---|
| Mutant or
wild-type | 0.914 | 0.025 | 0.912a |
| Age |
|
|
|
| Sex |
|
|
|
| Histological
subtype |
|
|
|
| Tumor grade |
|
|
|
| R2 | 0.832 |
|
|
| Adjusted
R2 |
| 0.832 |
|
| F change in
R2 |
|
|
1,291.813a |
|
| B, Model
2 |
|
|
Variable | B | SE.B | β |
|
| Mutant or
wild-type | 0.917 | 0.026 | 0.916a |
| Age | −0.031 | 0.027 | −0.030 |
| Sex | −0.017 | 0.026 | −0.017 |
| Histological
subtypes |
|
|
|
| Tumor grade |
|
|
|
| R2 | 0.832 |
|
|
| Adjusted
R2 |
| 0.832 |
|
| F change in
R2 |
|
| 0.795 |
|
| C, Model
3 |
|
|
Variable | B | SE.B | β |
|
| Mutant or
wild-type | 0.915 | 0.025 | 0.914a |
| Age | −0.037 | −0.037 | −0.037 |
| Sex | −0.013 | 0.026 | −0.013 |
| Histological
subtypes | −0.074 | 0.033 | −0.057a |
| Tumor grade | 0.014 | 0.017 | 0.022 |
| R2 | 0.836 |
|
|
| Adjusted
R2 |
| 0.836 |
|
| F change in
R2 |
|
| 3.251b |
The second model included mutant or wild-type
status, age and sex as predictors. Mutant or wild-type status
remained a significant predictor (B=0.917; β=0.916; P<0.001),
with only a slight increase in its effect size compared to model 1.
Age (B=−0.031; β=−0.030; P>0.05) and sex (B=−0.017; β=−0.017;
P>0.05) were not significant predictors. Model 2 accounted for
83.2% of the variance in KRAS, as seen in model 1
(R2=0.832; adjusted R2=0.832), but there was
no significant increase in R2 compared to model 1
(F-change=0.795; P>0.05).
The third model included histological subtypes and
tumor grade as predictors alongside mutant or wild-type status, age
and sex. Mutant or wild-type status remained a significant
predictor (B=0.915; β=0.914; P<0.001). Age (B=−0.037; β=−0.037;
P>0.05) and sex (B=−0.013; β=−0.013; P>0.05) were not
statistically significant predictors. Histological subtype was a
significant predictor (B=−0.074; β=−0.057; P<0.001); however,
the tumor grade was not significant (B=0.014; β=0.022; P>0.05).
The model accounted for 83.6% of the variance in KRAS
(R2=0.836; adjusted R2=0.836), with a small
but significant increase in explanatory power compared with model 2
(F-change=3.251; P<0.05). One-way ANOVA as the out-put from
multivariate hierarchical regression confirmed the statistical
significance of all three models: Model 1, F(1,
261)=1,291.813, P<0.001; model 2, F(3,
259)=430.455, P<0.001; and model 3, F(6,
256)=222.487, P<0.001 (numbers in brackets represent
degree of freedom and sample number) (Table VI).
 | Table VI.ANOVA results for predictive models
of KRAS mutation variance based on genetic, demographic and
histological factors. |
Table VI.
ANOVA results for predictive models
of KRAS mutation variance based on genetic, demographic and
histological factors.
| Model | Sum of squares | df | Mean square | F | P-value |
|---|
| 1 | 54.475 | 1 | 54.475 | 1,291.813 | <0.001 |
| 2 | 54.542 | 3 | 18.181 | 430.455 | <0.001 |
| 3 | 54.943 | 6 | 9.157 | 222.487 | <0.001 |
For the NRAS gene, in the first model, the mutant or
wild-type status was a significant predictor (B=0.72; β=0.187;
P<0.05). The model accounted for 3.5% of the variance in NRAS
(R2=0.035; adjusted R2=0.031; F=9.462;
P<0.05), which indicated a significant contribution of mutant or
wild-type (Table VII).
 | Table VII.A multivariate hierarchical
regression analysis of the NRAS gene. |
Table VII.
A multivariate hierarchical
regression analysis of the NRAS gene.
| A, Model 1 |
|---|
|
|---|
| Variable | B | SE.B | β |
|---|
| Mutant or
wild-type | 0.72 | 0.023 | 0.187a |
| Age |
|
|
|
| Sex |
|
|
|
| Histological
subtypes |
|
|
|
| Tumor grade |
|
|
|
| R2 | 0.035 |
|
|
| Adjusted
R2 |
| 0.031 |
|
| F change in
R2 |
|
| 9.462a |
|
| B, Model
2 |
|
|
Variable | B | SE.B | β |
|
| Mutant or
wild-type | 0.07 | 0.024 | 0.182a |
| Age | 0.02 | 0.025 | 0.05 |
| Sex | 0.013 | 0.024 | 0.034 |
| Histological
subtypes |
|
|
|
| Tumor grade |
|
|
|
| R2 | 0.038 |
|
|
| Adjusted
R2 |
| 0.027 |
|
| F change in
R2 |
|
| 0.435 |
|
| C, Model
3 |
|
|
Variable | B | SE.B | β |
|
| Mutant or
wild-type | 0.072 | 0.023 | 0.186a |
| Age | 0.023 | 0.025 | 0.057 |
| Sex | 0.010 | 0.024 | 0.026 |
| Histological
subtypes | 0.054 | 0.030 | 0.112 |
| Tumor grade | 0.053 | 0.030 | 0.107 |
| R2 | 0.063 |
|
|
| Adjusted
R2 |
| 0.031 |
|
| F change in
R2 |
|
| 2.253a |
The second model incorporated age and sex as
predictors alongside the mutant or wild-type variable. Mutant or
wild-type status was a significant predictor of NRAS (B=0.07;
β=0.182; P<0.05), with only a slight decrease in its effect size
compared with model 1. Age (B=0.02; β=0.05; P>0.05) and sex
(B=0.013; β=0.034; P>0.05) were not significant predictors of
NRAS. Model 2 accounted for 3.8% of the variance in NRAS
(R2=0.038; adjusted R2=0.027), with a
non-significant overall F-statistic (F=0.435; P>0.05).
The third model included histological subtypes and
tumor grade as additional predictors. Mutant or wild-type status
remained a significant predictor of NRAS (B=0.072; β=0.186;
P<0.05). Age (B=0.023; β=0.057; P>0.05) and sex (B=0.010;
β=0.026; P>0.05) were not significant predictors. Similarly,
histological subtypes (B=0.054; β=0.112; P>0.05) and tumor grade
(B=0.053; β=0.107; P>0.05) were not significantly associated
with NRAS. Model 3 accounted for 6.3% of the variance in NRAS
(R2=0.063; adjusted R2=0.031), with a
significant F-statistic (F=2.253; P<0.05). However, the increase
in variance accounted for model 3 compared with the model 2 was a
modest increase (∆R2=0.025).
ANOVA confirmed that the three models were
statistically significant: Model 1, F(1, 261)=9.462;
P<0.05; model 2, F(3, 259)=3.431, P<0.05; model 3,
F(6, 256)=2.867; P<0.05. These results demonstrate
that each model represented a statistically significant improvement
over the null model (Table
VIII).
 | Table VIII.ANOVA results for predictive models
of NRAS mutation variance using genetic, demographic and
histological predictors. |
Table VIII.
ANOVA results for predictive models
of NRAS mutation variance using genetic, demographic and
histological predictors.
| Model | Sum of squares | Degree of freedom
(df) | Mean square | F | P-value |
|---|
| 1 | 0.338 | 1 | 0.338 | 9.462 | <0.05 |
| 2 | 0.369 | 3 | 0.123 | 3.431 | <0.05 |
| 3 | 0.608 | 6 | 0.101 | 2.867 | <0.05 |
Discussion
CRC is a highly prevalent malignancy with highly
variable occurrence and fatality rates globally (2). In 2023, an estimated 153,020
individuals received a diagnosis of CRC, with 52,550 fatalities
attributed to the disease, encompassing 19,550 cases and 3,750
deaths among those <50 years of age (32).
The impact of the RAS mutation pattern on anticancer
therapy orientation has been extensively documented (33). Patients with CRC tumors mutations in
exon 2 of the KRAS gene (specifically in codons 12 and 13) do not
experience a clinical response from anti-EGFR-based therapies
(34). Notably, some patients with
CRC with wild-type KRAS exon 2 do not exhibit a favorable response
to anti-EGFR therapy, indicating that the presence of other RAS
mutations (namely, KRAS exons 3 and 4 or NRAS exons 2, 3 and 4) and
BRAF or PIK3CA mutations can serve as a negative indicator for the
effectiveness of anti-EGFR treatment (23). A case study on brain metastasis in a
patient with KRAS wild-type CRC treated with cetuximab alongside
chemotherapy (capecitabine and oxaliplatin) demonstrated a
substantial reduction in brain metastases with the cetuximab and
chemotherapy regimen before any radiation intervention (35). However, to the best of our
knowledge, no real-world data has been published for patients with
CRC receiving anti-EGFR treatment and cetuximab.
In the present retrospective study, the RAS and BRAF
gene mutation rates in patients with mCRC were investigated. In
addition, the associations between these genetic alterations and
clinicopathological characteristics were examined. Results from the
present study were consistent with previous studies where >50%
of cases had CRC mutations (36).
The prevalence of KRAS mutations in patients with mutations was
48.5%, close to a results of a previous study conducted in Jordan
(44%) (37). For comparison,
Western Europe has a KRAS mutation rate of 44.7% (38), Indonesia has a rate of 41% (39) and China has a rate of 36.1%
(40).
In contrast to KRAS, NRAS changes are infrequent and
there is currently limited evidence on the incidence of these
mutations (41). In the present
study, the NRAS mutation rate was 3.82%. Zhang et al
(42) reported a rate of 3.69% in a
Chinese study, and studies on Italian and Indian patients indicated
frequencies of 6 and 6.3%, respectively (43). By contrast, Greek and Romanian
patients had a higher incidence rate of 9.57% (44). According to the present study, BRAF
mutations were detected in 0.76% of Jordanian patients, which is
lower compared with the percentage for Western countries (9.2%) and
Asian countries (4.9%) (45). The
KRAS, NRAS and BRAF frequency variations between studies are likely
due to several factors, including ethnicity, geographical
dispersion and utilization of distinct methods/assays to evaluate
the presence of these mutations.
The present study examined KRAS mutations in
patients with mCRC, namely those located in exon 2 and those
outside of exon 2. The prevalence of KRAS mutations was ~45.8%,
consistent with findings from a prior study in Jordan (37). Another previous study reported
mutational frequencies in this exon ranging from 15–46% based on
country (46). According to the
present study, among the KRAS mutations, G12V has the highest
prevalence, followed by G12D, G12A, G13D, G13A, G12S and G12C. The
findings indicate minor variations compared with research conducted
on Western populations, implying that ethnicity may influence the
patterns of KRAS mutations (47).
In the present study, among the NRAS mutations (3.9%), 1.9% of the
samples had a mutation in exon 2 (codon 12 or 13), which is
comparable to the Chinese population (32). The prevalence of BRAF mutations
ranges from 1.1 to 25% globally (41). Notably, the frequency of the V600E
mutation in the present study (0.76%) was detected in exon 15,
which is lower than the frequency seen in previous Asian studies
(48,49). The clinical significance of the KRAS
mutations, except those of codons 12 and 13, remains unclear.
Loupakis et al (50)
reported that a patient with mCRC and KRAS A146 mutation was
resistant to cetuximab. In another study, it was found that NRAS
mutation carriers showed a significantly lower response rate
compared with patients with wild-type KRAS when treated with
cetuximab (23). Nevertheless,
there is an ongoing debate over the association between BRAF
mutations and the effectiveness of anti-EGFR treatment (51). For example, a previous study
reported that RAS status was fully determined and a thorough
analysis of the association between clinicopathological and
molecular features indicated no statistically significant
association (52). Secondly, the
association between RAS subtypes and BRAF with patients'
clinicopathologicalfeatures was analyzed and the findings were
contradictory. Previous findings have indicated that KRAS mutations
are associated with different clinicopathological criteria,
although this association is not universally observed in all
studies (36,41,53).
In the present study, a multivariate analysis
demonstrated a significant association between KRAS mutations and
histological subtypes (41).
Additionally, there are numerous discrepancies and variations
regarding KRAS mutations and tumor locations or histological
subtypes (45). For example,
correlations were found between KRAS mutation and tumors in the
right colon (54) or rectal tumors
(28), whereas other studies
reported an association between KRAS mutations and a patient's age
(55), tumor site and
differentiation (55). The
association between NRAS mutations and clinicopathological features
was investigated in the present study. The results showed an
association with tumor grades. NRAS mutations were more frequent in
moderately differentiated tumors compared with poorly
differentiated tumors. However, a previous study established a
significant association between NRAS mutations and a patient's age
(56). Another study found an
association between NRAS mutations and the initial phases of cancer
and the lack of lymph node metastases (55). While other study found that NRAS
mutations are more commonly found in left-sided malignancies and
women (57), whereas Chang et
al (58) identified a link
between NRAS mutations and men. In addition, Shen et al
(59) reported that these mutations
occurred more frequently in distant metastatic tumors and their
occurrence varied depending on the different phases of the tumor.
Another study did not identify any associations (60). Notably, previous research conducted
on Western populations reported an association between BRAF-mutant
CRC and the female sex (61).
However, the present study did not find a statistically significant
association between BRAF mutations and sex. In contrast to the
study by Zhang et al (41),
which demonstrated a significant correlation between BRAF mutations
and right-sided colon cancer, the present study did not demonstrate
any significant association between BRAF mutations and tumor site
or any other clinicopathological features. However, it is important
to note that the findings of the present study are constrained by
the limited number of patients with a BRAF mutation, which amounted
to only 2 patients.
The present study has certain limitations, including
the limited sample size and single-centered nature of the present
study, which made it difficult to draw broader conclusions. The
association between KRAS, NRAS and BRAF mutations and the
clinicopathological features of patients with CRC may be further
understood in future investigations with larger patient groups.
The retrospective nature of the present study was
designed to demonstrate the association between RAS mutations and
clinicopathological variables regardless of the possible survival
benefits, and may help improve the life expectancy of patients
receiving anti-EGFR treatment with a wild-type RAS genotype. It can
be considered that the samples being studied inherently exhibit
variability in response to treatment. The therapeutic regimens
differed across the patients, which led to heterogeneity and may
have impacted the median overall survival, particularly in patients
with wild-type KRAS. This is because not all patients received
cetuximab and panitumumab as their initial treatment due to limited
availability and were thus treated with other chemotherapeutic
regimens. However, future work will focus on treatment efficacy and
overall patient survival, with predictive significance in selecting
patients for anti-EGFR therapy.
In conclusion, the present study examined the
association between clinicopathological characteristics and the
mutational landscape of mCRC in a cohort of patients. The results
emphasize the diversity of CRC and the need for individualized
treatment approaches, and highlight novel research opportunities,
particularly in comprehending the distinctive characteristics of
CRC in particular communities. Studies such as these serve a key
role in influencing clinical practice, determining future research
paths, and developing improved and personalized treatments for CRC
based on individual patient characteristics as a key objective in
the field of oncology. Furthermore, multicenter studies are
currently in the design phase to validate the generalizability in a
wide range of patients. These studies will additionally determine
the validity on patients of differing demographics, improving
reliability and accuracy in clinical settings worldwide. The
research may then be linked with clinical decision-making to help
drive personalized medicine and evidence-based therapeutics. Future
research is warranted to define the predictive significance of the
RAS and BRAF mutation status, and to discover any variations that
may benefit from different treatments based on patients' prognostic
values.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
RawA and RazA conceptualized the present study,
contributed to the drafting of the manuscript, and revised it
critically for important intellectual content. RoaA conducted the
formal analysis, contributed substantially to the acquisition and
interpretation of data, wrote parts of the manuscript, and
participated in revising the manuscript critically for important
intellectual content. AAl was involved in the design of the study,
validated the experimental procedures, contributed to the drafting
of the manuscript, and revised it critically for important
intellectual content. EQ used the SPSS software, entered the data
for analysis, and assisted in interpreting the data outcomes as
part of the manuscript's drafting and critical revision process.
RawA validated the study, curated the data, wrote the original
draft, and revised the manuscript critically. MO and AAb were
involved in the conceptualization and design of the study,
critically evaluated and interpreted the experimental data,
contributed to the drafting and critical revision of the manuscript
for important intellectual content, ensured the integrity and
accuracy of the work, and wrote, reviewed and edited the
manuscript, ensuring the interpretation and application of data
were appropriately conducted. AAb and RazA confirm the authenticity
of all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present investigation is characterized as an
observational retrospective study. All patients were treated
according to standard clinical protocols. The present study
received approval from the Institutional Review Board (IRB) of The
Hashemite University (Zarqa, Jordan; approval no. 13/9/2021/2022)
and Jordanian Royal Medical Services (Amman, Jordan). The
requirement for informed consent was waived by the Jordanian Royal
Medical Services (Amman, Jordan; IRB approval no. 2/2024, dated
12/2/2024).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
mCRC
|
metastatic CRC
|
|
EGFR
|
epidermal growth factor receptor
|
|
KRAS
|
Kirsten rat sarcoma viral oncogene
homolog
|
|
NRAS
|
neuroblastoma RAS viral oncogene
homolog
|
|
BRAF
|
v-Raf murine sarcoma viral oncogene
homolog B
|
References
|
1
|
Bonnot PE and Passot G: RAS mutation: Site
of disease and recurrence pattern in colorectal cancer. Chin Clin
Oncol. 8:55. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xi Y and Xu P: Global colorectal cancer
burden in 2020 and projections to 2040. Transl Oncol.
14:1011742021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jordan Cancer Registry, . Cancer Incidence
in Jordan. Minist Heal Jordan. 11:15–17. 2018.
|
|
5
|
Sharkas GF, Arqoub KH, Khader YS, Tarawneh
MR, Nimri OF, Al-Zaghal MJ and Subih HS: Colorectal cancer in
Jordan: Survival rate and its related factors. J Oncol.
2017:31807622017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saoudi González N, Salvà F, Ros J,
Baraibar I, Rodríguez-Castells M, García A, Alcaráz A, Vega S,
Bueno S, Tabernero J and Elez E: Unravelling the complexity of
colorectal cancer: Heterogeneity, clonal evolution, and clinical
implications. Cancers (Basel). 15:40202023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dienstmann R, Connor K and Byrne AT:
Precision therapy in RAS mutant colorectal cancer.
Gastroenterology. 158:806–811. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gong J, Cho M and Fakih M: RAS and BRAF in
metastatic colorectal cancer management. J Gastrointest Oncol.
7:6872016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsuchida N, Murugan AK and Grieco M:
Kirsten ras oncogene: Significance of its discovery in human cancer
research. Oncotarget. 7:467172016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lakatos G, Köhne CH and Bodoky G: Current
therapy of advanced colorectal cancer according to RAS/RAF
mutational status. Cancer Metastasis Rev. 39:1143–1157. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takane K, Akagi K, Fukuyo M, Yagi K,
Takayama T and Kaneda A: DNA methylation epigenotype and clinical
features of NRAS-mutation(+) colorectal cancer. Cancer Med.
6:1023–1035. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cercek A, Braghiroli MI, Chou JF, Hechtman
JF, Kemeny N, Saltz L, Capanu M and Yaeger R: Clinical features and
outcomes of patients with colorectal cancers harboring NRAS
mutations. Clin Cancer Res. 23:4753–4760. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Loree JM, Yu C, Tschautscher M,
Overman MJ, Broaddus R and Grothey A: Distinct impacts of KRAS,
NRAS and BRAF mutations on survival of patients with metastatic
colorectal cancer. J Clin Oncol. 36:3513. 2018. View Article : Google Scholar
|
|
14
|
Vigil D, Cherfils J, Rossman KL and Der
CJ: Ras superfamily GEFs and GAPs: Validated and tractable targets
for cancer therapy? Nat Rev Cancer. 10:842–857. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khan AQ, Kuttikrishnan S, Siveen KS,
Prabhu KS, Shanmugakonar M, Al-Naemi HA, Haris M, Dermime S and
Uddin S: RAS-mediated oncogenic signaling pathways in human
malignancies. Semin Cancer Biol. 54:1–13. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Colussi D, Brandi G, Bazzoli F and
Ricciardiello L: Molecular pathways involved in colorectal cancer:
Implications for disease behavior and prevention. Int J Mol Sci.
14:16365–16385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Newhall K, Price T, Peeters M, Kim TW, Li
J, Cascinu S, Ruff P, Suresh AS, Thomas A, Tjulandin S, et al:
Frequency of S492R mutations in the epidermal growth factor
receptor: Analysis of plasma dna from metastatic colorectal cancer
patients treated with panitumumab or cetuximab monotherapy. Ann
Oncol. 25:II1092014. View Article : Google Scholar
|
|
18
|
Karapetis CS, Khambata-Ford S, Jonker DJ,
O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD,
Robitaille S, et al: K-ras mutations and benefit from cetuximab in
advanced colorectal cancer. N Engl J Med. 359:1757–1765. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Meng M, Zhong K, Jiang T, Liu Z, Kwan HY
and Su T: The current understanding on the impact of KRAS on
colorectal cancer. Biomed Pharmacother. 140:1117172021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Richman SD, Seymour MT, Chambers P,
Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH and Quirke P:
KRAS and BRAF mutations in advanced colorectal cancer are
associated with poor prognosis but do not preclude benefit from
oxaliplatin or irinotecan: Results from the MRC FOCUS trial. J Clin
Oncol. 27:5931–5937. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nat.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li ZN, Zhao L, Yu LF and Wei MJ: BRAF and
KRAS mutations in metastatic colorectal cancer: Future perspectives
for personalized therapy. Gastroenterol Rep. 8:192–205. 2020.
View Article : Google Scholar
|
|
23
|
De Roock W, Claes B, Bernasconi D, De
Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V,
Papamichael D, Laurent-Puig P, et al: Effects of KRAS, BRAF, NRAS,
and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy
in chemotherapy-refractory metastatic colorectal cancer: A
retrospective consortium analysis. Lancet Oncol. 11:753–762. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kopetz S, Desai J, Chan E, Hecht JR,
O'Dwyer PJ, Lee RJ, Nolop KB and Saltz L: PLX4032 in metastatic
colorectal cancer patients with mutant BRAF tumors. J Clin Oncol.
28 (Suppl 15):S3534. 2010. View Article : Google Scholar
|
|
25
|
Avădănei ER, Căruntu ID, Nucă I, Balan RA,
Lozneanu L, Giusca SE, Pricope DL, Dascalu CG and Amalinei C: KRAS
mutation status in relation to clinicopathological characteristics
of romanian colorectal cancer patients. Curr Issues Mol Biol.
47:1202025. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li WQ, Kawakami K, Ruszkiewicz A, Bennett
G, Moore J and Iacopetta B: BRAF mutations are associated with
distinctive clinical, pathological and molecular features of
colorectal cancer independently of microsatellite instability
status. Mol Cancer. 5:22006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Koochak A, Rakhshani N, Karbalaie Niya MH,
Tameshkel FS, Sohrabi MR, Babaee MR, Rezvani H, Bahar B, Imanzade
F, Zamani F, et al: Mutation analysis of KRAS and BRAF genes in
metastatic colorectal cancer: A First large scale study from iran.
Asian Pacific J Cancer Prev. 17:603–608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kawazoe A, Shitara K, Fukuoka S, Kuboki Y,
Bando H, Okamoto W, Kojima T, Fuse N, Yamanaka T, Doi T, et al: A
retrospective observational study of clinicopathological features
of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with
metastatic colorectal cancer. BMC Cancer. 15:2582015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stintzing S, Tejpar S, Gibbs P, Thiebach L
and Lenz HJ: Understanding the role of primary tumour localisation
in colorectal cancer treatment and outcomes. Eur J Cancer.
84:69–80. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li C, Wang Q and Jiang KW: What is the
best surgical procedure of transverse colon cancer? An evidence map
and minireview. World J Gastrointest Oncol. 13:3912021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA;
WHO Classification of Tumours Editorial Board, : The 2019 WHO
classification of tumours of the digestive system. Histopathology.
76:182–188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Siegel RL, Wagle NS, Cercek A, Smith RA
and Jemal A: Colorectal cancer statistics, 2023. CA Cancer J Clin.
73:233–254. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pirvu EE, Severin E, Niţă I and Toma
Ștefania A: The impact of RAS mutation on the treatment strategy of
colorectal cancer. Med Pharm Rep. 96:5–15. 2023.PubMed/NCBI
|
|
34
|
Dempke WC and Heinemann V: Ras mutational
status is a biomarker for resistance to EGFR inhibitors in
colorectal carcinoma. Anticancer Res. 30:4673–4677. 2010.PubMed/NCBI
|
|
35
|
Ibrahimi AKH, Al-Hussaini M, Laban DA,
Ammarin R, Wehbeh L and Al-Mousa A: Cetuximab plus XELOX show
efficacy against brain metastasis from colorectal cancer: A case
report. CNS Oncol. 12:CNS972023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rimbert J, Tachon G, Junca A, Villalva C,
Karayan-Tapon L and Tougeron D: Association between
clinicopathological characteristics and RAS mutation in colorectal
cancer. Mod Pathol. 31:517–526. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sughayer WM and Sughayer MA: KRAS
mutations and subtyping in colorectal cancer in Jordanian patients.
Oncol Lett. 4:705–710. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Afrǎsânie VA, Marinca MV, Alexa-Stratulat
T, Gafton B, Păduraru M, Adavidoaiei AM, Miron L and Rusu C: KRAS,
NRAS, BRAF, HER2 and microsatellite instability in metastatic
colorectal cancer-practical implications for the clinician. Radiol
Oncol. 53:265–274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Levi M, Prayogi G, Sastranagara F,
Sudianto E, Widjajahakim G, Gani W, Mahanadi A, Agnes J, Khairunisa
BH and Utomo AR: Clinicopathological associations of K-RAS and
N-RAS mutations in indonesian colorectal cancer cohort. J
Gastrointest Cancer. 49:124–131. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ye ZL, Qiu MZ, Tang T, Wang F, Zhou YX,
Lei MJ, Guan WL and He CY: Gene mutation profiling in Chinese
colorectal cancer patients and its association with
clinicopathological characteristics and prognosis. Cancer Med.
9:745–756. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang J, Zheng J, Yang Y, Lu J, Gao J, Lu
T, Sun J, Jiang H, Zhu Y, Zheng Y, et al: Molecular spectrum of
KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer
patients: Analysis of 1,110 cases. Sci Rep. 5:186782015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang X, Ran W, Wu J, Li H, Liu H, Wang L,
Xiao Y, Wang X, Li Y and Xing X: Deficient mismatch repair and RAS
mutation in colorectal carcinoma patients: A retrospective study in
Eastern China. PeerJ. 6:e43412018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jauhri M, Bhatnagar A, Gupta S, Manasa BP,
Minhas S, Shokeen Y and Aggarwal S: Prevalence and coexistence of
KRAS, BRAF, PIK3CA, NRAS, TP53, and APC mutations in Indian
colorectal cancer patients: Next-generation sequencing-based cohort
study. Tumor Biol. 39:10104283176922652017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Negru S, Papadopoulou E, Apessos A,
Stanculeanu DL, Ciuleanu E, Volovat C, Croitoru A, Kakolyris S,
Aravantinos G, Ziras N, et al: KRAS, NRAS and BRAF mutations in
Greek and Romanian patients with colorectal cancer: A cohort study.
BMJ Open. 4:e0046522014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Al-Shamsi HO, Jones J, Fahmawi Y, Dahbour
I, Tabash A, Abdel-Wahab R, Abousamra AO, Shaw KR, Xiao L, Hassan
MM, et al: Molecular spectrum of KRAS, NRAS, BRAF, PIK3CA, TP53,
and APC somatic gene mutations in Arab patients with colorectal
cancer: Determination of frequency and distribution pattern. J
Gastrointest Oncol. 7:882–902. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ouerhani S, Bougatef K, Soltani I,
Elgaaied ABA, Abbes S and Menif S: The prevalence and prognostic
significance of KRAS mutation in bladder cancer, chronic myeloid
leukemia and colorectal cancer. Mol Biol Rep. 40:4109–4114. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Neumann J, Zeindl-Eberhart E, Kirchner T
and Jung A: Frequency and type of KRAS mutations in routine
diagnostic analysis of metastatic colorectal cancer. Pathol Res
Pract. 205:858–862. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Baldus SE, Schaefer KL, Engers R, Hartleb
D, Stoecklein NH and Gabbert HE: Prevalence and heterogeneity of
KRAS, BRAF, and PIK3CA mutations in primary colorectal
adenocarcinomas and their corresponding metastases. Clin Cancer
Res. 16:790–799. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Therkildsen C, Bergmann TK,
Henrichsen-Schnack T, Ladelund S and Nilbert M: The predictive
value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment
in metastatic colorectal cancer: A systematic review and
meta-analysis. Acta Oncol (Madr). 53:852–864. 2014. View Article : Google Scholar
|
|
50
|
Loupakis F, Ruzzo A, Cremolini C, Vincenzi
B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E,
et al: KRAS codon 61, 146 and BRAF mutations predict resistance to
cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type
metastatic colorectal cancer. Br J Cancer. 101:715–721. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Karapetis CS, Jonker D, Daneshmand M,
Hanson JE, O'Callaghan CJ, Marginean C, Zalcberg JR, Simes J, Moore
MJ, Tebbutt NC, et al: PIK3CA, BRAF, and PTEN status and benefit
from cetuximab in the treatment of advanced colorectal
cancer-results from NCIC CTG/AGITG CO.17. Clin Cancer Res.
20:744–753. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mounjid C, El Agouri H, Mahdi Y, Laraqui
A, Chtati E, Ech-charif S, Khmou M, Bakri Y, Souadka A and Souadka
BE: Assessment of KRAS and NRAS status in metastatic colorectal
cancer: Experience of the national institute of oncology in rabat
morocco. Ann Cancer Res Ther. 30:80–84. 2022. View Article : Google Scholar
|
|
53
|
Zanatto RM, Santos G, Oliveira JC,
Pracucho EM, Nunes AJF, Lopes-Filho GJ and Saad SS: Impact of kras
mutations in clinical features in colorectal cancer. Arq Bras Cir
Dig. 33:e15242020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Taïeb J, Emile JF, Malicot K, Zaanan A,
Tabernero J, Mini E, Rougier P, Van Laethem JL, Bridgewater JA,
Folprecht G, et al: Prognostic value of KRAS exon 2 gene mutations
in stage III colon cancer: Post hoc analyses of the PETACC8 trial.
J Clin Oncol. 32:3549. 2014. View Article : Google Scholar
|
|
55
|
Ounissi D, Weslati M, Boughriba R, Hazgui
M and Bouraoui S: Clinicopathological characteristics and
mutational profile of KRAS and NRAS inTunisian patients with
sporadic colorectal cancer. Turkish J Med Sci. 51:148–158.
2021.PubMed/NCBI
|
|
56
|
Mahdi Y, Khmou M, Souadka A, Agouri H El,
Ech-charif S, Mounjid C and Khannoussi B El: Correlation between
KRAS and NRAS mutational status and clinicopathological features in
414 cases of metastatic colorectal cancer in Morocco: The largest
North African case series. BMC Gastroenterol. 23:1932023.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Irahara N, Baba Y, Nosho K, Shima K, Yan
L, Dias-Santagata D, Iafrate AJ, Fuchs CS, Haigis KM and Ogino S:
NRAS mutations are rare in colorectal cancer. Diagnostic Mol
Pathol. 19:157–163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chang YY, Lin PC, Lin HH, Lin JK, Chen WS,
Jiang JK, Yang SH, Liang WY and Chang SC: Mutation spectra of RAS
gene family in colorectal cancer. Am J Surg. 212:537–544.e3. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shen Y, Wang J, Han X, Yang H, Wang S, Lin
D and Shi Y: Effectors of epidermal growth factor receptor pathway:
The genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in
colorectal cancer characteristics and personalized medicine. PLoS
One. 8:e816282013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jouini R, Ferchichi M, BenBrahim E, Ayari
I, Khanchel F, Koubaa W, Saidi O, Allani R and Chadli-Debbiche A:
KRAS and NRAS pyrosequencing screening in Tunisian colorectal
cancer patients in 2015. Heliyon. 5:e013302019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Tran B, Kopetz S, Tie J, Gibbs P, Jiang
ZQ, Lieu CH, Agarwal A, Maru DM, Sieber O and Desai J: Impact of
BRAF mutation and microsatellite instability on the pattern of
metastatic spread and prognosis in metastatic colorectal cancer.
Cancer. 117:4623–4632. 2011. View Article : Google Scholar : PubMed/NCBI
|