Introduction
Osteosarcoma (OS) is an aggressive primary malignant
bone tumor originating from abnormal osteogenic cells, which
predominantly affects adolescents and young adults (1,2).
Globally, it accounts for ~2% of pediatric cancers and 20% of
primary bone tumors (3). Despite
its relatively low incidence, the high mortality rate associated
with OS is a cause for grave concern. For patients with localized
OS, the 5-year survival rate is 60–70%, whereas for patients with
metastatic OS, the 5-year survival rate is only 15–30% (4). Current treatment strategies primarily
involve aggressive surgical resection, often necessitating
limb-sparing (5) or amputation
procedures (6), in conjunction with
adjuvant chemotherapy (7). However,
the toxicity of chemotherapy and the potential for drug resistance
are formidable challenges in the management of this malignancy.
Cancer cells exploit changes in metabolic pathways
as a key strategy to acquire the characteristics necessary for
progression and metastasis (8). The
tricarboxylic acid cycle is aided by propionate metabolism, which
is a downstream process of lipid metabolism that serves as a source
of energy (9). In atherosclerosis,
propanoic acid, generated through intestinal cholesterol
metabolism, has been shown to reduce low-density lipoprotein and
total cholesterol levels in the blood, thereby alleviating
atherosclerotic conditions (10).
Research has suggested that flaxseed oil and vitamin E improve
sperm quality and embryo development by modulating the
4-aminobutyrate aminotransferase (ABAT) gene, which plays a crucial
role in propionate metabolism (11). In addition, the dysregulation of
propionate metabolism in breast and lung cancer cells has been
found to be associated with invasive characteristics and increased
metastatic potential (12).
However, the role and mechanisms of propionate metabolism in OS
remain poorly understood.
The aim of the present study was to identify novel
prognostic indicators for OS based on gene expression profiles and
clinical data. First, bioinformatics methods were employed to
explore propionate metabolism-related genes associated with OS.
These genes were used to establish a prognostic model to gain
deeper insights into the associations between OS and the immune
microenvironment, as well as interacting pathways. Second,
functional assays were performed to study the impact of key
propionate metabolism-related genes in OS on cell proliferation,
migration and invasion. Using this multifaceted approach, combining
bioinformatics analysis with functional experiments, the present
study sought to elucidate the role of propionate metabolism in
OS.
Materials and methods
Data acquisition
The GSE12865 dataset from the Gene Expression
Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) was used for
differential gene expression analysis (13). The GSE12865 gene expression data,
comprising 12 tumor and 2 normal tissues, were derived from the
GPL6244 platform. The GSE162454 single-cell RNA data, comprising
primary OS tissues from six patients, were derived from the
GPL24676 platform (14). In
addition, clinical information and RNA sequencing results for
patients with OS were obtained from the TARGET database (https://www.cancer.gov/ccg/research/genome-sequencing/target/about).
Also, the MSigDB (https://www.gsea-msigdb.org/gsea/index.jsp) was
searched using the keyword ‘METABOLISM’, and the
‘KEGG_PROPANOATE_METABOLISM’ collection was retrieved. The original
datasets were formatted for analysis in R (version 4.0.4,
r-project.org/), and the limma package was employed for data
normalization and differentially expressed gene (DEG)
identification within the GEO dataset. The DEG selection criteria
were: Upregulation, fold change (FC) >1.3, P<0.05, and
downregulation, FC <0.77, P<0.05.
Functional enrichment analysis
To elucidate protein-protein interactions (PPIs),
the STRING database (version 10.5, string-db.org/) was used. Genes
with interaction scores >0.4 were used to construct a PPI
network among the propionate metabolism-related genes and DEGs,
which were visualized and analyzed using Cytoscape software
(version 3.4). DAVID (version 6.8; http://david.ncifcrf.gov/) was employed for Gene
Ontology (GO) (15) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) analyses, with the
threshold set at P<0.05.
Cox proportional hazards model
construction
Cox univariate regression was performed on the
datasets of patients with OS from the TARGET database using the
survival package in R. The glmnet package was then used to conduct
regression analysis via the least absolute shrinkage and selection
operator (LASSO)-Cox method, combining information on gene
expression, survival status and time. The overall survival risk
score was calculated as follows: Risk score=∑βi ×
expGenei, where expGene is the gene expression level and
β is the regression coefficient obtained from the model. The median
risk score was then determined to classify patients into high- and
low-risk groups. The proportional hazards assumption of the Cox
proportional hazards model was estimated using Kaplan-Meier
analysis. Receiver operating characteristic (ROC) curves were drawn
in R using the survival ROC package, and the area under the curve
(AUC) was calculated. In addition, the data for each patient were
analyzed and visualized using R software.
Clinical and pathological factor
analysis
Using samples and clinical information of the OS
cohort from the TARGET database, clinicopathological factors
including the risk model, age and sex were incorporated into the
risk model. Univariate and multivariate Cox analysis was
subsequently performed using these clinicopathological parameters.
The rms and survival packages were used to design a nomogram to
predict the survival duration of of patients with OS. Finally,
calibration curves and the Harrell concordance index (C-index) were
evaluated to assess the predictive accuracy of the nomogram.
Characterization of the tumor
microenvironment (TME)
The CIBERSORT algorithm was employed to compute the
abundance of 22 types of tumor-infiltrating immune cells (TIICs)
based on OS expression profiles from the TARGET database. The
correlations of the levels of these TIICs with the expression
levels of key genes were subsequently analyzed. In addition, the
ESTIMATE tool was used to assess the stromal and immune cell
infiltration of malignant tumors, and provide stromal,
immunological and ESTIMATE scores for the OS TME.
Tumor immune dysfunction and exclusion
(TIDE) analysis
The TIDE algorithm was applied to calculate TIDE
scores (16). A lower TIDE score
indicates a higher likelihood of responding to immunotherapy.
Common immune checkpoint expression profiles were downloaded and
examined for variations in expression among molecular subtypes
(17).
Gene set enrichment analysis (GSEA)
for risk subgroups
GSEA was employed to identify significantly enriched
gene sets. The clusterProfiler package was used to perform the
GSEA, with all candidate gene sets from the Hallmark database
serving as the background set. The cutoff for significance was set
at P<0.05.
Tumor immune single-cell hub (TISCH)
database
The TISCH database is a repository for data on
TME-related genes, which aggregates data from 27 tumor datasets
across 76 cancer types, encompassing nearly 20,000 single-cell
transcriptomes (18). TISCH was
employed to comprehensively study the heterogeneity of the TME in
diverse cell types and cancer categories.
Cell lines and maintenance
The osteocyte cell line hFOB1.19 (GNHu14) and four
OS cell lines (U2OS, SCSP-5030; HOS, TCHu167; MG63, TCHu124; and
SAOS-2, TCHu114) were purchased from The Cell Bank of Type Culture
Collection of The Chinese Academy of Sciences and cultured in DMEM
(cat. no. GNM31051-2; Gino Life & Health Holding Group Ltd.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin G (MedChemExpress) and 100 µg/ml streptomycin
(Sigma-Aldrich). The cells were maintained at 37°C with 5%
CO2 in a humidified incubator.
Reverse transcription-quantitative PCR
(RT-qPCR)
Using TRIzol® reagent (Invitrogen,
Beijing, China), cellular RNA was collected from the five cell
lines. The RNA percentage was determined using a NanoDrop
spectrophotometer (Thermo Fisher Scientific, Inc.). According to
the manufacturer's instructions, cDNA was synthesized from the RNA
using a PrimeScript II 1st strand cDNA Synthesis Kit (6210A; Takara
Bio, Inc.). qPCR was performed on an ABI PRISM 7900HT Sequence
Detection System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with SYBR™ Green Universal Master Mix (4344463; Thermo Fisher
Scientific, Inc.). The thermocycling conditions were 4 min at 94°C,
followed by 40 cycles of 30 sec at 94°C, 30 sec at the annealing
temperature and 30 sec at 72°C. The transcript levels of ABAT were
normalized to those of GAPDH. The mRNA expression levels were
calculated using the 2−ΔΔCq method (19). The primer sequences (Sangon Biotech
Co., Ltd.) are listed in Table
I.
 | Table I.Primers for reverse
transcription-quantitative PCR. |
Table I.
Primers for reverse
transcription-quantitative PCR.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| ABAT |
CTTCCGTCTTCATCAGAGGC |
CAGCTTCCAGCACAGCTACC |
| GAPDH |
CATGAGAAGTATGACAACAGCCT |
AGTCCTTCCACGATACCAAAGT |
Small interfering RNA (siRNA)
transfection
siRNAs targeting ABAT (si-ABAT-1, si-ABAT-2 and
si-ABAT-3; 10 nM) and a negative control siRNA (si-NC) were
obtained from Guangzhou RiboBio Co., Ltd. The siRNA sequences are
listed in Table II. Transfection
of OS cells was performed using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc) with 1 µg siRNA per well. After
transfection for 6 h (37°C), the cell medium was replaced, and the
cells were cultured for an additional 48 h before being collected
for subsequent experiments. The transfection effectiveness was
evaluated via RT-qPCR.
 | Table II.siRNA sequences. |
Table II.
siRNA sequences.
| Name | Sense (5′-3′) | Antisense
(5′-3′) |
|---|
| si-ABAT-1 |
UGGAAGAGUUUGUGAAAGAUU |
UCUUUCACAAACUCUUCCAUU |
| si-ABAT-2 |
CGGAGAACUUUGUGGAGAAUU |
UUCUCCACAAAGUUCUCCGUU |
| si-ABAT-3 |
GGGAGGACCUGCUAAAUAAUU |
UUAUUUAGCAGGUCCUCCCUU |
| NC |
UUCUCCGAACGUGUCACGUTT |
ACGUGACACGUUCGGAGAATT |
Protein extraction and western
blotting (WB)
The transfected OS cells were lysed using RIPA
buffer (P0013, Beyotime, Shanghai, China), and the total protein
concentration of the lysate was determined using a BCA protein
assay kit (Beyotime Institute of Biotechnology). The proteins (50
µg) were then separated by 10% SDS-PAGE and transferred to PVDF
membranes (Bio-Rad Laboratories, Inc.). After blocking with 3%
skimmed milk at room temperature for 1 h, the membranes were
incubated with anti-ABAT (1:1,000; ab216465, Abcam) and anti-GAPDH
(1:1,000; ab9485, Abcam) primary antibodies at 4°C overnight. After
washing, the membranes were further incubated at room temperature
for 1 h with secondary HRP-conjugated goat anti-rabbit antibodies
(1:5,000; ab6721, Abcam). Odyssey imaging equipment (LI-COR
Biosciences) was used to scan the blots, and Odyssey v2.0 software
(LI-COR Biosciences) was used to evaluate the results using GAPDH
as the internal control.
Cell viability assay
Following transient transfection for 24 h, the cells
were placed in 48-well plates (4,000 cells/well). On days 1, 2, 3,
4 and 5 of culture, Cell Counting Kit-8 (CCK-8) solution (cat. no.
P0038, Beyotime Institute of Biotechnology) was added to the cells
and the plates were incubated at 37°C for 2 h The optical density
was subsequently assessed at 450 nm using a Synergy 3 microplate
reader (Biotek; Agilent Technologies, Inc.).
Transwell assays
Transwell chambers (pores, 8 µm; BD Biosciences)
were used to evaluate the migration and invasion of the transfected
OS cells. The transfected cells (5×104) were seeded into
the upper chamber of the Transwell chamber. The upper chamber was
supplemented with serum-free culture medium, whereas the medium in
the lower chamber was supplemented with 10% FBS. For invasion
experiments, a Matrigel (cat. no. 356234, Biolead, Beijing, China)
coating was applied to the upper chamber. The Matrigel was allowed
to solidify at 37°C for 30 min before the cells were added to the
top chamber. The cells were cultured for 24 h at 37°C in a 5%
CO2 atmosphere, fixed, stained with crystal violet
(0.1%) for 15 min (room temperature) and observed under a light
microscope (magnification, ×200) to assess migration and invasion.
Quantification was performed using Image J software (version
1.53t).
Statistical analysis
All assays were repeated no less than three times to
ensure reproducibility. Statistical analyses were performed using
GraphPad Prism 8.0 (Dotmatics). Data are presented as the mean ±
standard deviation. Comparisons between two groups were conducted
using unpaired Student's t-tests, and multiple group comparisons
were made using one-way ANOVA followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
result.
Results
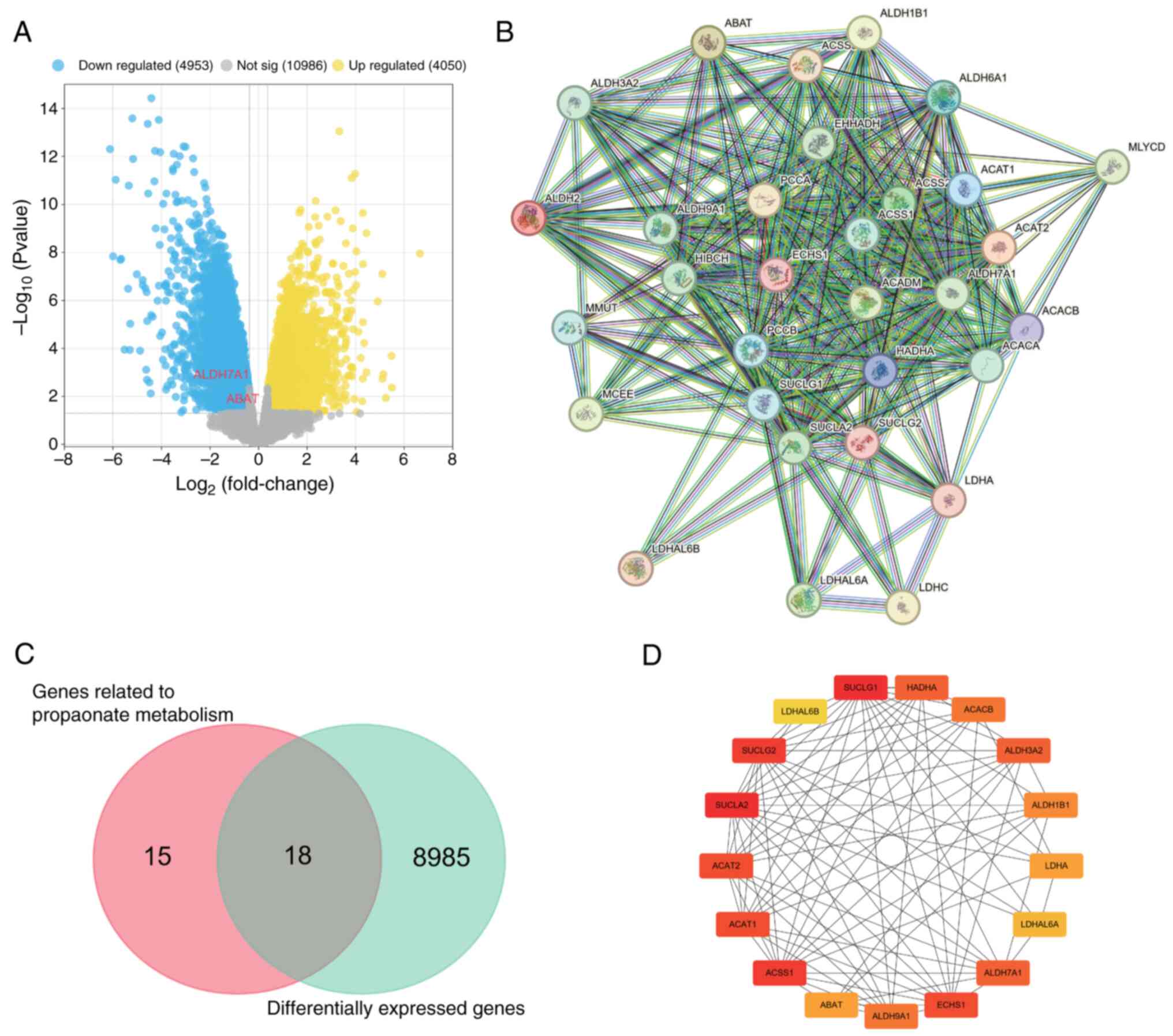
OS-DEGs associated with propionate
metabolism
In the GSE12865 gene set, the differential gene
expression analysis revealed a total of 9,003 DEGs, including 4,050
upregulated and 4,953 downregulated genes (Fig. 1A). Using the MSigDB dataset, a set
of 33 genes associated with propionate metabolism was obtained, and
a PPI network was constructed (Fig.
1B). Venn diagram analysis identified 18 DEGs in the GSE12865
dataset that are associated with propionate metabolism (Fig. 1C). Since these genes displayed
significant differences in expression between tumor and normal
tissues and were associated with propionate metabolism, they were
considered suitable for in-depth analysis. Using Cytoscape, a PPI
network incorporating hub genes was constructed based on the
aforementioned 18 genes. The hub genes comprised SUCLA2, SUCLG1,
succinate-CoA ligase GDP-forming b subunit (SUCLG2), ACSS1, ACAT2,
ACAT1, ECHS1, HADHA, aldehyde dehydrogenase 7 family member A1
(ALDH7A1), ALDH3A2, ACACB, aldehyde dehydrogenase 9 family member
A1 (ALDH9A1), ALDH1B1, LDHA, ABAT, LDHAL6A and LDHAL6B (Fig. 1D).
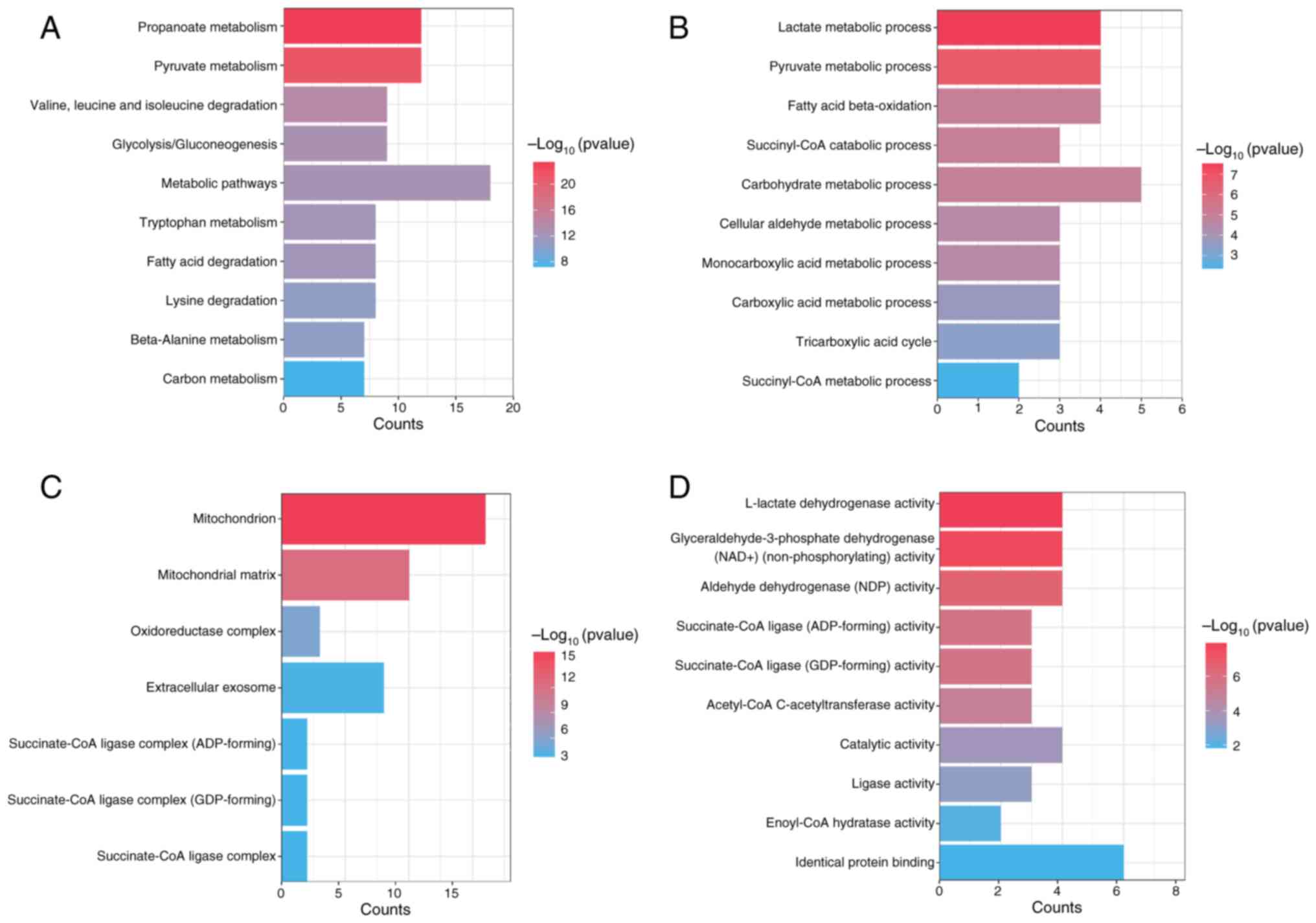
Functional enrichment analysis reveals
insights into DEG functions and mechanisms
To elucidate the mechanisms of the DEGs, functional
enrichment analysis was performed. KEGG analysis revealed
significant associations between propionate metabolism-related DEGs
and several pathways, including ‘carbon metabolism’, ‘metabolic
pathways’, ‘pyruvate metabolism’ and ‘propanoate metabolism’
(Fig. 2A). GO functional enrichment
analysis of the propionate metabolism related-DEGs highlighted
their significant involvement in various biological processes, such
as the ‘succinyl-CoA metabolic process’, ‘carbohydrate metabolic
process’ and ‘fatty acid beta-oxidation’ (Fig. 2B). Additionally, these genes were
associated with cellular components, including ‘succinate-CoA
ligase complex’, ‘extracellular exosome’, ‘oxidoreductase complex’,
‘mitochondrial matrix’ and ‘mitochondrion’ (Fig. 2C). Furthermore, GO molecular
function enrichment analysis revealed the involvement of these
genes in functions including ‘identical protein binding’,
‘enoyl-CoA hydratase activity’, ‘ligase activity’, ‘aldehyde
dehydrogenase (NAD) activity’ and ‘L-lactate dehydrogenase
activity’ (Fig. 2D). These findings
provide insights into the functional roles and mechanisms of the
DEGs.
Identification of prognostic genes and
development of the risk model
Four genes were identified as prognostic genes using
single-factor Cox analysis: ABAT, ALDH7A1, ALDH9A1 and SUCLG2
(Fig. 3A). The selection of these
four genes with a λ-value of 0.05 was subsequently confirmed by
LASSO Cox regression (Fig. 3B and
C). Multivariate stepwise Cox analysis ultimately selected two
genes, ABAT and ALDH7A1, for inclusion in a propionate
metabolism-related gene signature. The formula for the computation
of patient-specific risk scores was as follows: Risk
score=(−0.2968) × ABAT + (−0.1483) × ALDH7A1. The median risk score
was subsequently applied to classify patients into high- and
low-risk groups (Fig. 3D). Survival
curves and scatterplots of risk scores and survival curves
demonstrated that deceased patients were predominantly concentrated
in the high-risk group (Fig. 3D).
Kaplan-Meier analysis revealed significant differences in overall
survival outcomes between the two patient risk groups, indicating a
worse prognosis in the high-risk group (Fig. 3E). Time-dependent ROC curves for 1-,
3- and 5-year overall survival demonstrated AUC values of 0.76,
0.75 and 0.75, respectively (Fig.
3F), highlighting the strong prognostic potential of the risk
model.
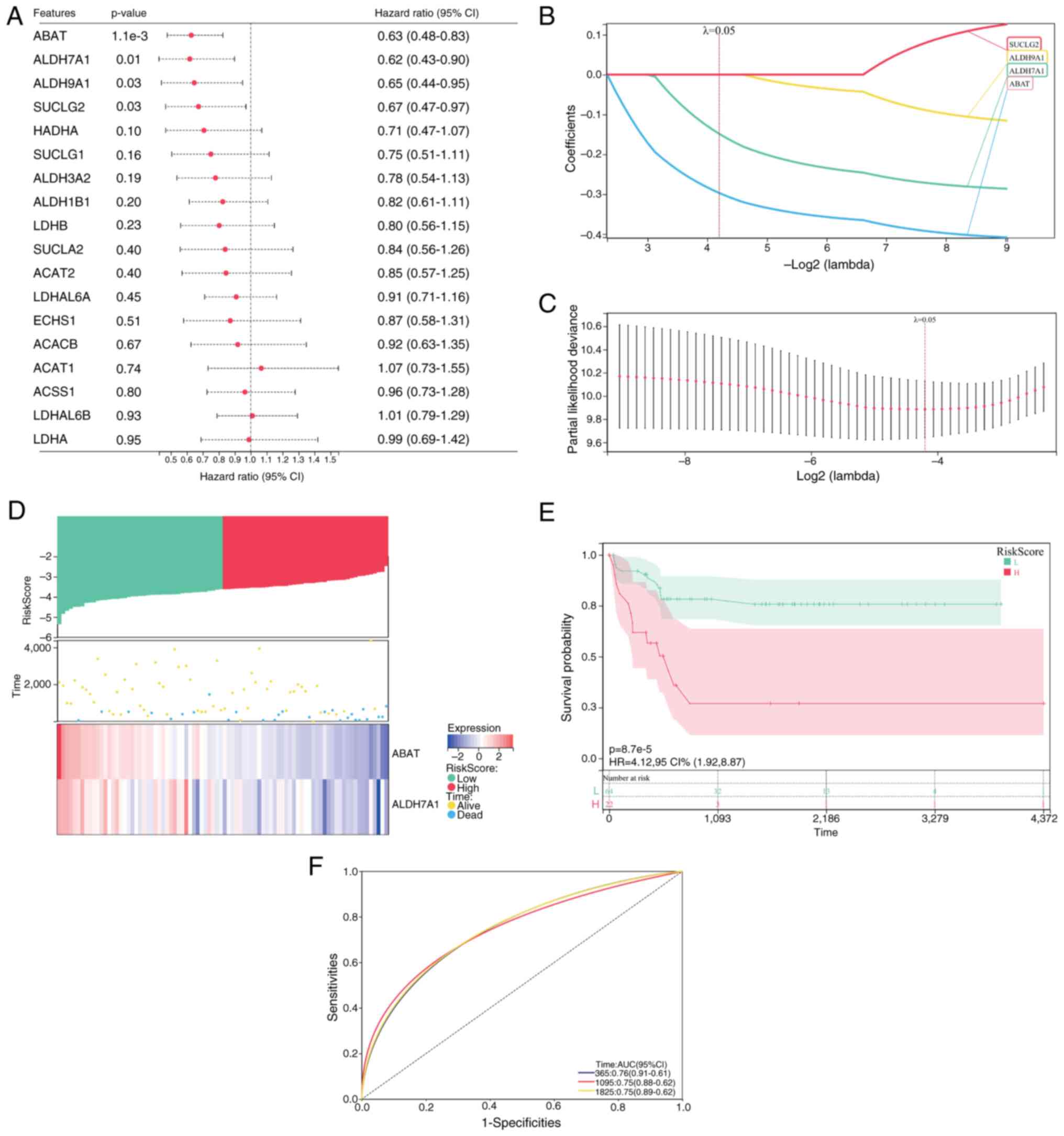 | Figure 3.Construction of the propionate
metabolism-related gene signature based on data of patients with OS
obtained from the TARGET database. (A) Cox univariate analysis of
18 propionate metabolism-related genes. (B) LASSO coefficient
profiles of four propionate metabolism-related genes identified
from the univariate analysis. (C) Relationship between partial
likelihood deviance and log2(λ) in LASSO regression. (D)
Patient risk scores, survival status and distribution of ABAT and
ALDH7A1 gene expression levels. (E) Kaplan-Meier survival curve
analysis of overall survival for patients with OS based on the risk
score. (F) Receiver operating characteristic curve analysis of the
2-gene signature. OS, osteosarcoma; LASSO, least absolute shrinkage
and selection operator; ABAT, 4-aminobutyrate aminotransferase;
ALDH7A1, aldehyde dehydrogenase 7 family member A1; ALDH9A1,
aldehyde dehydrogenase 9 family member A1; SUCLG2, succinate-CoA
ligase GDP-forming b subunit; HR, hazard ratio; AUC, area under the
curve; 95% CI, 95% confidence interval. |
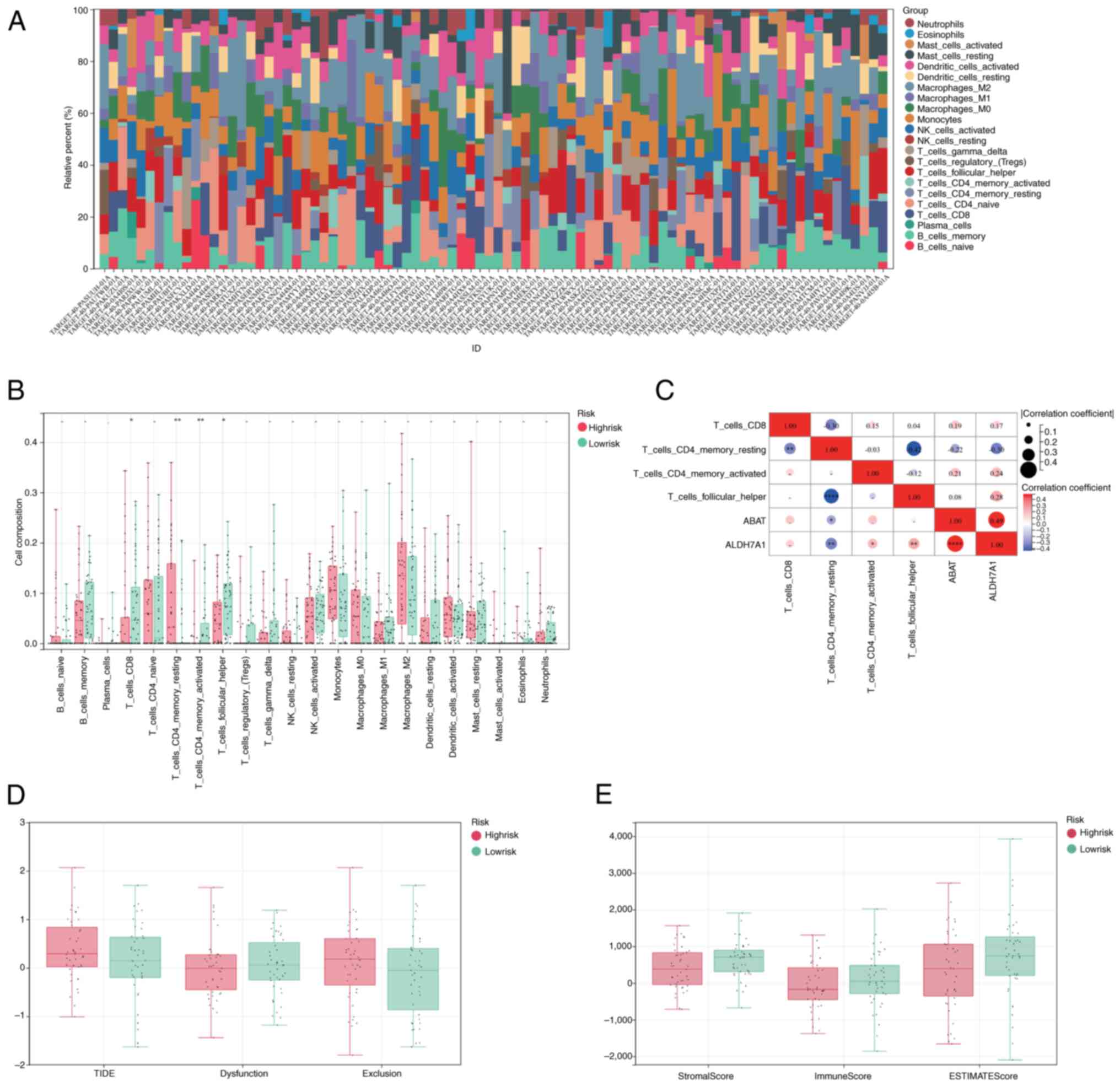
Immune infiltration and TIDE
analysis
CIBERSORT was used to calculate the percentages of
different immune cell types in the OS cohort from the TARGET
database. Following profiling of the immune infiltration patterns
of patients with OS, a marked difference in immune cell
infiltration was detected between the high- and low-risk groups
(Fig. 4A). In the TARGET-OS cohort,
statistically significant differences in the infiltration of
CD8+ T cells, resting and activated CD4+
memory cells, and follicular helper cells were observed between the
two groups (Fig. 4B). Furthermore,
ABAT levels were correlated with the infiltration of resting
CD4+_memory_T cells, and ALDH7A1 was correlated with the
infiltration of resting and activated CD4+ memory T
cells, CD8+ T cells and follicular_helper cells
(Fig. 4C). Patients in the
high-risk group presented higher TIDE and exclusion scores and
lower dysfunction scores compared with those in the low-risk group
(Fig. 4D). In addition, higher
stromal, immune and ESTIMATE scores were associated with the
low-risk group (Fig. 4E).
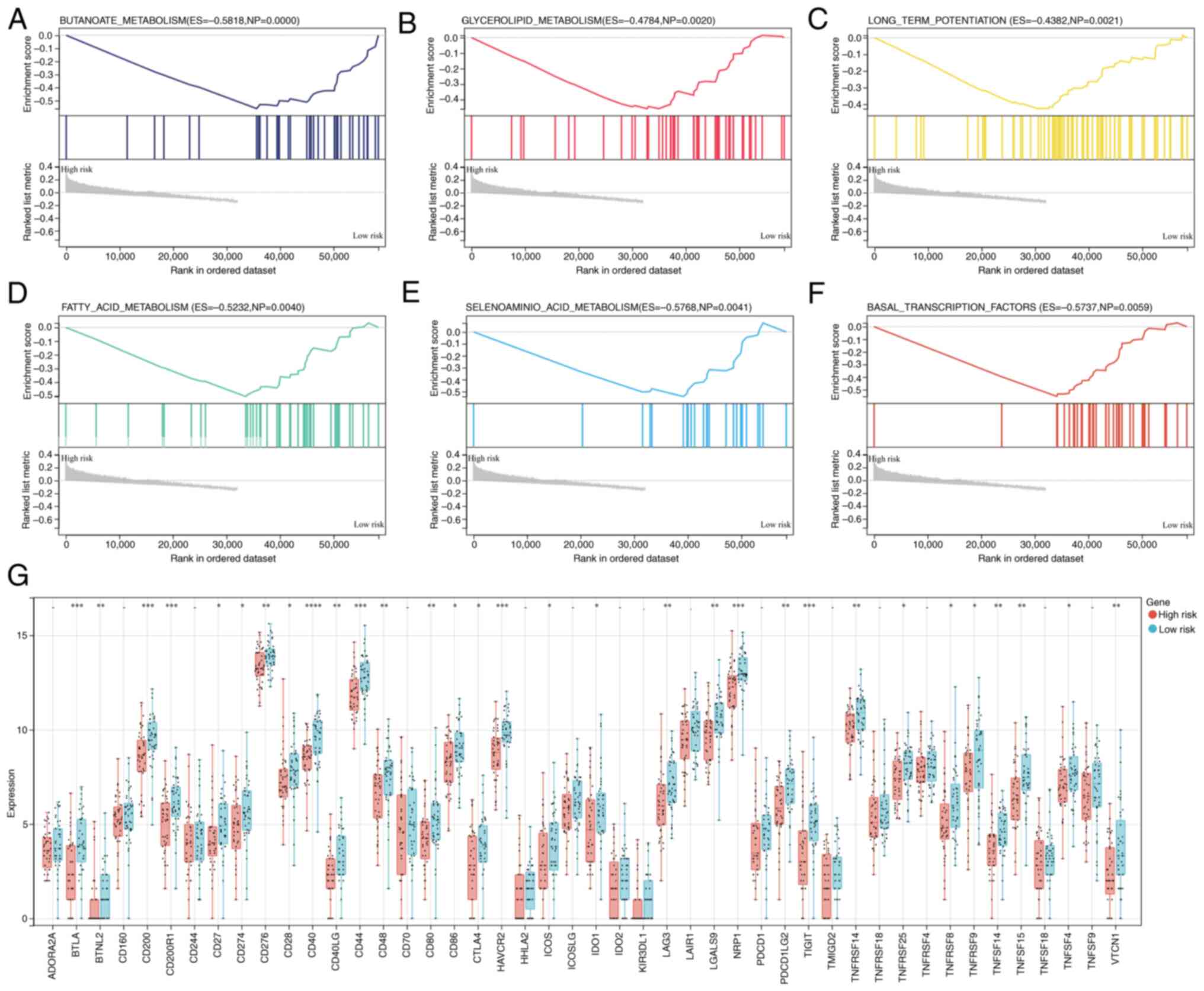
Association of the gene signature with
immune features
Following the initial demonstration of the
relationship between prognosis and the constructed gene signature
for OS, the impact of the gene signature on cancer immune features
was further explored. GSEA revealed that the low-risk group was
closely associated with the KEGG pathways ‘BUTANOATE_METABOLISM’,
‘GLYCEROLIPID_METABOLISM’, ‘LONG_TERM_POTENTIATION’,
‘FATTY_ACID_METABOLISM’, ‘SELENOAMINO_ACID_METABOLISM’ and
‘BASAL_TRANSCRIPTION_FACTORS’ (Fig.
5A-F). In addition, the levels of immune checkpoint molecules
in the two groups were also investigated. The low-risk group
presented increased expression of numerous genes, including BTNL2,
ICOS and TNFRSF9 (Fig. 5G).
Prognostic factors and calibration of
the risk score model
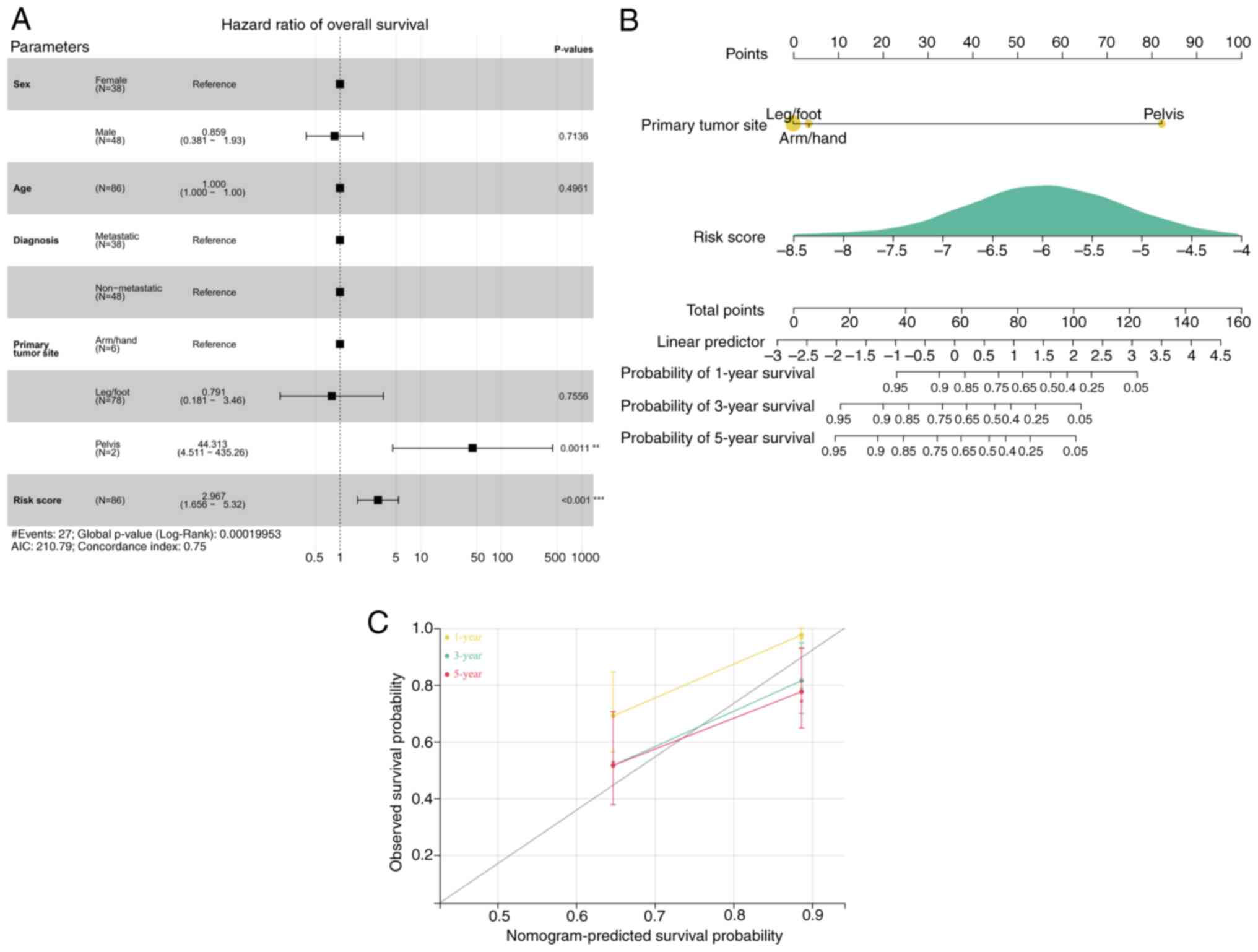
Cox regression analysis demonstrated that the risk
score and primary tumor site were prognostic parameters for OS
(Fig. 6A). A nomogram was
subsequently generated based on the risk model and primary tumor
site. According to a previous report, the c-index is 0.75,
indicating a reasonable predictive value (20). Our results also demonstrate a
reasonable predictive value (Fig.
6B). The calibration curves of the predictions at years 1, 3
and 5 closely aligned with the observed outcomes (Fig. 6C).
Single-cell expression analysis of
ABAT and ALDH7A1
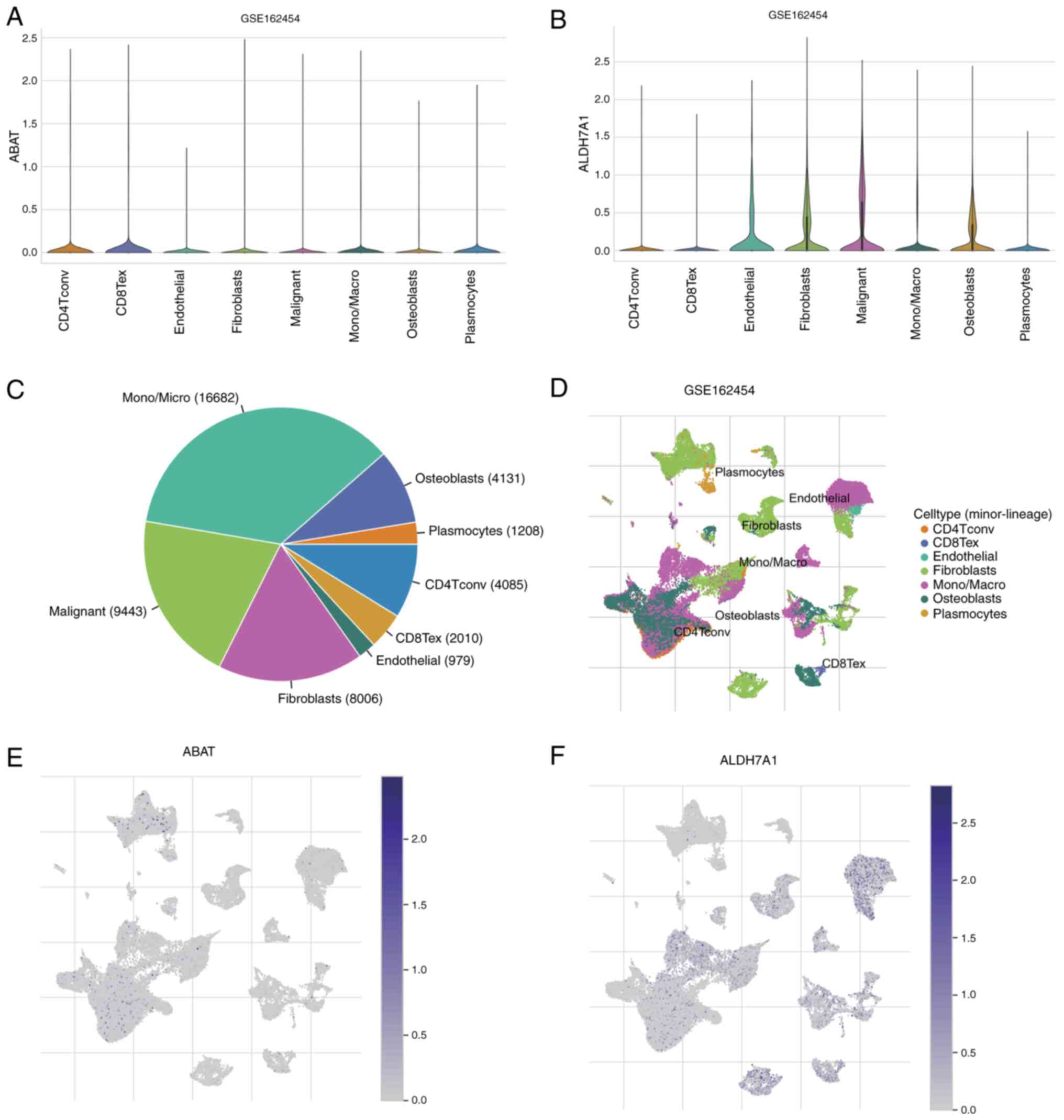
The distributions of ABAT and ALDH7A1 expression in
different types of cell in patients with OS from the GSE162454
dataset are shown in Fig. 7A and B.
Further analysis of this dataset, showing the cell counts of seven
cell types, is depicted in Fig. 7C,
and the percentages and quantities of these TME-related cells are
represented in Fig. 7D. t-SNE was
applied to visualize the cell clusters, with cells having similar
characteristics grouped together (Fig.
7D). These findings revealed that the predominant immune cells
are monocytes/macrophages (mono/macro cells; n=16,682). In the
dataset, ABAT was primarily concentrated in plasmocytes and
CD4+ T conventional (CD4Tconv) cell clusters (Fig. 7E), whereas ALDH7A1 was mainly
localized in mono/macro cells, endothelial cells, fibroblasts and
CD8+ T exhausted cell clusters (Fig. 7F). This indicates that ABAT and
ALDH7A1 may have functions in immune cells as well as in cancer
cells.
ABAT knockdown significantly promotes
OS cell growth
To study the role of ABAT in OS, RT-qPCR analysis
was performed on a panel of OS cell lines. As shown in Fig. 8A, ABAT expression was downregulated
in the SAOS-2, HOS and U2OS OS cell lines compared with that in the
hFOB1.19 osteoblast cell line, with the lowest expression observed
in SAOS-2 cells. Therefore, follow-up experiments were conducted
using the HOS and U2OS cell lines. Subsequently, RT-qPCR analysis
indicated that ABAT expression in OS cells was silenced by ABAT
siRNA, and WB results were consistent with these findings (Fig. 8B-E). siABAT-2 exhibited the highest
transfection efficiency and so was used in subsequent experiments.
CCK-8 analysis revealed that ABAT knockdown significantly increased
OS cell viability compared with that in the si-NC group (Fig. 8F and G). In addition, Transwell
assays demonstrated that ABAT knockdown markedly promoted OS cell
migration (Fig. 8H) and invasion
(Fig. 8I) compared with that in the
si-NC group. In summary, these results indicated that ABAT
knockdown promoted OS cell progression.
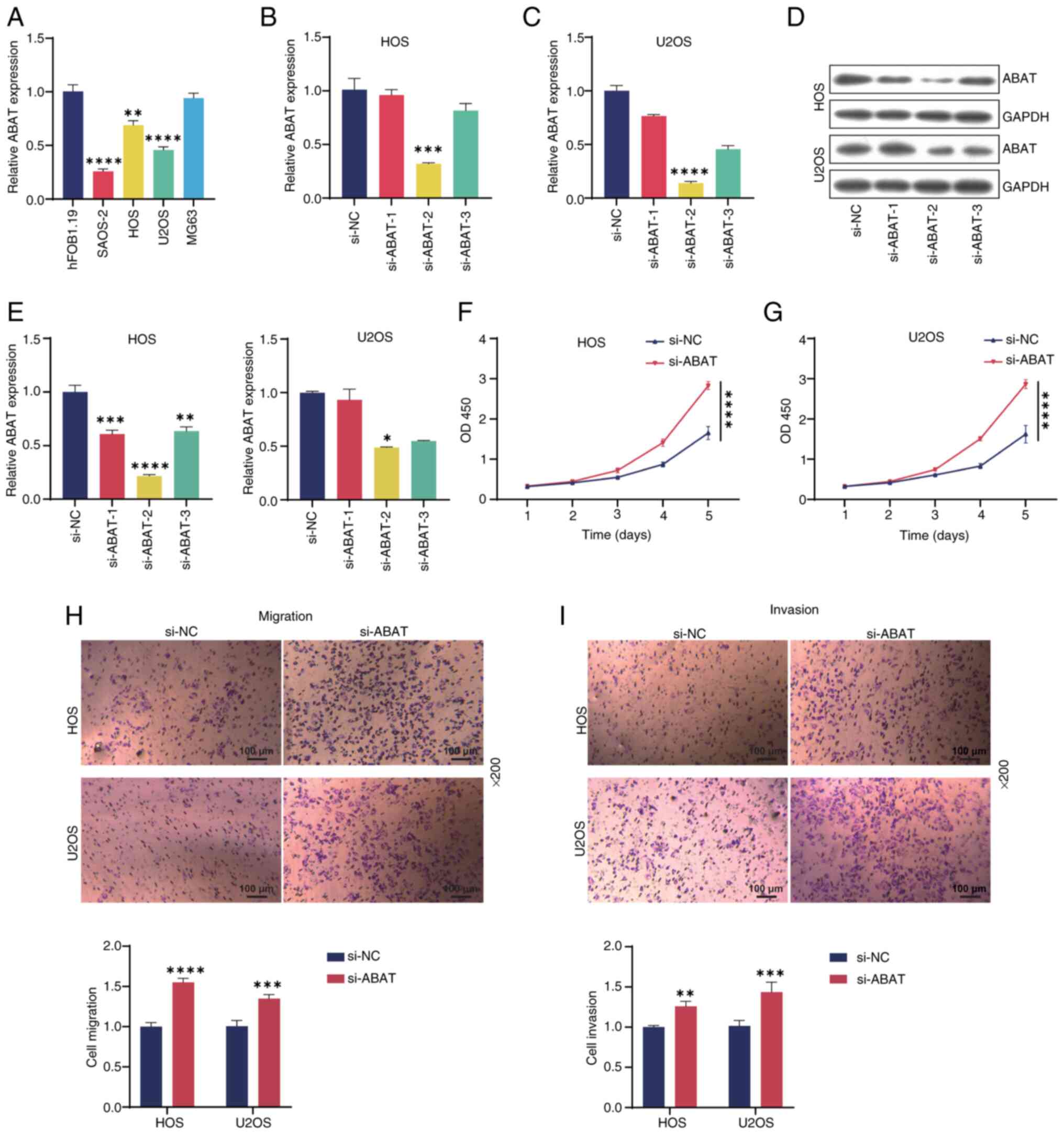 | Figure 8.Silencing ABAT inhibits the
proliferation, migration and invasion of OS cells. (A) Expression
of ABAT in OS or osteoblast cell lines detected by RT-qPCR. (B) HOS
and (C) U2OS cells were transfected with si-ABAT-1/2/3 or si-NC for
24 h, and the expression of ABAT was evaluated by RT-qPCR. (D,E)
ABAT expression levels in the two cell lines after transfection
were also tested by western blotting. Viability of (F) HOS and (G)
U2OS OS cells measured by Cell Counting Kit-8 assay. (H) Migration
and (I) invasion of HOS and U2OS cells as evaluated by Transwell
assays. Magnification, ×200. *P<0.05, **P<0.01, ***P<0.001
and ****P<0.0001, vs. siNC. ABAT, 4-aminobutyrate
aminotransferase; OS, osteosarcoma; RT-qPCR, reverse
transcription-quantitative PCR; si, small interfering RNA; NC,
negative control; OD450, optical density at 450 nm. |
Discussion
Substantial advancements have been made in the
comprehension of propionate metabolism, with researchers uncovering
its pivotal role in tumorigenesis. For example, Gomes et al
(21) reported that methylmalonic
acid (MMA), a byproduct of propionate metabolism, was upregulated
in the serum of elderly individuals and acted as a mediator of
tumor progression. These researchers also demonstrated that MMA
levels are increased via changes in propionate metabolism in cancer
cells, particularly in aggressive cancers such as triple-negative
breast cancers, enabling the cells to undergo prometastatic
reprogramming (12). However, the
role of propionate metabolism in OS cells has not been fully
elucidated.
In the present study, 18 propionate
metabolism-related DEGs were identified from the GSE12865 dataset.
To substantiate the molecular functions of these aberrantly
expressed DEGs, comprehensive functional enrichment analyses were
conducted. These DEGs were found to be predominantly associated
with processes associated with carbon, b-alanine and propanoate
metabolism. Consistent with this, abnormal cellular metabolism in
bone is known to be a key factor in the development of bone-related
diseases such as OS (22).
Furthermore, the DEGs were significantly enriched in GO categories
including ‘tricarboxylic acid cycle’, ‘extracellular exosome’ and
‘mitochondrial matrix’. Notably, in OS research, the recruitment of
bone marrow-derived macrophages has been shown to facilitate
pulmonary metastasis (23).
Researchers have also recognized that mitochondrial dysfunction is
a crucial factor contributing to cellular metabolic disturbances in
OS (24). These findings suggest
that the dysregulation of energy metabolism is a prominent feature
in patients with OS and is characterized by substantial
downregulation of the tricarboxylic acid cycle (25).
To assess the associations between the propionate
metabolism-related DEGs and clinical outcomes in patients with OS,
the 18 DEGs were subjected to Cox univariate regression analysis,
which led to the selection of four candidate genes. Further
analysis led to the construction of a risk model comprising two of
these genes: ABAT and ALDH7A1. Patients were classified into high-
and low-risk categories according to this risk score, and
Kaplan-Meier survival analysis revealed a striking separation in
their survival curves. It is notable that among the multitude of
analyzed genes associated with propionate metabolism, only two
genes were identified as pivotal contributors to the construction
of this gene signature. This signature has the potential to predict
the prognosis and clinical outcomes of patients with OS, offering
insights into the TME and therapeutic response.
Immunophenotypic analysis has been demonstrated to
be valuable for the diagnosis, differential diagnosis and
classification of OS cells (26).
In the present study, activated CD4+ memory T cells and
follicular_helper cells were present in elevated proportions in the
low-risk group. Conversely, a greater proportion of resting
CD4+ memory T cells was observed in the high-risk group.
In addition, based on various scores calculated for the two groups
and immune checkpoint marker levels, patients in the low-risk group
were indicated to have a greater likelihood of responding favorably
to immune checkpoint blockade therapy. Notably, previous research
by Fritzsching et al (27)
revealed an improvement in the survival period among patients with
a relatively high CD8+ T cell/FOXP3+ T cell
ratio in the OS microenvironment. Moreover, Lu et al
(28) conducted an analysis of
peripheral blood samples from patients with OS and found an
association between follicular helper T cell activity levels and
adverse prognosis.
At the single-cell level, the analysis performed in
the present study revealed that mono/macro cells are predominant in
OS. Prior research has elucidated the composition of the OS
microenvironment, which is primarily composed of tumor-associated
macrophages (TAMs) (29). TAMs play
pivotal roles in the promotion of tumor growth and angiogenesis by
upregulating tumorigenic and angiogenic factors (30). The enrichment of ABAT in plasmocytes
and CD4Tconv cells suggests that ABAT may be involved in the
regulation of immune responses and antibody production. Plasmocytes
are key cells for antibody generation, whereas CD4Tconv cells are
crucial for assisting immune responses and activating other immune
cells (31,32). Downregulation of ABAT may impair the
function of these cells, thereby weakening the host antitumor
immune response. By contrast, the localization of ALDH7A1 in
mono/macro cells, endothelial cells and fibroblasts suggests that
it may be involved in TME remodeling and immune suppression.
Macrophages in the TME often exhibit an M2 phenotype, which is
associated with immunosuppression, the promotion of angiogenesis,
tissue remodeling and the facilitation of tumor cell growth
(33). Monocytes are recruited into
the TME and differentiate into TAMs, which contribute to
immunosuppression and the formation of a supportive
microenvironment for tumor progression (34). Furthermore, endothelial cells and
fibroblasts are critical in tumor progression and immune evasion
(35). These findings highlight the
potential of ALDH7A1 as a key regulator of immune modulation and
suggest that it may serve as a target for novel immunotherapeutic
strategies. On the basis of the findings of the current study, the
prognosis of OS appears to be intricately associated with the
expression levels of ABAT and ALDH7A1. ABAT has previously been
implicated as a suppressor of cancer behavior, as its low
expression level in hepatocellular carcinoma effectively restrains
tumorigenesis (36). In addition,
ABAT modulates lactate production, which inhibits the
tumorigenicity of clear cell renal cell carcinoma (37). By contrast, ALDH7A1 is upregulated
in pancreatic ductal adenocarcinoma and promotes tumor formation
(38). ALDH7A1 also promotes the
clonogenicity and migratory capabilities of human prostate cancer
cells, playing a functional role in the formation of bone
metastasis (39). In addition, the
present study demonstrated that ABAT silencing promoted OS cell
metastasis and growth, suggesting that ABAT is a suppressor gene in
OS tumorigenesis.
The present study has several limitations. First,
while key genes related to propionate metabolism in OS were
identified through bioinformatics analysis, their regulatory roles
in propionate metabolism require further experimental validation.
Second, the role of ALDH7A1 in OS progression and its potential
effects on tumor cell behavior were not experimentally verified. In
future research, experiments on ALDH7A1 are planned to further
validate the findings of the present study. In addition, the
specific roles of BAT and ALDH7A1 in monocytes and macrophages
within the TME warrant further exploration.
In summary, the present study provides a prognostic
model based on propionate metabolism-related genes, which generates
unique risk scores associated with clinical outcomes and the TME.
This risk model exhibits potential for use in further research into
the clinical prognosis and immunotherapeutic options for patients
with OS. Two key genes, ABAT and ALDH7A1, associated with
propionate metabolism in OS were identified and found to correlate
with immune cell infiltration in patients with OS. These findings
offer valuable insights for improving the overall management of
OS.
Acknowledgements
Not applicable.
Funding
This study was supported by University Natural Science
Foundation of Anhui Province (grant no, 2023AH053173) and Research
Fund of Anhui Institute of Translational Medicine (grant no.
2023zhyx-C74).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
WL, ZX, ML and NZ were responsible for conception
and design of the study. SZ and JZ provided administrative support.
WL and ML provided study materials. NZ, QZ, SZ and JZ collected and
assembled the data. WL, ML and SZ performed data analysis and
interpretation. WL, ML, NZ, SZ and JZ wrote the manuscript. WL, ZX,
ZL and ML confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rothzerg E, Xu J and Wood D: Different
subtypes of osteosarcoma: Histopathological patterns and clinical
behaviour. J Mol Pathology. 4:99–108. 2023. View Article : Google Scholar
|
|
2
|
Moukengue B, Lallier M, Marchandet L,
Baud'huin M, Verrecchia F, Ory B and Lamoureux F: Origin and
therapies of osteosarcoma. Cancers. 14:35032022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Al-Dasuqi K, Cheng R, Moran J, Irshaid L,
Maloney E and Porrino J: Update of pediatric bone tumors:
Osteogenic tumors and osteoclastic giant cell-rich tumors. Skeletal
Radiol. 52:671–685. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ouyang H and Wang Z: Predictive value of
the systemic immune-inflammation index for cancer-specific survival
of osteosarcoma in children. Front Public Health. 10:8795232022.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Séguin B, Pinard C, Lussier B, Williams D,
Griffin L, Podell B, Mejia S, Timercan A, Petit Y and Brailovski V:
Limb-sparing in dogs using patient-specific,
three-dimensional-printed endoprosthesis for distal radial
osteosarcoma: A pilot study. Vet Comp Oncol. 18:92–104. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Papakonstantinou E, Stamatopoulos A, I
Athanasiadis D, Kenanidis E, Potoupnis M, Haidich AB and Tsiridis
E: Limb-salvage surgery offers better five-year survival rate than
amputation in patients with limb osteosarcoma treated with
neoadjuvant chemotherapy. A systematic review and meta-analysis. J
Bone Oncol. 25:1003192020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsukamoto S, Mavrogenis AF, Angelelli L,
Righi A, Filardo G, Kido A, Honoki K, Tanaka Y, Tanaka Y and Errani
C: The effect of adjuvant chemotherapy on localized extraskeletal
osteosarcoma: A systematic review. Cancers. 14:25592022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
El Hassouni B, Granchi C, Vallés-Martí A,
Supadmanaba IGP, Bononi G, Tuccinardi T, Funel N, Jimenez CR,
Peters GJ, Giovannetti E and Minutolo F: The dichotomous role of
the glycolytic metabolism pathway in cancer metastasis: Interplay
with the complex tumor microenvironment and novel therapeutic
strategies. Semin Cancer Biol. 60:238–248. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Adams SH, Hoppel CL, Lok KH, Zhao L, Wong
SW, Minkler PE, Hwang DH, Newman JW and Garvey WT: Plasma
acylcarnitine profiles suggest incomplete long-chain fatty acid
β-oxidation and altered tricarboxylic acid cycle activity in type 2
diabetic African-American women. J Nutr. 139:1073–1081. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xue H, Chen X, Yu C, Deng Y, Zhang Y, Chen
S, Chen X, Chen K, Yang Y and Ling W: Gut microbially produced
indole-3-propionic acid inhibits atherosclerosis by promoting
reverse cholesterol transport and its deficiency is causally
related to atherosclerotic cardiovascular disease. Circ Res.
131:404–420. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuan C, Zhang K, Wang Z, Ma X, Liu H, Zhao
J, Lu W and Wang J: Dietary flaxseed oil and vitamin E improve
semen quality via propionic acid metabolism. Front Endocrinol
(Lausanne). 14:11397252023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gomes AP, Ilter D, Low V, Drapela S,
Schild T, Mullarky E, Han J, Elia I, Broekaert D, Rosenzweig A, et
al: Altered propionate metabolism contributes to tumour progression
and aggressiveness. Nat Metab. 4:435–443. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sadikovic B, Yoshimoto M, Chilton-MacNeill
S, Thorner P, Squire JA and Zielenska M: Identification of
interactive networks of gene expression associated with
osteosarcoma oncogenesis by integrated molecular profiling. Hum Mol
Genet. 18:1962–1975. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Y, He M, Tang H, Xie T, Lin Y, Liu S,
Liang J, Li F, Luo K, Yang M, et al: Single-cell and spatial
transcriptomics reveal metastasis mechanism and microenvironment
remodeling of lymph node in osteosarcoma. BMC Med. 22:2002024.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sherman BT, Hao M, Qiu J, Jiao X, Baseler
MW, Lane HC, Imamichi T and Chang W: DAVID: A web server for
functional enrichment analysis and functional annotation of gene
lists (2021 update). Nucleic Acids Res. 50:W216–W221. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Q, Li M, Yang M, Yang Y, Song F,
Zhang W, Li X and Chen K: Analysis of immune-related signatures of
lung adenocarcinoma identified two distinct subtypes: Implications
for immune checkpoint blockade therapy. Aging (Albany NY).
12:3312–3339. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Danilova L, Ho WJ, Zhu Q, Vithayathil T,
De Jesus-Acosta A, Azad NS, Laheru DA, Fertig EJ, Anders R, Jaffee
EM and Yarchoan M: Programmed cell death ligand-1 (PD-L1) and CD8
expression profiling identify an immunologic subtype of pancreatic
ductal adenocarcinomas with favorable survival. Cancer Immunol Res.
7:886–895. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han Y, Wang Y, Dong X, Sun D, Liu Z, Yue
J, Wang H, Li T and Wang C: TISCH2: Expanded datasets and new tools
for single-cell transcriptome analyses of the tumor
microenvironment. Nucleic Acids Res. 51:D1425–D1431. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen W, Lin Y, Jiang M, Wang Q and Shu Q:
Identification of LARS as an essential gene for osteosarcoma
proliferation through large-scale CRISPR-Cas9 screening database
and experimental verification. J Transl Med. 20:3552022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gomes AP, Ilter D, Low V, Endress JE,
Fernández-García J, Rosenzweig A, Schild T, Broekaert D, Ahmed A,
Planque M, et al: Age-induced accumulation of methylmalonic acid
promotes tumour progression. Nature. 585:283–287. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Si J, Wang C, Zhang D, Wang B and Zhou Y:
Osteopontin in bone metabolism and bone diseases. Med Sci Monit.
26:e9191592020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhong L, Liao D, Li J, Liu W, Wang J, Zeng
C, Wang X, Cao Z, Zhang R, Li M, et al: Rab22a-NeoF1 fusion protein
promotes osteosarcoma lung metastasis through its secretion into
exosomes. Signal Transduct Target Ther. 6:592021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng MH, Pan CY, Chen NF, Yang SN, Hsieh
S, Wen ZH, Chen WF, Wang JW, Lu WH and Kuo HM: Piscidin-1 induces
apoptosis via mitochondrial reactive oxygen species-regulated
mitochondrial dysfunction in human osteosarcoma cells. Sci Rep.
10:50452020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sreedhar A and Zhao Y: Dysregulated
metabolic enzymes and metabolic reprogramming in cancer cells.
Biomed Rep. 8:3–10. 2018.PubMed/NCBI
|
|
26
|
Fx G: Osteosarcoma cell immunophenotype
and heterogeneity. Zhonghua Bing Li Xue Za Zhi. 22:285–287.
1993.(In Chinese). PubMed/NCBI
|
|
27
|
Fritzsching B, Fellenberg J, Moskovszky L,
Sápi Z, Krenacs T, Machado I, Poeschl J, Lehner B, Szendrõi M,
Bosch AL, et al: CD8+/FOXP3+-ratio in osteosarcoma microenvironment
separates survivors from non-survivors: A multicenter validated
retrospective study. Oncoimmunol. 4:e9908002015. View Article : Google Scholar
|
|
28
|
Lu J, Kang X, Wang Z, Zhao G and Jiang B:
The activity level of follicular helper T cells in the peripheral
blood of osteosarcoma patients is associated with poor prognosis.
Bioengineered. 13:3751–3759. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cersosimo F, Lonardi S, Bernardini G,
Telfer B, Mandelli GE, Santucci A, Vermi W and Giurisato E:
Tumor-associated macrophages in osteosarcoma: From mechanisms to
therapy. Int J Mol Sci. 21:52072020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang Q, Liang X, Ren T, Huang Y, Zhang H,
Yu Y, Chen C, Wang W, Niu J, Lou J and Guo W: The role of
tumor-associated macrophages in osteosarcoma
progression-therapeutic implications. Cell Oncol. 44:525–539. 2021.
View Article : Google Scholar
|
|
31
|
Nutt SL, Hodgkin PD, Tarlinton DM and
Corcoran LM: The generation of antibody-secreting plasma cells. Nat
Rev Immunol. 15:160–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ning S, Wu J, Pan Y, Qiao K, Li L and
Huang Q: Identification of CD4+ conventional T cells-related lncRNA
signature to improve the prediction of prognosis and immunotherapy
response in breast cancer. Front Immunol. 13:8807692022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Boutilier AJ and Elsawa SF: Macrophage
polarization states in the tumor microenvironment. Int J Mol Sci.
22:69952021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ugel S, Canè S, De Sanctis F and Bronte V:
Monocytes in the tumor microenvironment. Annu Rev Pathol.
16:93–122. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fang J, Lu Y, Zheng J, Jiang X, Shen H,
Shang X, Lu Y and Fu P: Exploring the crosstalk between endothelial
cells, immune cells, and immune checkpoints in the tumor
microenvironment: New insights and therapeutic implications. Cell
Death Dis. 14:5862023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han H, Zhou S, Chen G, Lu Y and Lin H:
ABAT targeted by miR-183-5p regulates cell functions in liver
cancer. Int J Biochem Cell Biol. 141:1061162021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu J, Chen Z, Zhao H, Dong H, Zhu L, Zhang
Y, Wang J, Zhu H, Cui Q, Qi C, et al: ABAT and ALDH6A1, regulated
by transcription factor HNF4A, suppress tumorigenic capability in
clear cell renal cell carcinoma. J Transl Med. 18:1012020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tan M, Meng J, Sun X, Fu X and Wang R:
EPS8 supports pancreatic cancer growth by inhibiting BMI1 mediated
proteasomal degradation of ALDH7A1. Exp Cell Res. 407:1127822021.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
van den Hoogen C, van der Horst G, Cheung
H, Buijs JT, Lippitt JM, Guzmán-Ramírez N, Hamdy FC, Eaton CL,
Thalmann GN, Cecchini MG, et al: High aldehyde dehydrogenase
activity identifies tumor-initiating and metastasis-initiating
cells in human prostate cancer. Cancer Res. 70:5163–5173. 2010.
View Article : Google Scholar : PubMed/NCBI
|