Introduction
Osteosarcoma is the most prevalent malignant bone
tumor in children and adolescents, with an estimated annual
incidence of 3–5 cases per million individuals under 20 years of
age globally. It originates from primitive bone mesenchymal cells,
typically manifests in the metaphysis of long bones, and exhibits
rapid growth and progression (1).
Advances in treatment techniques for osteosarcoma, including
surgery combined with neoadjuvant radiotherapy and chemotherapy,
have led to a significant improvement in the 5-year overall
survival rate of patients with osteosarcoma (2). However, recurrent metastasis remains a
persistent challenge, and the molecular mechanisms underlying
osteosarcoma remain poorly understood (3). Consequently, it is urgently necessary
to elucidate the molecular mechanisms underlying the progression of
osteosarcoma and identify novel therapeutic targets.
N6-methyladenosine (m6A) is
the most prevalent internal chemical modification of mRNA (4) and a key post-transcriptional
modification of RNA. The effects of this modification on the
regulation of RNA depend on dynamic interactions between
methyltransferases, demethylation enzymes and binding proteins,
which are also known as writers, erasers and readers, respectively
(5). Recently,
m6A-associated enzymes have been reported to perform
roles in several types of tumor, including hepatocellular carcinoma
(6), gastric cancer (7), lung cancer (8), glioma (9) and osteosarcoma (10). KIAA1429, also known as virus-like
m6A methyltransferase associated, is a component of the
intact m6A methyltransferase complex, and the largest
protein within this complex (11).
KIAA1429 facilitates m6A methylation by recruiting the
core components methyltransferase-like 3 (METTL3), METTL14 and
Wilms tumor 1-associated protein (WTAP) to specific regions of
target RNA. KIAA1429 has also been reported to play a role in
several types of tumor (12).
KIAA1429 mediates the m6A modification of carbohydrate
sulfotransferase 11 (CHST11) mRNA, leading to the recruitment of
YTH domain-containing family (YTHDF) protein 2 (YTHDF2), which
reduces the stability and expression of CHST11, thereby promoting
the proliferation of diffuse large B-cell lymphoma cells (13). Additionally, KIAA1429 is highly
expressed in hepatocellular carcinoma, where it facilitates tumor
proliferation by reducing the stability of Rho family GTPase 3. In
osteosarcoma, KIAA1429 has been identified as a key factor that
promotes the proliferation, migration and invasion of osteosarcoma
cells through the KIAA1429/JAK2/STAT3 signaling pathway (14). However, the precise role of KIAA1429
in osteosarcoma has yet to be fully elucidated; therefore, further
comprehensive evaluation of KIAA1429 is necessary.
Long non-coding RNAs (lncRNAs) are a class of
transcripts that are >200 nucleotides in length (15). Due to the absence of open reading
frames, they do not encode polypeptides or proteins (16). However, there is a substantial body
of evidence showing that lncRNAs have a pivotal role in the
regulation of gene expression. Aberrant RNA expression has been
shown to regulate gene expression by either silencing or activation
(17). m6A modifications
can regulate the function of lncRNAs by altering their structure,
thereby inducing the binding of RNA-binding proteins. Furthermore,
m6A modifications can influence the triple-helical
structure of lncRNAs, which may affect their ability to interact
with DNA (18,19). KIAA1429 has been demonstrated to
upregulate the expression of LINC01106 via m6A
modification, thereby facilitating the proliferation of lung
adenocarcinoma cells (20). Friend
of GATA family member 2-antisense 1 (ZFPM2-AS1) is a lncRNA
transcribed from the antisense strand of the ZFPM2 gene, which
encodes a zinc finger protein. It was initially identified to be
aberrantly overexpressed in gastric cancer.
Additionally, ZFPM2-AS1 has been reported to play a
role in a variety of other tumor types, including lung (21), esophageal (22) and thyroid (23) cancer. However, the function of
ZFPM2-AS1 in osteosarcoma, and the potential for m6A
modification of ZFPM2-AS1, have yet to be elucidated. Therefore,
the present study aimed to resolve these issues.
The present study evaluated the expression of
KIAA1429 and ZFPM2-AS1 in osteosarcoma. In addition, the effect of
KIAA1429 knockdown on the proliferation, migration and invasion of
143B and MG63 osteosarcoma cell lines was examined. Furthermore,
the impact of KIAA1429 overexpression on the m6A
modification level and stability of ZFPM2-AS1 was investigated.
Finally, the ability of ZFPM2-AS1 knockdown to attenuate
KIAA1429-mediated pro-proliferative, migratory and invasive effects
in osteosarcoma cells was assessed.
Materials and methods
Bioinformatics analysis and software
availability
The expression levels of KIAA1429 and ZFPM2-AS1 in
sarcomas and normal tissue were analyzed using data from The Cancer
Genome Atlas (TCGA) via the UALCAN tool (https://ualcan.path.uab.edu/index.html) (24). The association between the
expression of ZFPM2-AS1 in sarcoma tissues and the overall survival
of patients was analyzed using TCGA data via the Gene Expression
Profiling Interactive Analysis 2 (GEPIA2) database (http://gepia2.cancer-pku.cn/#index) (25). The expression of KIAA1429 in 18
pairs of osteosarcoma and non-tumoral tissues was analyzed using
the GSE99671 osteosarcoma-related dataset from the Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html).
The correlation between the expression of KIAA1429 and other genes
in 87 osteosarcoma tissue samples from TCGA-TARGET-osteosarcoma
(OS) database was analyzed using PDX for Childhood Cancer
Therapeutics online analysis software. The 20 genes identified as
having the highest expression correlation were subsequently
downloaded and collated. The correlation between the expression of
KIAA1429 (probe ID: 25962) and ZFPM2-AS1 (probe ID: 102723356) was
analyzed in 52 samples from the GEO osteosarcoma-related dataset
GSE87624. Sarcoma data were also downloaded from TCGA database to
analyze the correlation between KIAA1429 and ZFPM2-AS1 expression
in 260 sarcoma tissue samples. The gene sequence of ZFPM2-AS1
(NR_125796.1) was retrieved from the National Center for
Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/gene/?term=ZFPM2-AS1).
Finally, m6A modification sites in the ZFPM2-AS1
sequence were predicted using SRAMP (http://www.cuilab.cn/sramp/) online bioinformatics
software (26).
Patients and tissue samples
A total of 20 pairs of surgically resected
osteosarcoma and paraneoplastic tissue samples were collected from
the Central Hospital Affiliated to Shenyang Medical College
(Shenyang, China) and Liaoning Provincial Cancer Hospital
(Shenyang, China). The samples were collected from 8 male and 12
female patients (median age, 16 years; age range, 8–22 years) who
underwent surgery between April 2016 and April 2022. All patients
were clinically and pathologically diagnosed with osteosarcoma.
Written informed consent was obtained from all adult participants
and from the parents or legal guardians of participants <18
years of age. The study protocol was approved by the Medical Ethics
Committee of the Central Hospital Affiliated to Shenyang Medical
College (approval no. 2022DEC12-7).
Cell culture
The hFOB1.19 human osteoblast cell line and the 143B
and MG63 osteosarcoma cell lines were obtained from The Cell Bank
of Type Culture Collection of The Chinese Academy of Sciences.
Characterization of the three cell lines using short tandem repeat
analysis demonstrated that these cells were not cross-contaminated.
The hFOB1.19 osteoblasts were cultured in Gibco®
DMEM/F12 medium, the MG63 cells were cultured in Minimal Essential
Medium, and the 143B cells were cultured in RPMI-1640 medium (all
from Thermo Fisher Scientific, Inc.). The media were supplemented
with 10% (v/v) Gibco fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) and a solution of penicillin and streptomycin
(penicillin, 100 U/ml; streptomycin, 0.1 mg/ml). All three cell
lines were cultured in a cell culture incubator in an atmosphere
containing 5% CO2. The hFOB1.19 cells were maintained at
34°C, while the 143B and MG63 cells were cultured at 37°C.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells and tissues using
an Invitrogen TRIzol® Plus RNA Purification Kit (Thermo
Fisher Scientific, Inc.) following the manufacturer's instructions.
Reverse transcription was performed with the PrimeScript RT Master
Mix (Takara Bio, Inc., cat. no. RR036A) in a 20 µl reaction
containing 1 µg total RNA, 5X RT Master Mix, and RNase-free water.
The thermal protocol included 37°C for 15 min, 85°C for 5 sec, and
a final hold at 4°C. qPCR was carried out using the TB Green Premix
Ex Taq II kit (cat. no. RR820A; Takara Bio, Inc.) under the
following conditions: Initial denaturation at 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. GAPDH
served as the internal reference gene, and relative gene expression
was calculated using the 2−ΔΔCq method. All primers were
synthesized by Takara Bio, Inc., and sequences are listed in
Table SI.
Western blot analysis
Proteins were extracted from cells and tissues using
a protein extraction kit (cat. no. GM1001; Wuhan Servicebio
Technology Co., Ltd.). Protein concentrations were determined using
a BCA protein assay kit (Wuhan Servicebio Technology Co., Ltd.). A
total of 25 µg protein per lane was denatured in a water bath at
100°C for 5 min, followed by separation using 8% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; cat. no.
G2176; Wuhan Servicebio Technology Co., Ltd.) at a constant voltage
of 120 V. Proteins were transferred to polyvinylidene fluoride
(PVDF) membranes (cat. no. G6044-0.45; Wuhan Servicebio Technology
Co., Ltd.) at a constant current of 400 mA. Membranes were blocked
with NcmBlot Rapid Blocking Buffer (cat. no. P30500; NCM Biotech)
at room temperature (25°C) for 15 min. Primary antibodies against
KIAA1429 (cat. no. ab271136; Abcam; 1:1,000 dilution) and GAPDH
(cat. no. ab9485; Abcam; 1:2,500 dilution) were incubated with the
membranes at 4°C overnight. After washing with Tris-buffered saline
containing 0.1% Tween 20 (TBST), membranes were incubated with
horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG
secondary antibody (cat. no. ab205718; Abcam; 1:10,000 dilution) at
room temperature for 1 h. Protein bands were visualized using an
ultra-sensitive ECL chemiluminescence detection kit (cat. no.
G2074; Wuhan Servicebio Technology Co., Ltd.) and imaged with a gel
documentation system (ChemiScope 6100; Shanghai Qinxiang Scientific
Instrument Co., Ltd.). Protein expression levels were quantified
using ImageJ software (Version 1.53; National Institutes of Health)
and normalized to GAPDH.
Cell transfection
Two short hairpin RNAs (shRNAs) targeting the
KIAA1429-specific junction region (shKIAA1429-1 and shKIAA1429-2),
their corresponding negative control shRNAs (shNCs), a KIAA1429
overexpression plasmid (oeKIAA1429), and the empty plasmid vector
pCMV6-Entry were designed and synthesized by Hanheng Biotechnology
(Shanghai) Co., Ltd. shRNAs (final concentration: 50 nM) and
plasmids (final concentration: 2 µg/ml) were transfected into 143B,
MG63, and hFOB1.9 cells using the Invitrogen
Lipofectamine® 3000 reagent (cat. no. L3000001; Thermo
Fisher Scientific, Inc.) under standard conditions (37°C for 6 h),
followed by replacement with complete culture medium. Additionally,
a small interfering RNA (siRNA) targeting ZFPM2-AS1 (siZFPM2-AS1)
and its negative control siRNA (siNC) were designed by Guangzhou
RiboBio Co., Ltd. and transiently transfected into osteosarcoma
cells at a final concentration of 50 nM using the
RiboFect® CP Transfection Kit (cat. no. C10511-05;
Guangzhou RiboBio Co., Ltd.), according to the manufacturer's
protocol. Cells were harvested 48 h post-transfection for
subsequent experiments. The sequences of shRNAs, siRNAs, and
oeKIAA1429 are listed in Table
SI.
Cell Counting Kit-8 (CCK-8) assay
143B and MG63 cells were inoculated into 96-well
plates at a density of 2×103 cells/well, and cultured in
a cell culture incubator at 37°C in an atmosphere containing 5%
CO2. At 24, 48, 72 and 96 h, 10 µl CCK-8 solution (cat.
no. G4103; Wuhan Servicebio Technology Co., Ltd.) was added to each
well for 2 h. Following this, the 96-well plates were removed from
the incubator and the absorbance at 450 nm was measured using a
microplate reader (Multiskan FC Photometer; Thermo Fisher
Scientific, Inc.).
Transwell assay
Different densities of the 143B and MG63 cells were
used, according to whether the Transwell assay was used to assess
migration (4×104 cells) or invasion (8×104
cells). The cells were cultured in the upper chamber of the
Transwell apparatus (Corning, Inc.). After 24 h at 37°C, the cells
in the upper chamber were scraped off and those on the lower side
of the Transwell membrane were fixed with anhydrous ethanol prior
to staining with 0.1% crystal violet staining solution (cat. no.
G1014; Wuhan Servicebio Technology Co., Ltd.) for 30 min at room
temperature. Following a rinse with running water, the migrated or
invaded cells were observed under an inverted microscope (Olympus
Corporation). For invasion assays, the upper chambers of Transwell
inserts (Corning, Inc.) were pre-coated with Matrigel Basement
Membrane Matrix (Corning, cat. no. 354234) diluted 1:8 in
serum-free medium, followed by polymerization at 37°C for 1 h. Cell
densities were adjusted according to assay type: 4×104
cells/well for migration and 8×104 cells/well for
invasion. Cells were suspended in serum-free RPMI-1640 medium (for
143B) or MEM (for MG63) and added to the upper chamber. The lower
chamber contained complete medium with 10% fetal bovine serum
(Thermo Fisher Scientific, Inc.) as a chemoattractant. After 24 h
incubation at 37°C in 5% CO2, non-invaded cells on the
upper membrane surface were removed using a cotton swab. Cells on
the lower side were fixed with anhydrous ethanol for 15 min,
stained with 0.1% crystal violet (Wuhan Servicebio, cat. no. G1014)
for 30 min, and rinsed with PBS. Images were captured using an
inverted microscope (Olympus Corporation) at 200× magnification.
Cell counts were quantified from five random fields per
membrane.
m6A-methylated RNA
immunoprecipitation-qPCR (MeRIP-qPCR) assay
The assay was conducted using an
m6A-Methylated RNA Immunoprecipitation Kit (cat. no.
11096.6; Guangzhou Ribobio Co., Ltd.) in accordance with the
manufacturer's instructions. In brief, the collected RNA was
fragmented using RNA Fragmentation Buffer. A fraction (one-tenth)
of the fragmented RNA was retained as an input control group for
subsequent analysis, while the remainder was used for
m6A immunoprecipitation (IP). Anti-m6A
magnetic beads were prepared using m6A antibody (cat.
no. ab151230; dilution, 1:500). The fragmented RNA was added to the
MeRIP reaction mixture, and incubated with anti-m6A
magnetic beads. After incubation, the mixture was centrifuged at
1,000 × g for 3 min at 4°C and the supernatant was discarded. The
magnetic beads were then washed, followed by elution of the bound
RNA. The eluted RNA fragments were recovered and purified using the
GeneJET RNA Purification Kit (cat. no. K0731; Thermo Fisher
Scientific, Inc.). The recovered RNA was designated as the IP
group. Subsequently, the input and IP groups were analyzed by
RT-qPCR.
Actinomycin D assay
The collected cells (2×106 cells) were
transferred to 6-well plates and cultured to a confluency of 80%.
The cells were then treated with 2 µg/ml actinomycin D reagent
(cat. no. 50-76-0; MedChemExpress). Total cellular RNA was
extracted at 0, 4, 8 and 12 h and analyzed by RT-qPCR.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 8.0 software (Dotmatics). Results are expressed as the mean ±
standard deviation. For comparative analyses involving two groups,
paired tests were used to evaluate results from paired patient
tissue samples: a paired t-test was applied for normally
distributed data, whereas the Wilcoxon signed-rank test was applied
for non-normally distributed data. Unpaired t-tests were used for
the comparison of two independent samples with normally distributed
means. In datasets comprising three or more groups, one-way
analysis of variance followed by Tukey's post hoc test was used for
normally distributed data, and Kruskal-Wallis test was performed
followed by Dunn's post hoc analysis for non-normally distributed
data. Linear correlation analyses were performed, with Pearson
correlation coefficients calculated for normally distributed data
and Spearman correlation coefficients for non-normally distributed
data. Each experiment was replicated thrice. P<0.05 was
considered to indicate a statistically significant difference.
Results
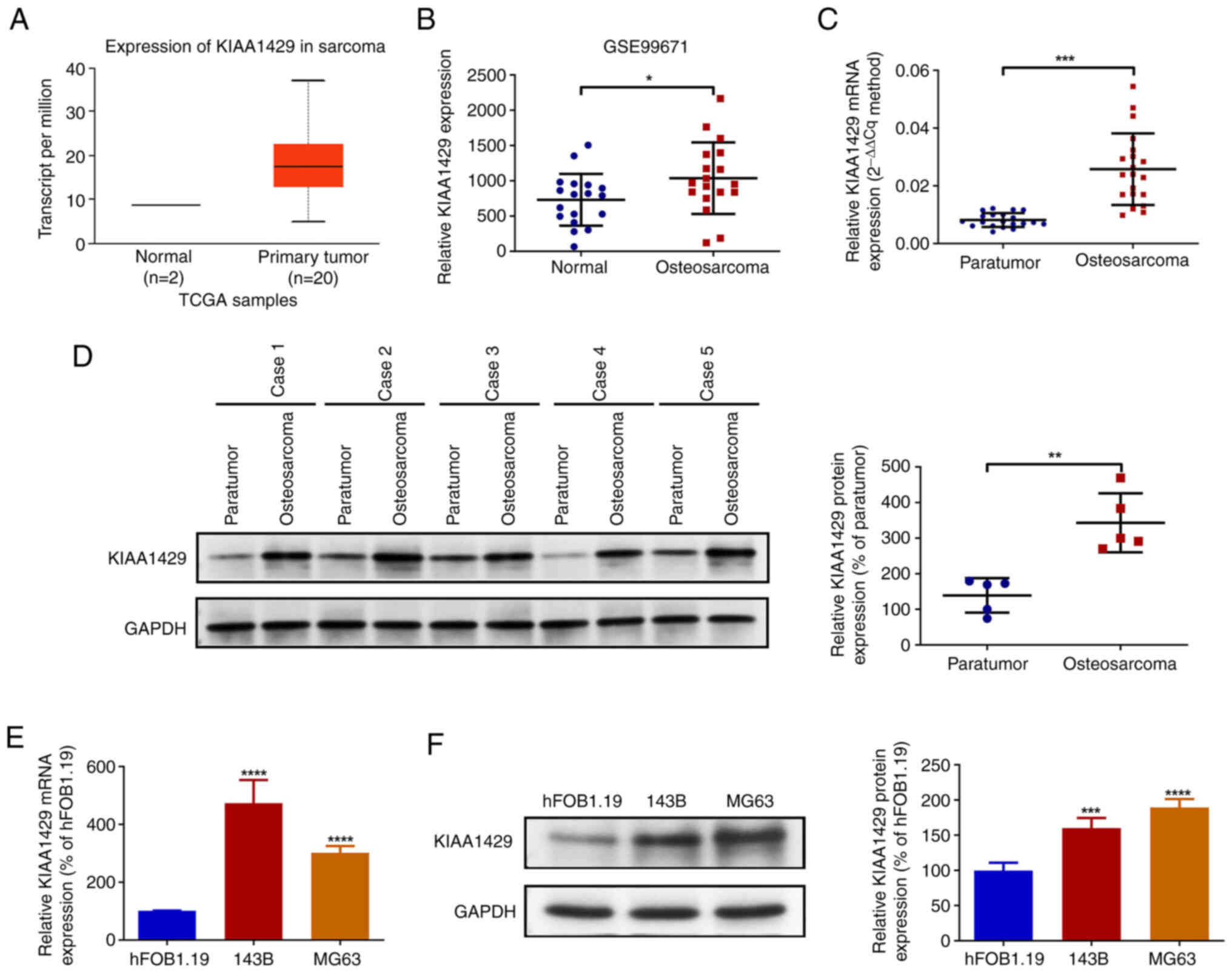
KIAA1429 is upregulated in
osteosarcoma tissues and cell lines
Bioinformatics analysis was performed to investigate
the expression of KIAA1429 in osteosarcoma and normal tissues.
Analysis of TCGA data revealed that the expression of KIAA1429 in
sarcoma was markedly elevated compared with that in normal tissues.
The expression of KIAA1429 was also analyzed in the GSE99671
dataset from the GEO database, which revealed that the expression
of KIAA1429 in sarcoma tissues was significantly higher compared
with that in the corresponding normal tissues (Fig. 1A and B). Subsequently, 20 pairs of
osteosarcoma and paraneoplastic tissues collected from patients
were examined by RT-qPCR analysis, which revealed a significant
upregulation of KIAA1429 expression in the osteosarcoma tissues
(Fig. 1C). In addition, the western
blot analysis of five pairs of osteosarcoma and paraneoplastic
tissues confirmed that expression of KIAA1429 protein was
significantly upregulated in the osteosarcoma tissues (Fig. 1D). Subsequently, normal osteoblast
and osteosarcoma cell lines were analyzed using RT-qPCR and western
blotting. The results demonstrated that KIAA1429 mRNA and protein
expression levels in the 143B and MG63 osteosarcoma cells were
significantly higher compared with those in the hFOB1.19 cells
(Fig. 1E and F). The findings of
the bioinformatics analysis, tissue and cellular experiments
collectively demonstrate that KIAA1429 expression is upregulated in
osteosarcoma.
KIAA1429 knockdown inhibits the
proliferation, migration and invasion of osteosarcoma cells
The impact of KIAA1429 on osteosarcoma cell
proliferation, migration and invasion was investigated in
functional assays of 143B and MG63 cells transfected with
shKIAA1429-1, shKIAA1429-2 and shNC. The successful establishment
of KIAA1429 knockdown was verified in the two cell lines via
RT-qPCR and western blotting (Fig.
2A-D). A CCK-8 cell proliferation assay was then performed to
assess the impact of KIAA1429 knockdown on cell proliferation. The
results demonstrated that KIAA1429 knockdown significantly
inhibited the proliferative capability of the 143B and MG63 cells
compared with that of the corresponding shNC controls (Fig. 2E and F). Subsequently, Transwell
assays were performed to evaluate the migration and invasion of the
143B and MG63 cells. The results revealed that the migratory and
the invasive capabilities of the cells were significantly
diminished following the knockdown of KIAA1429 (Fig. 2G and H). Together, these findings
suggest that KIAA1429 promotes the proliferation, migration and
invasion of 143B and MG63 osteosarcoma cells.
KIAA1429-mediated m6A
modification is associated with ZFPM2-AS1 stability in osteosarcoma
cells
Expression data from 87 osteosarcoma tissues in
TCGA-TARGET-OS database were analyzed to identify genes whose
expression levels significantly correlated with KIAA1429 expression
levels. Of the top 20 genes whose expression correlated with that
of KIAA1429, ZFPM2-AS1 was the only lncRNA identified (Table SII and Fig. 3A). The online tool SRAMP was then
used to predict the presence of theoretical m6A
modification sites (RRACH sequences) in the ZFPM2-AS1 sequence, and
six such sites were identified (Fig.
S1A). Based on this finding, the potential regulation of
ZFPM2-AS1 by KIAA1429 was investigated. ZFPM2-AS1 expression levels
were found to correlate with KIAA1429 expression levels in the
GSE87624 osteosarcoma-related dataset as well as in sarcoma tissues
in TCGA database (Fig. 3B and C).
Similarly, reduced expression levels of ZFPM2-AS1 were observed
following KIAA1429 knockdown in 143B and MG63 cells (Fig. 3D and E). hFOB1.19 cells
overexpressing KIAA1429 were successfully constructed, as confirmed
by western blotting (Fig. S1B).
RT-qPCR results revealed a corresponding increase in the expression
level of ZFPM2-AS1, further supporting a positive association
between KIAA1429 and ZFPM2-AS1 expression (Fig. S1C). Although ZFPM2-AS1 has been
characterized as an oncogene in numerous types of tumor, its
specific role in osteosarcoma remains unclear. Analysis of data
from TCGA revealed that ZFPM2-AS1 expression is upregulated in
sarcoma tissue compared with normal tissue, and high ZFPM2-AS1
expression is associated with a poorer overall survival profile in
patients with sarcoma (Fig. 3F and
G). Subsequently, the expression levels of ZFPM2-AS1 were
assessed in 20 pairs of osteosarcoma and paraneoplastic tissues via
RT-qPCR analysis. The results demonstrated that the expression of
ZFPM2-AS1 in osteosarcoma tissues was upregulated compared with
that in paraneoplastic tissues (Fig.
3H). Furthermore, analysis of the expression of ZFPM2-AS1 in
hFOB1.19, 143B and MG63 cells. revealed that ZFPM2-AS1 expression
in the osteosarcoma cell lines was elevated compared with that in
the normal osteoblast cell line (Fig.
3I). Taken together, these results provide evidence to support
a correlation between the expression levels of ZFPM2-AS1 and
KIAA1429, and demonstrate that the expression of ZFPM2-AS1 is
upregulated in osteosarcoma tissues and cells.
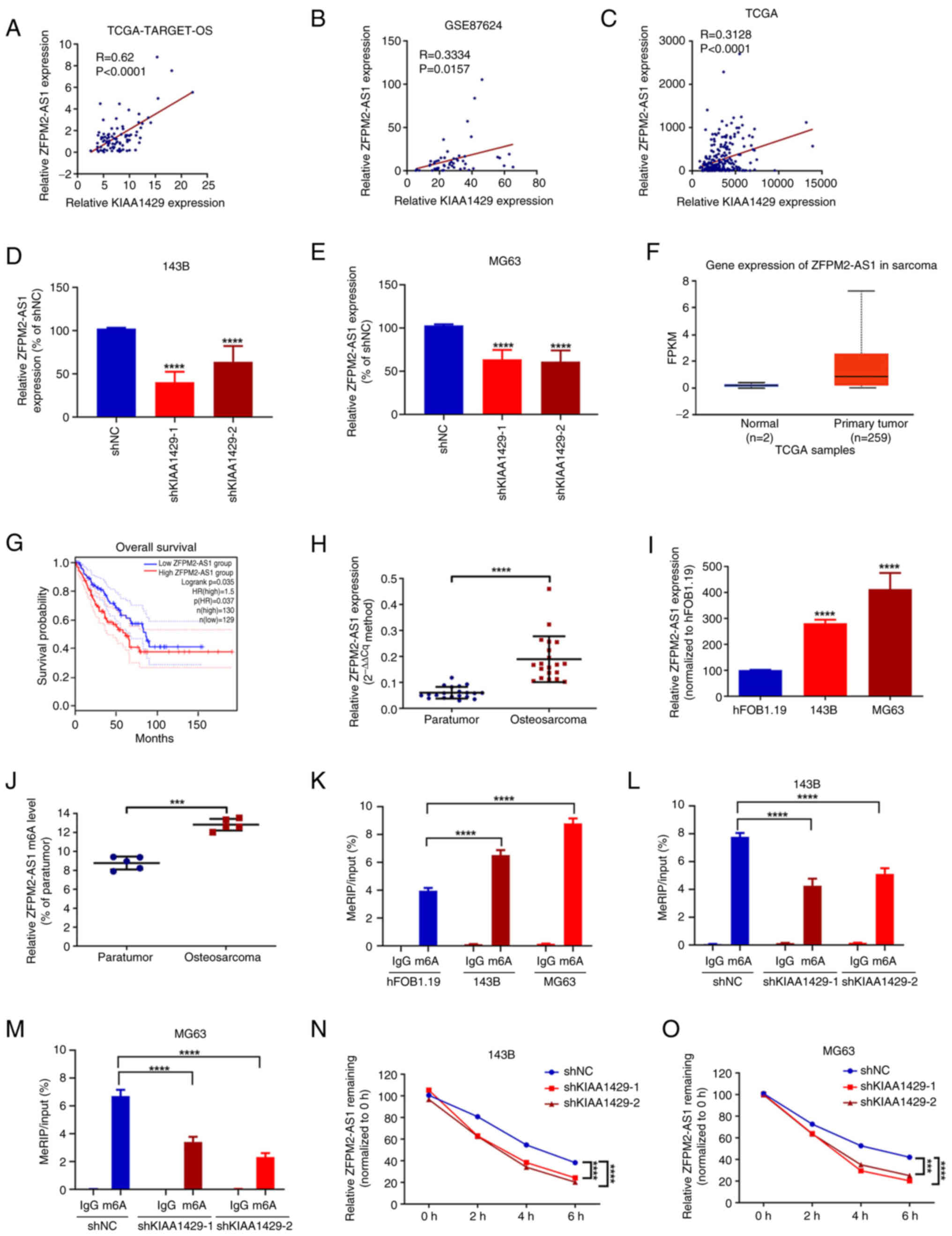 | Figure 3.KIAA1429-mediated m6A
modification is associated with ZFPM2-AS1 stability in osteosarcoma
cells. Correlation of KIAA1429 and ZFPM2-AS1 expression in (A) 87
osteosarcoma tissues from TCGA-TARGET-OS database, (B) 52
osteosarcoma tissues from the GSE87624 osteosarcoma-associated
dataset and (C) 260 sarcoma tissues from TCGA. RT-qPCR detection of
ZFPM2-AS1 expression in (D) 143B and (E) MG63 cells with KIAA1429
knockdown. ****P<0.0001 vs. shNC. (F) ZFPM2-AS1 expression in
sarcoma tissues and normal tissues from TCGA database, analyzed
using UALCAN. P<0.001 primary tumor vs. normal. (G) Kaplan-Meier
analysis of the overall survival of patients with sarcoma based on
ZFPM2-AS1 expression levels. (H) ZFPM2-AS1 expression in 20 pairs
of osteosarcoma and paraneoplastic tissues analyzed by RT-qPCR.
****P<0.0001 as indicated. (I) Expression of ZFPM2-AS1 in
hFOB1.19, 143B and MG63 cells detected by RT-qPCR. ****P<0.0001
vs. hFOB1.19. m6A levels of ZFPM2-AS1 in (J) 5 pairs of
osteosarcoma and paraneoplastic tissues, (K) hFOB1.19, 143B and
MG63 cells, and KIAA1429 knockdown (L) 143B and (M) MG63 cells
determined by MeRIP-qPCR assay. ***P<0.001 and ****P<0.0001
as indicated. Expression levels of ZFPM2-AS1 determined by RT-qPCR
assay in KIAA1429 knockdown (N) 143B and (O) MG63 cells at various
time points after the addition of actinomycin D. ***P<0.001 and
****P<0.0001 as indicated. All data are presented as the mean ±
SD from three independent experiments. m6A,
N6-methyladenosine; ZFPM2-AS1, friend of GATA family
member 2-antisense 1; TCGA, The Cancer Genome Atlas; OS,
osteosarcoma; RT-qPCR, reverse transcription-quantitative PCR;
shNC, negative control shRNA; shKIAA1429, shRNA targeting KIAA1429;
shRNA, short hairpin RNA; FPKM, fragments per kilobase of
transcript per million mapped reads; MeRIP,
m6A-methylated RNA immunoprecipitation. |
SRAMP predicted m6A modification sites at
positions 579, 613, 624, 750, 780 and 833 of the ZFPM2-AS1
sequence, spanning ~260 base pairs. (Fig. S1A). Primers were designed based on
the gene regions containing these predicted modification sites, and
the m6A methylation level of ZMIZ1-AS1 in osteosarcoma
tissues and cells was determined by MeRIP-qPCR. The results
revealed that the m6A methylation level of ZFPM2-AS1 was
significantly elevated in osteosarcoma tissues and cells in
comparison with the levels observed in paraneoplastic tissues and
hFOB1.19 cells, respectively (Fig. 3J
and K). To further investigate the regulatory role of KIAA1429,
the m6A methylation levels of ZFPM2-AS1 were examined
via MeRIP-qPCR assay in 143B and MG63 cells following KIAA1429
knockdown. The knockdown of KIAA1429 was observed to result in a
significant reduction in the m6A methylation level of
ZFPM2-AS1 compared with that in the shNC group (Fig. 3L and M). In addition, an actinomycin
D assay revealed that the knockdown of KIAA1429 shortened the
half-life of ZFPM2-AS1, indicating that its stability was reduced
(Fig. 3N and O). Considered
together, these findings suggest that m6A methylation
levels of ZFPM2-AS1 are elevated in osteosarcoma tissues and cells.
Furthermore, based on the observed reduction following KIAA1429
knockdown, the results suggest that KIAA1429 may increase the
m6A methylation levels of ZFPM2-AS1 and promote its
stability.
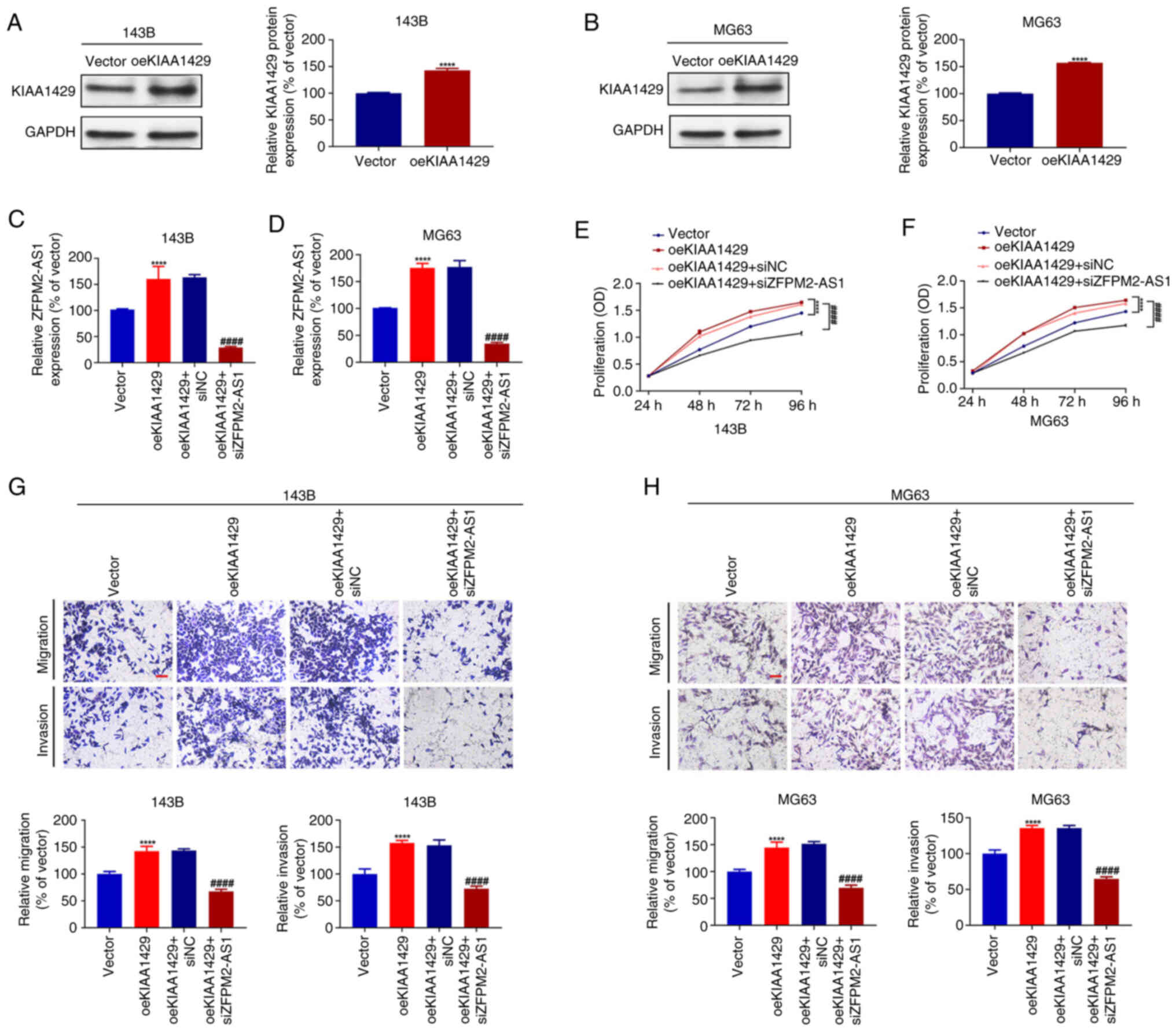
ZFPM2-AS1 knockdown attenuates the
promotive effect of KIAA1429 on the proliferation, migration and
invasion of osteosarcoma cells
Finally, the impact of ZFPM2-AS1 on
KIAA1429-mediated osteosarcoma proliferation, migration and
invasion was investigated. First, a cell model with stable
overexpression of KIAA1429 was established in 143B and MG63 cells,
and the successful construction of the model was verified via
western blotting (Fig. 4A and B).
Subsequently, siZFPM2-AS1 was transfected into 143B and MG63 cells,
resulting in significantly reduced ZFPM2-AS1 expression levels
compared with those in the respective siNC-transfected cells, as
validated by RT-qPCR (Fig. S1D and
E). Then, siZFPM2-AS1 was transfected into 143B and MG63 cells
overexpressing KIAA1429, and ZFPM2-AS1 expression was quantified by
RT-qPCR. The expression level of ZFPM2-AS1 was observed to increase
following the overexpression of KIAA1429 compared with that in the
empty vector control group, which was consistent with the observed
reduction in ZFPM2-AS1 expression following KIAA1429 knockdown
(Fig. 4C and D). However, this
oeKIAA1429-induced increase in ZFPM2-AS1 expression was attenuated
by co-transfection with siZFPM2-AS1 (Fig. 4C and D). Subsequently, CCK-8 and
Transwell functional assays were performed. The overexpression of
KIAA1429 significantly promoted cell proliferation, migration and
invasion compared with that of the vector control cells. However,
these effects of KIAA1429 were attenuated following the knockdown
of ZFPM2-AS1 in KIAA1429 overexpressing cells (Fig. 4E and H). These findings suggest that
the knockdown of ZFPM2-AS1 attenuates the effects of KIAA1429 on
the proliferation, migration and invasion of 143B and MG63
cells.
Discussion
m6A modification is a dynamic and
reversible process regulated by m6A
methylation-associated enzymes, which are classified into three
categories: Writers, erasers and readers. The methylation reaction
is catalyzed by a methyltransferase complex composed of
m6A writers such as METTL3, along with METTL14, WTAP and
KIAA1429, which performs a key role in the m6A
methylation process. By contrast, two demethylating enzymes, namely
AlkB homolog 5, which is an RNA-specific demethylase, and fat mass
and obesity-associated protein, function as erasers, reversing the
methylation of m6A. Furthermore, m6A-binding
proteins, including YTHDF1, YTHDF2, YTHDF3 and YTHDC1, act as
readers that regulate downstream processes including the
translation and degradation of m6A-modified RNAs. The
m6A methyltransferases, METTL3, METTL14 and WTAP, have
been extensively studied in various types of tumors, including
osteosarcoma. KIAA1429 serves as a scaffold within the
m6A methyltransferase complex and is its largest known
component. It helps coordinate the positioning of the core
METTL3/METTL14/WTAP component on RNA substrates for site-specific
m6A methylation, particularly near to the
3′-untranslated region and stop codon. In the present study,
analysis of data from the GEO and TCGA databases, combined with
mRNA and protein profiling, revealed that KIAA1429 expression is
upregulated in osteosarcoma tissues and cells. In addition, the
knockdown of KIAA1429 inhibited the proliferation, migration and
invasion of osteosarcoma cells. Previous studies have demonstrated
that KIAA1429 can mediate the m6A modification of
lncRNAs, including LINC00667 (27),
LINC00958 (28), POU6F2-AS1
(29) and LINC01106 (20), contributing to tumorigenesis. The
present study focused on the role of KIAA1429 in the modification
of ZFPM2-AS1 in osteosarcoma.
ZFPM2-AS1 was first reported to play a role in
gastric cancer in 2018 (30), and
has since been implicated in colorectal cancer (31), esophageal squamous cell carcinoma
(22), lung adenocarcinoma
(32) and several other tumor types
(33). However, to the best of our
knowledge, its specific role in osteosarcoma has not previously
been reported. In the present study, a positive correlation between
KIAA1429 and ZFPM2-AS1 expression was identified in osteosarcoma
via bioinformatic and RT-qPCR analyses. ZFPM2-AS1 expression was
found to be upregulated in osteosarcoma and the high expression of
ZFPM2-AS1 was significantly associated with a poor prognosis in
patients with sarcoma. Regarding the underlying mechanism by which
ZFPM2-AS1 exerts its pro-cancer effects, previous studies have
focused on its function as a competitive endogenous RNA. The
m6A modification of KIAA1429 has been shown to affect
the stability of lncRNAs, including LINC00958 (28). In the present study, six possible
m6A modification sites were identified in the ZFPM2-AS1
gene sequence, and KIAA1429 was found to increase both the
m6A level and stability of ZFPM2-AS1. Furthermore,
ZFPM2-AS1 knockdown partially attenuated the pro-proliferative,
migratory and invasive effects of KIAA1429 on osteosarcoma
cells.
The influence of m6A modification on RNA
stability is complex and involves numerous factors. m6A
modification can either enhance or reduce the recognition of RNA by
m6A-reading proteins, such as YTHDFs, YTHDC1 and
insulin-like growth factor 2 mRNA-binding proteins. The resulting
changes in RNA can lead to either stabilization or degradation
through distinct regulatory mechanisms (34). For example, a previous study
revealed that m6A-containing RNAs with heat-responsive
protein 12 (HRSP12)-binding sites near to RNase P/MRP-directed
cleavage sites are preferentially targeted for endoribonucleolytic
cleavage through the YTHDF2-HRSP12-RNase P/MRP axis (35). In addition, in acute myeloid
leukemia, the binding of YTHDC1 to nuclear m6A
transcripts promotes the formation of nuclear condensates, thereby
safeguarding nuclear mRNAs, such as Myc, from poly (A) tail exosome
targeting complex-mediated degradation (36). The actinomycin D experiments
performed in the present study demonstrated that the knockdown of
KIAA1429 shortened the half-life of the KIAA1429-modified lncRNA
ZFPM2-AS1, indicating that its stability was reduced. However,
further research is necessary to ascertain the precise underlying
mechanism of action.
In summary, in the present study, the expression
levels of KIAA1429 and ZFPM2-AS1 in osteosarcoma tissues and cells
were analyzed, and their roles in promoting the proliferation,
migration and invasion of osteosarcoma cells were revealed. The
findings suggest that KIAA1429 contributes to the stability of
ZFPM2-AS1 through m6A modification. However, more
in-depth studies are required to identify the specific
m6A modification sites that mediate the interaction
between KIAA1429 and ZFPM2-AS1, and to determine whether other
m6A-associated enzymes are involved in the regulation of
ZFPM2-AS1. Despite these remaining issues, the present study has
identified a potential novel therapeutic target for the molecular
therapy of osteosarcoma.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XB and YZ participated in experimental design
discussions. XB conceived and designed the study, supervised the
project and acquired funding. XB and YB performed the key
experiments. YB and YZ were responsible for data acquisition and
processing (YB validated data and YZ processed the preliminary data
while verifying raw data authenticity). HZ, CL, and HS performed
the statistical analysis, interpreted experimental data and
validated results. KG performed independent data analysis and
biological interpretation of key findings. YZ conducted the
literature analysis and prepared all figures and visualization
materials. YZ drafted the initial manuscript. KG and XB critically
revised the manuscript (KG optimized experimental protocols,
established data quality control criteria, validated data integrity
and reproducibility, and revised for methodological accuracy and
biological interpretation while XB revised the manuscript for
intellectual content). KG and XB confirm the authenticity of all
the raw data. All authors have read and approved the final version
of the manuscript.
Ethics approval and consent to
participate
The Medical Ethics Committee of Central Hospital
Affiliated to Shenyang Medical College approved the use of clinical
tissue specimens from Central Hospital Affiliated to Shenyang
Medical College and Liaoning Provincial Cancer Hospital in this
study (approval no, 2022DEC12-7). All adult participants and the
parents or legal guardians of participants <18 years of age
provided written informed consent for participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bielack SS, Kempf-Bielack B, Delling G,
Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M,
Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma
of the extremities or trunk: An analysis of 1,702 patients treated
on neoadjuvant cooperative osteosarcoma study group protocols. J
Clin Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment and a collaborative
pathway to success. J Clin Oncol. 33:3029–3035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cai Y, Feng R, Lu T, Chen X, Zhou X and
Wang X: Novel insights into the m(6)A-RNA methyltransferase METTL3
in cancer. Biomark Res. 9:272021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen D, Gu X, Nurzat Y, Xu L, Li X, Wu L,
Jiao H, Gao P, Zhu X, Yan D, et al: Writers, readers, and erasers
RNA modifications and drug resistance in cancer. Mol Cancer.
23:1782024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Xiu M, Wang J, Gao Y and Li Y:
METTL16-SENP3-LTF axis confers ferroptosis resistance and
facilitates tumorigenesis in hepatocellular carcinoma. J Hematol
Oncol. 17:782024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu Y, Yang YL, Chen XY, Chen ZY, Zhu JS
and Zhang J: Helicobacter pylori-enhanced hnRNPA2B1 coordinates
with PABPC1 to promote Non-m(6)A translation and gastric cancer
progression. Adv Sci (Weinh). 11:e23097122024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mao L, Wang L, Lyu Y, Zhuang Q, Li Z,
Zhang J, Gu Z, Lu S, Wang X, Guan Y, et al: Branch chain amino acid
metabolism promotes brain metastasis of NSCLC through EMT
occurrence by regulating ALKBH5 activity. Int J Biol Sci.
20:3285–3301. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo X, Qiu W, Li B, Qi Y, Wang S, Zhao R,
Cheng B, Han X, Du H, Pan Z, et al: Hypoxia-induced neuronal
activity in glioma patients polarizes microglia by potentiating RNA
m6A demethylation. Clin Cancer Res. 30:1160–1174. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei X, Feng J, Chen L, Zhang C, Liu Y,
Zhang Y, Xu Y, Zhang J, Wang J, Yang H, et al: METTL3-mediated m6A
modification of LINC00520 confers glycolysis and chemoresistance in
osteosarcoma via suppressing ubiquitination of ENO1. Cancer Lett.
August 29–2024.(Ebup ahead of print).
|
|
11
|
Knuckles P, Lence T, Haussmann IU, Jacob
D, Kreim N, Carl SH, Masiello I, Hares T, Villaseñor R, Hess D, et
al: Zc3h13/Flacc is required for adenosine methylation by bridging
the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery
component Wtap/Fl(2)d. Genes Dev. 32:415–429. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu W, Wang JZ, Wei JF and Lu C: Role of
m6A methyltransferase component VIRMA in multiple human cancers
(Review). Cancer Cell Int. 21:1722021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen X, Lu T, Cai Y, Han Y, Ding M, Chu Y,
Zhou X and Wang X: KIAA1429-mediated m6A modification of CHST11
promotes progression of diffuse large B-cell lymphoma by regulating
Hippo-YAP pathway. Cell Mol Biol Lett. 28:322023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo J, Wang X, Chen Z, Zhou H and Xiao Y:
The role and mechanism of JAK2/STAT3 signaling pathway regulated by
m6A methyltransferase KIAA1429 in osteosarcoma. J Bone Oncol.
39:1004712023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Goodall GJ and Wickramasinghe VO: RNA in
cancer. Nat Rev Cancer. 21:22–36. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang Y and Fullwood MJ: Roles, functions,
and mechanisms of long Non-coding RNAs in cancer. Genomics
Proteomics Bioinformatics. 14:42–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luongo M, Laurenziello P, Cesta G,
Bochicchio AM, Omer LC, Falco G, Milone MR, Cibarelli F, Russi S
and Laurino S: The molecular conversations of sarcomas: exosomal
non-coding RNAs in tumor's biology and their translational
prospects. Mol Cancer. 23:1722024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shaath H, Vishnubalaji R, Elango R,
Kardousha A, Islam Z, Qureshi R, Alam T, Kolatkar PR and Alajez NM:
Long non-coding RNA and RNA-binding protein interactions in cancer:
Experimental and machine learning approaches. Semin Cancer Biol.
86((Pt 3)): 325–345. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi Q, Chu Q, Zeng Y, Yuan X, Wang J,
Zhang Y, Xue C and Li L: Non-coding RNA methylation modifications
in hepatocellular carcinoma: Interactions and potential
implications. Cell Commun Signal. 21:3592023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu D, Wang Z and Li F: KIAA1429 Induces
m6A Modification of LINC01106 to enhance the malignancy of lung
adenocarcinoma cells via the JAK/STAT3 pathway. Crit Rev Immunol.
44:49–61. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Han S, Cao D, Sha J, Zhu X and Chen D:
LncRNA ZFPM2-AS1 promotes lung adenocarcinoma progression by
interacting with UPF1 to destabilize ZFPM2. Mol Oncol.
14:1074–1088. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun G and Wu C: ZFPM2-AS1 facilitates cell
growth in esophageal squamous cell carcinoma via up-regulating
TRAF4. Biosci Rep. 40:BSR201943522020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ren R, Du Y, Niu X and Zang R: ZFPM2-AS1
transcriptionally mediated by STAT1 regulates thyroid cancer cell
growth, migration and invasion via miR-515-5p/TUSC3. J Cancer.
12:3393–3406. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47((W1)): W556–W560.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou Y, Zeng P, Li YH, Zhang Z and Cui Q:
SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based
on sequence-derived features. Nucleic Acids Res. 44:e912016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ren S, Zhang Y, Yang X, Li X, Zheng Y, Liu
Y and Zhang X: N6-methyladenine-induced LINC00667 promoted breast
cancer progression through m6A/KIAA1429 positive feedback loop.
Bioengineered. 13:13462–13473. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang D, Chang S, Li F, Ma M, Yang J, Lv X,
Huangfu L and Jia C: m(6) A transferase KIAA1429-stabilized
LINC00958 accelerates gastric cancer aerobic glycolysis through
targeting GLUT1. IUBMB Life. 73:1325–1333. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu D and Chen A: lncRNA POU6F2-AS1
regulated by KIAA1429 contributes to colorectal cancer progression
in an m(6)A modification manner. Mol Biotechnol. 67:115–122. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kong F, Deng X, Kong X, Du Y, Li L, Zhu H,
Wang Y, Xie D, Guha S, Li Z, et al: ZFPM2-AS1, a novel lncRNA,
attenuates the p53 pathway and promotes gastric carcinogenesis by
stabilizing MIF. Oncogene. 37:5982–5996. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiao M, Liang Z and Yin Z: Long non-coding
RNA ZFPM2-AS1 promotes colorectal cancer progression by sponging
miR-137 to regulate TRIM24. Mol Med Rep. 23:982021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xue M, Tao W, Yu S, Yan Z, Peng Q, Jiang F
and Gao X: lncRNA ZFPM2-AS1 promotes proliferation via
miR-18b-5p/VMA21 axis in lung adenocarcinoma. J Cell Biochem.
121:313–321. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tan F: ZFPM2-AS1: An oncogenic long
non-coding RNA in multiple cancer types. Mini Rev Med Chem.
23:88–98. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Y, Xu Y, Bao Y, Luo Y, Qiu G, He M,
Lu J, Xu J, Chen B and Wang Y: N6-methyladenosine (m6A)
modification in osteosarcoma: expression, function and interaction
with noncoding RNAs-an updated review. Epigenetics. 18:22602132023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Park OH, Ha H, Lee Y, Boo SH, Kwon DH,
Song HK and Kim YK: Endoribonucleolytic cleavage of
m(6)A-Containing RNAs by RNase P/MRP complex. Mol Cell.
74:494–507.e8. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheng Y, Xie W, Pickering BF, Chu KL,
Savino AM, Yang X, Luo H, Nguyen DT, Mo S, Barin E, et al:
N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear
body that suppresses myeloid leukemic differentiation. Cancer Cell.
39:958–972.e8. 2021. View Article : Google Scholar : PubMed/NCBI
|