Introduction
DNA methylation plays an essential role in various
physiological processes such as mammalian development, genomic
imprinting, X chromosome inactivation and aging (1). Alterations in the DNA methylation
status were also found to be involved in the initiation and
progression of human cancer. Among these alterations, the aberrant
methylation of CpG islands located in the promoter regions or first
exons of various genes is the best-categorized epigenetic change
(2). By silencing tumor suppressor
genes or activating oncogenes, epigenetic modifications can affect
many important cellular processes, such as cell cycle control, DNA
repair and apoptosis. Previously, the aberrant methylation status
of specific genes was linked to the pathogenesis of various types
of cancer, and tumor-specific methylation changes were established
as prognostic markers in numerous tumor entities (3). Unlike genetic modifications, such as
mutations or genomic imbalances, epigenetic changes are potentially
reversible, making these changes particularly important therapeutic
targets in cancer and other diseases (4).
In recent years, different techniques have been
developed for the genome-wide screening of CGI methylation status.
Included is differential methylation hybridization (DMH) which is a
high-throughput DNA methylation screening tool that utilizes
methylation-sensitive restriction enzymes to profile-methylated
fragments by hybridizing them to a CpG island microarray. This
method was introduced by Huang and coworkers in 1999 (5), further developed and has since been
widely used (6).
Human hepatocellular carcinoma (HCC) is the sixth
most common type of cancer worldwide and is the second cause of
cancer-related death in China. The main obstacles to improving the
outcome of HCC patients is the high frequency of recurrence and
metastasis. Various genetic and epigenetic abnormalities in HCC
have been identified, suggesting a multi-step nature of
hepatocarcinogenesis (7). A number
of genetic and epigenetic alterations have been noted in HCC
(8). However, each alteration
appears to be implicated in a limited fraction of HCC. Thus, the
systematic analysis of the genetic and epigenetic alterations
underlying the origin and evolution of HCC has attracted the
interest of scientists worldwide (9–13). The
present study employed the DMH method to analyze the differential
methylation status of 12K CpG islands in a number of HCC cell
lines. Tumor cell lines are commonly used as experimental tools in
cancer research as such cell lines exhibit a number of the same
epigenetic and genetic aberrations that are noted in primary
carcinomas. The HCC cell lines selected in this study are of
different origin, and among these, the MHCC97 series cell lines
have the same genetic backgrounds but exhibit different metastatic
potential. These characteristics render them useful for screening
epigenetic alterations underlying the mechanism of metastasis.
Materials and methods
Cell culture
Human HCC cell lines HepG2, Hep3B, PLC/RPF/5 and
human liver cell line L02 were purchased from American Type Culture
Collection (ATCC). SMMC-7721 and BEL-7402 cell lines were purchased
from the Institute of Biochemistry and Cell Biology, Chinese
Academy of Sciences. The MHCC97-H, MHCC97-L, HCCLM3 and HCCLM6 cell
lines, which have similar genetic backgrounds but different
metastatic potential, were established at our institute. Cells were
cultured in high glucose DMEM supplemented with 10% fetal bovine
serum and penicillin-streptomycin under humidified conditions in 5%
CO2.
CpG island microarray hybridization and
data analysis
Total DNA of the cells was extracted using the
DNeasy Kit (Qiagen, Germany). The CpG island array containing
12,192 CpG island clones was purchased from Canada UHN Microarray
Center. Hybridization and signal scanning were conducted by the
Capitalbio Corporation of Beijing, China. The hybridization images
were analyzed using GenePix Pro 4.0 software (Axon Instruments).
The data were smoothed using the Lowess method which was
subsequently followed by cluster analysis. The ratios of Cy5 to Cy3
were calculated for each location on each microarray. A ratio
(Cy5/Cy3) of ≥2 or ≤0.5 was considered to indicate differential
methylation loci. Additionally, the hybridization output was
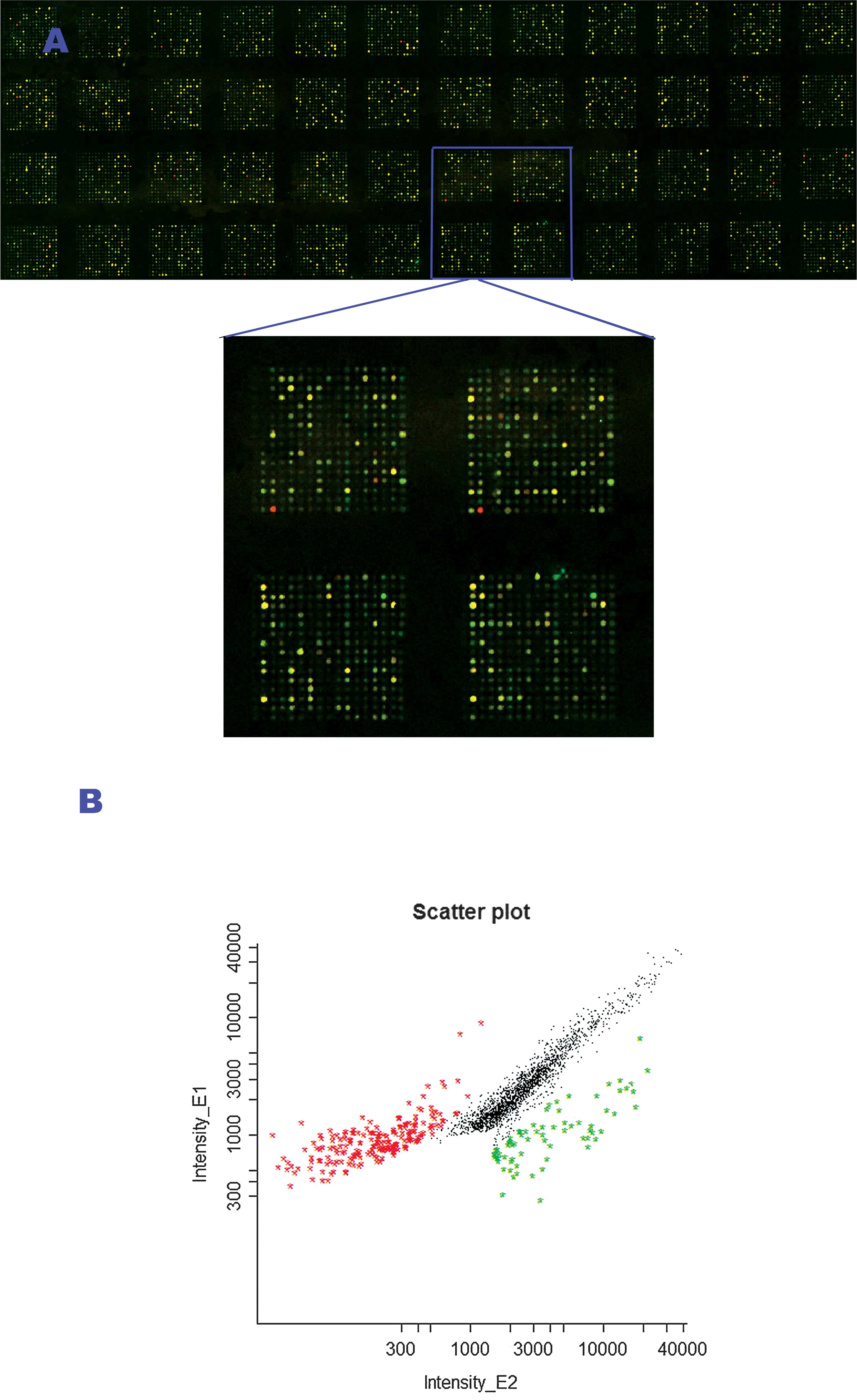
measured in intensities of the two fluorescence reporters (Fig. 1).
Analysis of the genes associated with the
differentially methylated CpG islands
The official website (http://data.microarrays.ca/cpg/) of the UHN human CpG
island database was accessed, and the upstream, within and
downstream genes associated with significant CpG islands were
located according to their ID numbers. The references were searched
and the associated genes were classified according to their
function.
Bisulfite treatment of DNA- and
methylation-specific PCR
The bisulfite treatment of DNA was adapted from
Frommer et al with slight modifications (14). Briefly, 2 μg DNA was diluted with 50
μl distilled H2O, and 5.5 μl 3 M NaOH was added.
Incubation was carried out at 37°C for 15 min to create
single-stranded DNA. Freshly prepared 10 mM hydroquinone (30 μl)
(Sigma) and 520 μl 3 M sodium bisulfite (pH 5.0) (Sigma, S-8890)
were added to each tube. After thorough mixing, a layer of 200 μl
mineral oil was added. Incubation was carried out at 50°C for 16 h,
and the oil was then removed. The modified DNA was purified using
the Promega Wizard Cleanup DNA kit and precipitated at −20°C
overnight. Centrifugation was performed at 12,000 rpm for 30 min at
4°C. The supernatant was removed using a pipette, and the
precipitate was washed with 500 μl 75% ethanol and centrifuged at
12,000 rpm for 5 min at 4°C, and repeated once. The supernatant was
then removed using a pipette, and the dried pellet was resuspended
in 30 μl distilled water and preserved at −20°C until use.
Two differential CpG islands were randomly selected
and methylation-specific PCR (MSP) was performed to verify the
microarray results. PCR primers were designed using MethPrimer
(http://www.ucsf.edu/urogene/methprimer/index1.html).
The PCR program involved an initial denaturation at 95°C for 10
min, followed by 35 cycles of amplification as follows:
denaturation at 95°C for 30 sec, annealing at 48°C for 30 sec, and
extension at 72°C for 30 sec, with an additional extension at 72°C
for 4 min. The PCR products were analyzed with agarose gel
electrophoresis and DNA sequencing.
Results
The DNA amplicons of different HCC cell lines, with
Chang’s liver cell line as a control, were fluorescently labeled
with Cy5 and Cy3, respectively, and cohybridized to a UHN 12K human
CpG island microarray. Findings showed obvious differentiation of
CpG island methylation between HCC and the control cell lines
(Fig. 1). After further cluster
analysis, we detected 58 differentially methylated CpG islands and
66 related tumor-associated genes that exhibited a similar trend in
alteration (6 of the 9 cell lines had an identical trend) in the 9
HCC cell lines compared with the normal control. Among these, 37
CpG islands showed hypermethylation (Table I) and 29 showed hypomethylation
(Table II). In addition, when
compared with the MHCC97-L cells, 16 differentially methylated CpG
islands that may be associated with metastatic potential were
detected in the MHCC97-H cells (24 related genes) (Table III).
 | Table IHypermethylated CpG islands and
associated genes in the HCC cell lines. |
Table I
Hypermethylated CpG islands and
associated genes in the HCC cell lines.
| ID of CpG island | Associated genes |
|---|
| UHNhscpg0011158 | PPP2CA
(upstream) |
| CDKL3
(downstream) |
| UHNhscpg0009735 | SRPK1
(downstream) |
| UHNhscpg0004629 | CP110
(downstream) |
| UHNhscpg0004752 | GJB2 (within) |
| GJB6
(downstream) |
| GJA3 (upstream) |
| UHNhscpg0004878 | BCL2
(downstream) |
| UHNhscpg0002925 | SMAD7
(downstream) |
| UHNhscpg0008205 | IRS4 (upstream) |
| UHNhscpg0008444 | FGFR2
(downstream) |
| UHNhscpg0003663 | LHX5 (within) |
| UHNhscpg0007707 | PIK3CB
(upstream) |
| UHNhscpg0004799 | VRK2 (upstream) |
| BCL11A
(downstream) |
| FANCL (within) |
| UHNhscpg0009327 | TM4SF5
(upstream) |
| UHNhscpg0008203 | PIM1 (upstream) |
| UHNhscpg0000104 | BCL11A
(upstream) |
| UHNhscpg0008444 | BRWD2 (upstream) |
| UHNhscpg0005488 | PDCD4
(downstream) |
| UHNhscpg0008226 | ST5 (downstream) |
| UHNhscpg0004835 | CBX4 (within) |
| CBX8 (upstream) |
| UHNhscpg0003460 | CDH18 (upstream) |
| UHNhscpg0009830 | EPHB4 (upstream) |
| UHNhscpg0003252 | ATF2 (within) |
| UHNhscpg0004910 | HNF4G
(downstream) |
| UHNhscpg0004957 | LRP1 (upstream) |
| UHNhscpg0004834 | GSTA4 (upstream) |
| ICK (within) |
| UHNhscpg0004596 | RYK (upstream) |
| UHNhscpg0003693 | FZD3 (upstream) |
| UHNhscpg0001904 | SPRY3
(downstream) |
| UHNhscpg0004926 | MAPK8IP3
(downstream) |
| UHNhscpg0011292 | SLIT2
(downstream) |
| UHNhscpg0003315 | HAND2 (within) |
 | Table IIHypomethylated CpG islands and
associated genes in the HCC cell lines. |
Table II
Hypomethylated CpG islands and
associated genes in the HCC cell lines.
| ID | Associated genes |
|---|
| UHNhscpg0006172 | SEC13L1
(upstream) |
| UHNhscpg0002224 | PRC1 (upstream) |
| UHNhscpg0008081 | RAE1 (within) |
| UHNhscpg0007289 | HNF4G (upstream) |
| UHNhscpg0002482 | CDCA7L
(upstream) |
| UHNhscpg0006623 | EPHA7 (within) |
| MAP3K7
(upstream) |
| UHNhscpg0005013 | PECAM1 (within) |
| UHNhscpg0003756 | PA2G4 (upstream) |
| UHNhscpg0002482 | CDCA7L
(upstream) |
|
UHNhscpg0001077 | EGR3
(downstream) |
|
UHNhscpg0002020 | NAB2
(downstream) |
|
UHNhscpg0007707 | PIK3CB
(upstream) |
|
UHNhscpg0002943 | PPIAL4
(upstream) |
|
UHNhscpg0009335 | MAF (upstream) |
|
UHNhscpg0010477 | ID2
(downstream) |
|
UHNhscpg0005863 | ZNF278
(within) |
|
UHNhscpg0001373 | MAPRE2
(downstream) |
|
UHNhscpg0005140 | PTPNS1
(downstream) |
|
UHNhscpg0000025 | VCL
(downstream) |
|
UHNhscpg0002244 | DPYSL2
(within) |
|
UHNhscpg0004830 | THBS1 (within) |
|
UHNhscpg0001262 | NR5A2 (within) |
|
UHNhscpg0009002 | DYNLRB2
(downstream) |
|
UHNhscpg0002482 | RAPGEF5
(downstream) |
|
UHNhscpg0004867 | RIN3
(downstream) |
|
UHNhscpg0009717 | IRS4
(upstream) |
|
UHNhscpg0008232 | |
|
UHNhscpg0001274 | ROBO3 (within) |
|
UHNhscpg0005005 | SV2B (within) |
 | Table IIIGenes associated with the
differentially methylated CpG islands in the MHCC97H and MHCC97L
cell linesa. |
Table III
Genes associated with the
differentially methylated CpG islands in the MHCC97H and MHCC97L
cell linesa.
| Cell line | Associated
genes |
|---|
| 97H/97L |
| With
hypermethylation in the CpG island | BCL11A, VRK2,
FANCL, GJB2, GJB6, GJA3, CBX4, CBX8, ICK, ATF2, LRP1, GSTA4,
TM4SF5 |
| With
hypomethylation in the CpG island | THBS1, ZNF278,
IRS4, HNF4G, CDCA7L, RAPGEF5, PPIAL4, PA2G4, PIK3CB, MAF,
DYNLRB2 |
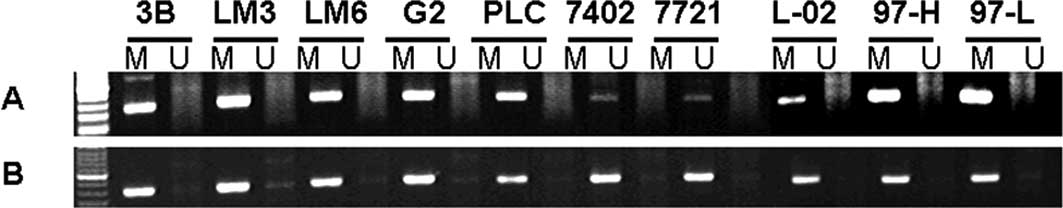
Two randomly selected differential CpG islands were
further analyzed with MSP and DNA sequencing in the HCC cell lines.
The results showed a high correlation with the microarray results
(Fig. 2). The methylation status of
a number of other significant CpG islands and expression of the
associated gene were also detected with MSP and RT-PCR with the aim
of analyzing the association of gene expression with DNA
methylation status.
Discussion
Tumorigenesis and tumor metastasis are complex,
multi-step processes involving genetic and epigenetic alterations.
The roles of epigenetic alterations, particularly that of DNA
methylation status, in the progression and metastasis of HCC
recently attracted the interest of numerous investigators. Due to
HCC sensitivity to the influence of external factors as opposed to
the DNA sequence, alterations in the DNA methylation status may be
the earliest event in tumorigenesis and metastasis. The reversible
characteristics of the DNA methylation status make it attractive in
tumor intervention (15,16).
The present study employed the differential
methylation hybridization method to assess the alteration of CpG
island methylation in a number of HCC cell lines. The results
showed that alteration of the CpG island methylation is a common
event in HCC cell lines of different origin and metastatic
potential. The genes associated with these CpG islands included
pro-oncogenes and oncogenes, tumor suppressor genes, apoptosis
genes, proliferation and cell cycle regulatory genes, as well as
tumor angiogenesis- and immuno-escape-associated genes. The concept
of a CpG island methylator phenotype (CIMP) was first proposed by
Issa et al (17) and is
currently widely accepted as a novel type of biomarker employed to
classify different types of cancer or cancers of different stages
(17,18). The present array-based study
profiled the CpG island methylation status in HCC cell lines of
different origin and metastatic potential. The potential of an
HCC-specific CIMP was also noted, particularly since numerous liver
cell-specific genes were involved in the differences. As a ‘marker
pattern’, CIMP has a marked superiority in reflecting the clinical
complexity and variability of cancer versus the use of a single
marker.
Recently, molecular alterations of various signaling
pathways (e.g., Wnt/β-catenin, Ras/Raf/MEK/ERK and PI3K/Akt/mTOR)
were identified in liver carcinogenesis, providing novel molecular
targets for new treatment modalities (19,20).
Sorafenib, the first FDA-approved drug for advanced liver cancer,
is a multi-kinase inhibitor that targets the Ras/Raf signaling
pathway (21). A DNA methylation
mechanism was involved in the modification of these signaling
pathways. Hypermethylation in the promoters of the Wnt pathway
associated with adenomatous polyposis coli and E-cadherin
was recently reported (22–24). Based on our data, a number of other
members involved in the Wnt pathway also displayed alterations in
the methylation status in associated CpG islands. These members
included frizzled 3 (FZD3), receptor tyrosine kinase (RYK), c-myc,
ID2 and MAPRE2, which showed hypomethylation in the associated CpG
islands in the HCC cell lines compared with the control cells,
suggesting that DNA methylation is a crucial mechanism that adjusts
the Wnt signaling pathway to favor HCC. On the other hand,
alterations in the methylation of factors involved in the Ras/Raf
pathway were also found in this study, including RAPGEF5, which
serves as a Ras activator and Ras and Rab interactor 3 (RIN3).
Numerous other important CpG islands and the
associated genes previously investigated in other solid tumors were
also analyzed and verified in our data. EPHB4 is a member of the
Eph family of receptor tyrosine kinases. Its interaction with the
ligand ephrinB2 contributes to the growth of various types of
tumors and can influence the clinical outcome of cancer patients
(25–27). According to our data, the
EPHB4-associated CpG island was hypermethylated, and the expression
of the EPHB4 gene was downregulated in the HCC cell lines. The role
of the EPHB4 gene in HCC progression warrants further
investigation. Slit and Roundabout (Robo) were first identified as
guidance molecules for neurons and leukocytes (28). More recently, their roles were
expanded to include the mediation of angiogenesis, heart
morphogenesis and tumor metastasis (29–32).
In vertebrates three Slit (Slit1, Slit2 and Slit3) and four Robo
(Robo1, Robo2, Robo3/Rig-1 and Robo4/Magic Robo) genes have been
identified (33). Certain members
of the Slit/Robo signaling pathway also showed frequent alterations
of the CpG island methylation status in our data. Further
investigations of the methylation status of CpG islands in the
promoter region and mRNA expression levels of Slit1, Slit2, Slit3,
Robo1 and Robo3 were subsequently carried out in several HCC cell
lines of different metastatic ability (data not shown). A number of
differences in the methylation and expression of the genes were
detected in different HCC cell lines. In particular, Slit2
expression may be associated with metastatic potential, indicating
a potential involvement of Slit/Robo in the progression or
metastasis of HCC.
Various genes of unknown function also appeared in
the list due to regular alterations between HCC and the control
cell lines. The potential roles of these genes in the progression
and metastasis of HCC warrant further investigation.
In conclusion, the differentially methylated CpG
islands and associated genes screened by microarray in the HCC cell
lines in this study have provided the foundation for understanding
HCC-specific CIMP and for developing potential biomarkers of
significance for the prognosis and metastasis of HCC. Furthermore,
the reversible characteristic of DNA methylation status offers
suitable targets for the interference therapy of HCC.
Acknowledgements
This study was supported by a grant from the
National Nature Science Foundation of China (no. 30500484) and
Shanghai Nature Science Foundation (no. 04ZR14025).
References
|
1
|
Scarano MI, Strazzullo M, Matarazzo MR and
D’Esposito M: DNA methylation 40 years later: Its role in human
health and disease. J Cell Physiol. 204:21–35. 2005.PubMed/NCBI
|
|
2
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
3
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar
|
|
4
|
Pfister S, Schlaeger C, Mendrzyk F, et al:
Array-based profiling of reference-independent methylation status
(aprimes) identifies frequent promoter methylation and consecutive
downregulation of zic2 in pediatric medulloblastoma. Nucleic Acids
Res. 35:e512007. View Article : Google Scholar
|
|
5
|
Huang TH, Perry MR and Laux DE:
Methylation profiling of cpg islands in human breast cancer cells.
Hum Mol Genet. 8:459–470. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yan PS, Potter D, Deatherage DE, Huang TH
and Lin S: Differential methylation hybridization: profiling DNA
methylation with a high-density cpg island microarray. Methods Mol
Biol. 507:89–106. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Herath NI, Leggett BA and MacDonald GA:
Review of genetic and epigenetic alterations in
hepatocarcinogenesis. J Gastroenterol Hepatol. 21:15–21. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park IY, Sohn BH, Yu E, Suh DJ, Chung YH,
Lee JH and Surzycki SJ: Aberrant epigenetic modifications in
hepatocarcinogenesis induced by hepatitis B virus X protein.
Gastroenterology. 132:1476–1494. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L, Wang WL, Zhang Y, Guo SP, Zhang J
and Li QL: Epigenetic and genetic alterations of pten in
hepatocellular carcinoma. Hepatol Res. 37:389–396. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calvisi DF, Ladu S, Gorden A, et al:
Mechanistic and prognostic significance of aberrant methylation in
the molecular pathogenesis of human hepatocellular carcinoma. J
Clin Invest. 117:2713–2722. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calvisi DF, Ladu S, Gorden A, et al:
Molecular pathogenesis of human hepatocellular carcinoma:
Mechanistic and prognostic significance of aberrant methylation.
AACR Meeting Abstracts. 2006:763a2006.
|
|
12
|
Thorgeirsson SS and Grisham JW: Molecular
pathogenesis of human hepatocellular carcinoma. Nat Genet.
31:339–346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pei Y, Zhang T, Renault V and Zhang X: An
overview of hepatocellular carcinoma study by omics-based methods.
Acta Biochim Biophys Sin. 41:1–15. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Frommer M, McDonald LE, Millar DS, et al:
A genomic sequencing protocol that yields a positive display of
5-methylcytosine residues in individual DNA strands. Proc Natl Acad
Sci USA. 89:1827–1831. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Humeniuk R, Mishra PJ, Bertino JR and
Banerjee D: Molecular targets for epigenetic therapy of cancer.
Curr Pharm Biotechnol. 10:161–165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schneider-Stock R and Ocker M: Epigenetic
therapy in cancer: Molecular background and clinical development of
histone deacetylase and DNA methyltransferase inhibitors. IDrugs.
10:557–561. 2007.
|
|
17
|
Issa JP: Cpg island methylator phenotype
in cancer. Nat Rev Cancer. 4:988–993. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen L, Catalano PJ, Benson AB III,
O’Dwyer P, Hamilton SR and Issa JP: Association between DNA
methylation and shortened survival in patients with advanced
colorectal cancer treated with 5-fluorouracil-based chemotherapy.
Clin Cancer Res. 13:6093–6098. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tommasi S, Pinto R, Pilato B and Paradiso
A: Molecular pathways and related target therapies in liver
carcinoma. Curr Pharm Des. 13:3279–3287. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pang RW and Poon RT: From molecular
biology to targeted therapies for hepatocellular carcinoma: the
future is now. Oncology. 72(Suppl 1): 30–44. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gollob JA, Wilhelm S, Carter C and Kelley
SL: Role of Raf kinase in cancer: therapeutic potential of
targeting the Raf/mek/erk signal transduction pathway. Semin Oncol.
33:392–406. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Prasad CP, Mirza S, Sharma G, et al:
Epigenetic alterations of cdh1 and apc genes: relationship with
activation of wnt/beta-catenin pathway in invasive ductal carcinoma
of breast. Life Sci. 83:318–325. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Banno K, Yanokura M, Susumu N, et al:
Relationship of the aberrant DNA hypermethylation of cancer-related
genes with carcinogenesis of endometrial cancer. Oncol Rep.
16:1189–1196. 2006.PubMed/NCBI
|
|
24
|
Supic G, Kozomara R, Brankovic-Magic M,
Jovic N and Magic Z: Gene hypermethylation in tumor tissue of
advanced oral squamous cell carcinoma patients. Oral Oncol.
45:1051–1057. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kumar SR, Singh J, Xia G, et al: Receptor
tyrosine kinase ephb4 is a survival factor in breast cancer. Am J
Pathol. 169:279–293. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Noren NK, Lu M, Freeman AL, Koolpe M and
Pasquale EB: Interplay between ephb4 on tumor cells and vascular
ephrin-b2 regulates tumor growth. Proc Natl Acad Sci USA.
101:5583–5588. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Berclaz G, Karamitopoulou E, Mazzucchelli
L, et al: Activation of the receptor protein tyrosine kinase ephb4
in endometrial hyperplasia and endometrial carcinoma. Ann Oncol.
14:220–226. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bashaw GJ and Goodman CS: Chimeric axon
guidance receptors: the cytoplasmic domains of slit and netrin
receptors specify attraction versus repulsion. Cell. 97:917–926.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stella MC, Trusolino L and Comoglio PM:
The slit/robo system suppresses hepatocyte growth factor-dependent
invasion and morphogenesis. Mol Biol Cell. 20:642–657. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu ZJ and Herlyn M: Slit-Robo: neuronal
guides signal in tumor angiogenesis. Cancer Cell. 4:1–2. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marlow R, Strickland P, Lee JS, et al:
Slits suppress tumor growth in vivo by silencing sdf1/cxcr4 within
breast epithelium. Cancer Res. 68:7819–7827. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang B, Xiao Y, Ding BB, et al: Induction
of tumor angiogenesis by slit-robo signaling and inhibition of
cancer growth by blocking robo activity. Cancer Cell. 4:19–29.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hohenester E: Structural insight into
slit-robo signalling. Biochem Soc Trans. 36:251–256. 2008.
View Article : Google Scholar : PubMed/NCBI
|